INTRODUCTION
Environmental pollution by antibiotics leads to the development of antibiotic-resistant bacteria, which subsequently contribute to the ineffectiveness of antibiotics. The adaptation of drug-resistant genetics in pathogenic microorganisms is triggered by nonlethal doses of therapeutic drugs, particularly antibiotics, and this adaptation poses major risks to the public’s health (Kraemer et al., 2019). Moreover, uncontrolled applications of antibiotics to the environment could be harmful to the environment. Amoxicillin is a veterinary drug administered to animals to overcome respiratory tract infections and secondary bacterial infections caused by viral illnesses. In livestock animals, amoxicillin has a recovery rate of 43.4% from the urine after the oral dose. The unmetabolized amoxicillin is then widely dispersed into the environment and becomes a pollutant. Several practical and costly methods are used for amoxicillin elimination, including adsorption, accelerated oxidation, photodegradation, and ion degradation. However, the current techniques for antibiotic eradication are not effective. Thus, as a promising alternative treatment, bioremediation by microorganisms may be a safer and cheaper alternative to removing amoxicillin from the environment. Previous studies have shown that many microorganisms (bacteria and fungi) can degrade xenobiotics. Furthermore, bacteria that secrete β-lactamase can break down amoxicillin. For example, Enterobacter sp., Corynebacterium sp. (43.0%), Bacillus subtilis (76.7%), Flavobacterium sp. (41.0%), Acinetobacter lwoffii (74.3%), and Pseudomonas aeruginosa (77.8%) are able to break down 60 ppm of amoxicillin. Numerous bacterial strains, including Escherichia coli, Achromobacter sp., Vibrio sp., and Micrococcus sp., have shown high potential to degrade amoxicillin. Research by Guardado et al. (2019) observed a degradation yield of the antibiotic amoxicillin (80.0%) by a novel laccase from Pycnoporus sanguineus CS43. Recent research by Wang et al. (2021) found that the Ochrobactrum tritici bacteria can degrade penicillin V. This study has demonstrated that amoxicillin is effectively degraded by both soil fungi, Aspergillus tamarii isolate 58 and Lichtheimia ramosa strain R, thereby, providing a new alternative to safely overcome antibiotic pollution.
MATERIALS AND METHODS
Soil sample collection
Three soil samples with triplicates were collected randomly from different sites within the depth of 20 cm at a dairy farm with global positioning system coordinates 2°59′28.8?N 101°44′00.5?E, where veterinary antibiotics have been used extensively. Different sites were chosen for sample collection to obtain diverse soil fungal cultures that have been exposed to veterinary antibiotics. A soil sample (500 g) was collected and stored in sterile plastic bags for the analysis of pH, temperature, and moisture at the laboratory.
Isolation of soil fungi
The soil samples were serially diluted. A 10fold serial dilution was prepared from the microbial suspension. Soil with three and four dilution factors was plated on potato dextrose agar (PDA) (Merck, Germany). Afterward, the PDA plates were incubated for 3 to 5 days at 28°C. A single colony was streaked on PDA plates in order to obtain pure cultures. The pure cultures were labeled Fungi A and B, respectively. The plates were incubated for 3 to 5 days and stored at 4°C (Muthulakshmi et al., 2011).
Morphological observations
The fungi were morphologically identified to evaluate their features. Texture, color, diameter, and structure were assessed with the naked eye. Regarding the texture and structure, the morphology, elevation, margin, and aerial mycelium were recorded, while color was observed on the plate’s surface and at its backside. The Olympus CX12 light microscope was used to study both fungi for microscopic identification. The fungi were placed on a clean microscope slide with the adhesive tape technique applied to the surface of the fungi. The samples were then examined after a drop of methylene blue was added.
Molecular identification
DNA extraction
The genomic DNA of the fungi was extracted using a NORGEN fungi/yeast genomic DNA isolation kit, and the recommended extraction protocol was followed. The collecting solution (NaCl) was applied in a volume of 5 ml to the fungal-growing plate, and the fungi were carefully scraped with a micropipette so as not to scratch PDA. The fungi spores were carefully collected. To create the cell pellet, 1 ml of the spores with mycelium fragments was agitated for a minute at 14,000 rpm after being pipetted in the microcentrifuge tube. The pellet was left inside to be used in the following step while the supernatant was carefully pipetted out.
Following that, 500 ml of Lysis Buffer L was added to the microcentrifuge tube containing a pellet and gently vortexed to lyse the fungus. As a safety measure, the mixture was transported to the bead tube and set on a flatbed vortex using masking tape. Next, the mixture was vortexed at its highest speed for 5 minutes. The bead tube was then put into a water bath set at 65°C for 10 minutes while being inverted three times. The tubes were briefly spun again, and the lysate was then pipetted into a clean microcentrifuge tube without removing the bead. A new microcentrifuge tube was used to transfer the supernatant after the tube had been agitated for 2 minutes at 14,000 rpm. The obtained lysate was gently vortexed with an equivalent volume of 100% ethanol. The tube was vortexed once again after adding 300 μl of Solution BX.
Approximately 650 μl of lysate was added to the collection tube once the spin column and tube were constructed. The tube was agitated at 10,000 rpm for another minute; then, the process was continued with the residual lysate to reach the end. The column was then filled with around 500 μl of Wash Solution A, and it was agitated for a minute at 10,000 rpm. Another agitation at high speed (14,000 rpm) was used to maintain the lysate flow for 2 minutes to dry the resin. Next, approximately 100 μl of Elution Tube B solution was added to the column as it was being transferred into a new 1.7 ml elution tube. The tube was then agitated at 10,000 rpm for 2 minutes. Successfully extracted DNA was preserved at −20°C.
DNA amplification and sequencing
Internal transcribe spacer (ITS) one and four were employed in the polymerase chain reaction (PCR) to amplify the remaining fungal DNA. The forward primer was designated ITS 1 (5′-TCC GTA GGT GAA CCT GCG-3′), and the reverse primer was designated ITS 4 (5′-TCC TCC GCT TAT TGA TAT GC-3′). There was a 350–880 bp yield from the fragments (Kamaruddin et al., 2022).
This approach was performed according to Hamid and Ariffin (2020), who amplified the DNA of fungi. First, the denaturation was set at a high temperature of 94°C for 3 minutes. Then 35 cycles of denaturation at 94°C for 30 seconds, annealing temperature at 55°C for 30 seconds, an extension at 72°C for a minute, and final extension at 72°C for 15 minutes were done to complete the procedure (Hamid and Ariffin, 2020).
The Next Gene Scientific Company received the PCR products and their primers to conduct the sequencing investigation of fungi. Finally, the National Centre for Biotechnology’s BLAST program was used to examine both sequence results (NCBI).
Fungal growth curve
The fungal growth curve was calculated using the dry weight of biomass produced by the fungi. A cork borer was used to remove a thin slice of solid medium culture from the PDA plate and place it into a 300 ml conical flask filled with 100 ml of potato dextrose broth (PDB). Five different flasks will be utilized for each sample, and the mixture was incubated for 10 days at 25°C at an orbital shaker speed of 120 rpm. The interval incubation times of 48, 96, 144, 192, and 240 hours shall be written on each flask. A symmetrical curve from the lag phase to the log phase was also obtained by adding 6, 12, and 18 hours. The mycelia sample was filtered using the Buchner apparatus every 2 days, and it was dried in the oven for 24 hours at 72°C. The weight of the filter paper was deducted from the total dry weight volume before the net dry weight of the mycelia was calculated. The growth curve of the fungus was subsequently created by graphing incubation time (hours) versus dry cell weight.
Biodegradation procedure
Biodegradation assay
Two sterilized PDBs, each containing 100 ml of PDB in a 300 ml conical flask were injected with 100 μl of 1 × 106 of both fungi under sterile conditions. For the biotic control, only fungi with PDB were used, for the abiotic control, only antibiotics with PDB were used at the same pH and temperature as the samples.
The abiotic and biotic controls were cultured for 4 days at ambient temperatures and agitated at 120 rpm on a rotary shaker to mark the beginning of the log phase. The experiment was conducted in triplicate. The log phase began on the 4h day of incubation, and the culture broth was spiked with an amoxicillin solution at a concentration of 50 mg/ml with a final concentration (1,000 μg/ml) of amoxicillin. At room temperature, the incubation procedure was carried out for another 96 hours on the rotary shaker at 120 rpm. The biotransformation products were then removed at 0, 24, 48, 72, and 96 hours, yielding approximately 6 ml each time. Before High-performance liquid chromatography (HPLC) analysis, the products went through a cleaning process.
Liquid-liquid extraction
The biodegradation products underwent liquid–liquid extraction every 24 hours of incubation, beginning with 0 hours (LLE). A chloroform solution and HPLC-grade water were chosen as the two solvents due to various polarity and miscibility values. As chloroform is denser than water, it descends to the bottom. Amoxicillin residues can then be segregated according to their polarity in the biodegradation product (1:1). The 250 ml separating funnel was constructed by mixing 50 ml of HPLC-grade water, 50 ml of chloroform, and 6 ml of biodegradation products. The valve was repeatedly opened, and the separating funnel was inverted to evolve the gas. The formation of two layers of solvents was noted to occur after the mixture had settled for 10 minutes. A 50 ml falcon tube was used to collect both layers, which were then concentrated at 100°C using a rotary evaporator. Since solutions get more concentrated as temperatures rise, all solvents were evaporated due to the differences in the boiling points of water (100°C) and chloroform (61.2°C) (Zulkifle et al., 2022).
Solid-phase extraction (SPE)
SPE was the next step in the clean-up procedure for biodegradation products. After preconditioning approximately 6 ml of 500 mg bed weight C18 Hypersep SPE cartridge with 5 ml of HPLC-grade acetonitrile, 5 ml of HPLC-grade water was added two times under vacuum condition. One drop was released every 15 seconds. Then, around 4 ml of the biodegradation products were filtered through a 0.45 μm filter, transferred into the column, and allowed to pass through. After that, the analytes were eluted out using a 3 ml mixture of 0.1 M phosphate buffer (KH2PO4) and HPLC-grade acetonitrile (30:70, v/v). The purified products were collected in a 5 ml glass vial for HPLC quantification analysis (Zulkifle et al., 2022).
Amoxicillin standard curve
A final concentration of 1,000 ppm of the standard amoxicillin solutions was prepared. All equipment used in this preparation was rinsed using HPLC-grade water to reduce noise in HPLC. A 100 ml shake flask containing 100 ml of 0.1 M KH2PO4 (pH 3.5) was filled with approximately 100 mg of amoxicillin powder. The mixture was then placed into a 50 ml falcon tube after being purified with a 0.45 μm nylon syringe filter. The tube was covered in aluminum foil to prevent photodegradation.
In this section, amoxicillin standard curves were produced using the earlier preparation of amoxicillin standard solutions. The antibiotic was serially diluted to achieve the desired concentration (ppm), which ranged from 1 to 100 ppm.
The standard was filtered using a 0.45 μm filter before being injected into the HPLC to remove impurities. The average area under the graph (mAU*s) versus the standard concentration of amoxicillin was used to construct the standard curve after all results were obtained. The coefficient of determination (R2) was used to evaluate the standard curve.
HPLC analysis
The residual antibiotics were examined using analytical high-performance liquid chromatography (Agilent 3395 Series). To achieve good resolution chromatographic peaks (Rs >1.5), the mobile phase’s initial 50:50 mixture of HPLC-grade water and phosphate buffer (KH2PO4) was altered to a ratio of 60:40. Table 1 summarizes the HPLC parameters for detecting amoxicillin residues.
Antimicrobial assay
An antimicrobial test approach was used to measure the efficiency of amoxicillin after it underwent biodegradation. Escherichia coli ATCC 25922, Staphylococcus aureus ATCC 25923, and B. subtilis ATCC 19615 were used in triplicate to test the amoxicillin residual. According to Ariffin et al. (2017), well diffusion methods were employed with a slight change in the number of wells. The cultures were grown in nutrient agar plates with wells for controls and biodegradation products. Approximately 20 μl of the biodegradation product (0–96 hours of fermentation) was added to the respective wells and incubated at 37°C. This experiment was supported by Mohammad et al. (2018). The diameter of the inhibitory zone was measured.
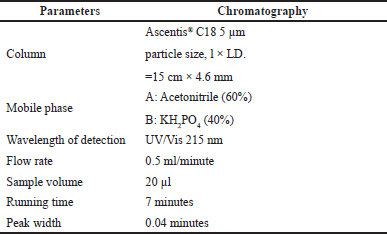 | Table 1. HPLC conditions for amoxicillin residual analysis by HPLC-photodiode-array detection (DAD). [Click here to view] |
Scanning electron microscope (SEM)
Before a SEM was used to observe the fungi, they were first cultured in broth. For 96 hours of degradation, 300 ml of two conical flasks containing approximately sterilized 100 ml PDB were used. After being spiked with 100 ml of fungi (A. tamarii and L. ramosa), both flasks were shaken at 120 rpm for 3days. The final concentration of PDB was then determined by injecting stock amoxicillin (10,000 ppm) into the broth. Two additional 300 ml conical flasks containing sterilized 100 ml PDBs were made for the 0-hour degradation, which spiked precisely before the SEM observation began. Following that, the four samples were all thoroughly dried in an oven at 75°C for 4 hours. The gold was coated using a gold-sputter machine after dipping the carbon tape onto dried fungi several times. The wet samples required longer coating times than the dry samples.
RESULTS AND DISCUSSION
Morphological and molecular identification
As illustrated in Figure 1a, after 4 days of incubation, Fungi A formed interval matcha dark and light green color stripes on the inner sides, and white color was noted at the colony’s edge at room temperature 27°C. Homa et al. (2019) stated that A. tamarii produces a white and green matcha color near the colony’s edges. Only a white colony with a little green tint was seen on the reverse plate, as shown in Figure 1b. Fungi A’s structure had a dry, powdery appearance, and no hyphae were visible on the colony. After 4 days of incubation, this fungus achieved an average diameter of about 5.7 cm, indicating that it is one of the fastest-growing fungi of Aspergillus species’ (Midorikawa et al., 2018).
Fungi B formed a grey–white colony on the top of the PDA after 4 days of incubation at ambient temperature (25°C). Additionally, the margin was described as a whole and had a circular shape with a greater elevation at the colony’s edge. Moreover, in wet conditions, the colony noticed a massive hyphal expansion (Fig. 1c). Lichtheimia ramosa generates a grey–white colony color with many hyphae on PDA when chloramphenicol is present, according to recent research by Imade et al. (2020). Figure 1d shows that the PDA’s reverse plate had a cloudy form. After 4 days of incubation, the Fungi B colony reached an average diameter of 6.9 cm, demonstrating a rapidly growing fungus with hyphae (Imade et al., 2020). The filamentous form of Fungi B had a moist and dense texture on the top of the colony, while the elevation and margin were classified as flat and filiform features.
DNA extraction and molecular identification were performed on both fungi. Figure 2 depicts the DNA bands for both fungi samples at the second and third wells. The isolates for Fungi A were about 700 bp in size, while those for Fungi B were about 800 bp. The outcomes demonstrate that both fungi fall within the 350 to 880 bp range for ITS1 and ITS4 amplicons (Kamaruddin et al., 2022). Both bands had more color intensity, indicating that the DNA samples were concentrated (Blount et al., 2022). After operating for 1 hour and 30 minutes at 70 Volts, the 100 bp ladder band parted. The forward and reverse primers complemented each other or self-anneal to form a hairpin loop, resulting in the emergence of primer dimers at the bottom of the agarose gel (Mohammad et al., 2018).
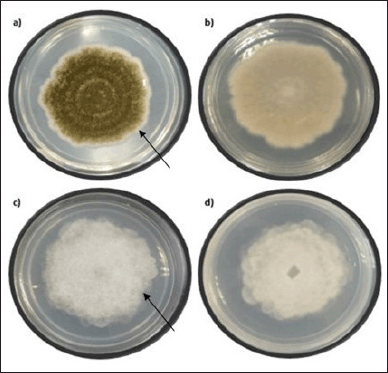 | Figure 1. After 4 days of incubation. (a) Top view of Fungi A with an arrow showing matcha dark and light green color stripes on the inner sides and white color at the colony’s edge. (b) Bottom view of Fungi A. (c) Top view of Fungi B. (d) Bottom view of Fungi B with an arrow shows a circular grey–white colony with a greater elevation at the colony’s edge. [Click here to view] |
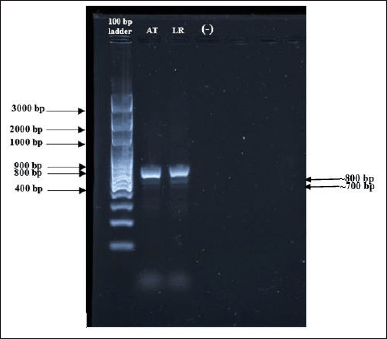 | Figure 2. The PCR products by gel electrophoresis. [100 bp DNA ladder (Solis Biodyne) as a marker. AT, LR, and negative control samples were used]. The isolates for Fungi A (AT) were about 700 bp in size, while those for Fungi B (LR) were about 800 bp. [Click here to view] |
Findings from DNA nucleotide–nucleotide basic local alignment search methods from NCBI’s BLAST were used to evaluate sequencing samples from both fungi. Table 2 includes a list of the identified fungi species. Sequencing results were then compared to data obtained using viable and nonviable fungal exposure assessment methods. ITS clone libraries were predominantly derived from the phylum Ascomycota in both air (68%) and dust (92%) samples and followed by the Basidiomycota and Zygomycota.
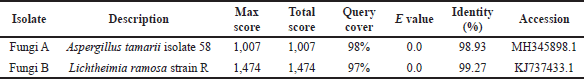 | Table 2. Summary of NCBI BLAST of the isolated fungi. [Click here to view] |
Biodegradation of amoxicillin by A. tamarii isolate 58 and L. ramosa strain R.
A modified analysis was used for amoxicillin residual detection by HPLC (Mohammad et al., 2018). According to the findings of the HPLC study, the amoxicillin concentration for A. tamarii isolate 58 and L. ramosa strain R effectively decreases as the incubation hours increase. Figure 3a depicts the concentration and percentage of amoxicillin residues during the biodegradation process by A. tamarii isolate 58. The amoxicillin concentration residues dropped significantly to 920 ppm at 24 hours from 0 hours. The amoxicillin concentration gradually dropped to 539 ppm after 48 hours, bringing the percentage of amoxicillin down to 46.1%. This was followed by 521 ppm at 72 hours (47.9%) and 490 ppm at 96 hours (51.0%), respectively. After 48 hours, the A. tamarii isolate 58 achieved the stationary phase, which caused the antibiotic concentration to decline gradually. Aspergillus tamarii produces xylanase, tannase, amylase, and protease enzymes to break down the enclosing components, as stated by Shanmugavel et al. (2018) and Midorikawa et al. (2018). This Aspergillus species produces tannase, which significantly improves ester bond hydrolysis and alters the structure of amoxicillin to make it less harmful to the environment (Venkitasamy et al., 2019).
Figure 3b depicts the concentration of amoxicillin residue after being incubated with L. ramosa strain R for 96 hours. After 48 hours, the amoxicillin residual concentration decreased to 550 ppm, and approximately over half of the amoxicillin percentage (45.0%) was degraded. The fungus then reached the stationary phase, which diminished nutrients in the PDB, and the amount of amoxicillin residual progressively increased to 60.0% at 96 hours of incubation. This fungus produces a variety of enzymes, including β-glucosidase, carboxymethylcellulose, and xylanase, to break down the surrounding materials and transform them into nutrition (de Andrade Silva et al., 2020).
The mechanism underlying amoxicillin degradation by A. tamarii isolate 58 and L. ramosa strain R
Amoxicillin is a type of β-lactam antibiotic with a distinctive molecule composed of an unstable, extremely strained, and reactive β-lactam amide bond. The degradation of amoxicillin occurs in various conditions, including alkaline or acidic environments, the presence of specific enzyme β-lactamase, and the treatment of weak nucleophiles such as water and metal ions (Mukhopadhyay et al., 2022). According to Senwan et al. (2017), the fusion of the thiazolidine and β-lactam rings leads to the formation of β-lactam antibiotics. This fusion triggers the antibiotic to react and is susceptible to weak nucleophiles due to the deformation of amide resonance. Additionally, unstable nonplanarity fragment structure based on torsional rotation and large string strain formation contributes to the deformation of amide resonance. A group of filamentous fungi, however, produces a variety of enzymes, including amidases and proteases, which have nucleophile groups, such as hydroxyl, amine, and sulfhydryl groups. As a result, this potent nucleophile group can attack the amide group in the β-lactam ring and lead to antibiotic degradation (Mukhopadhyay et al., 2022). The findings of this research further support the ability of A. tamarii isolate 58 and L. ramosa strain R to perform biotransformation.
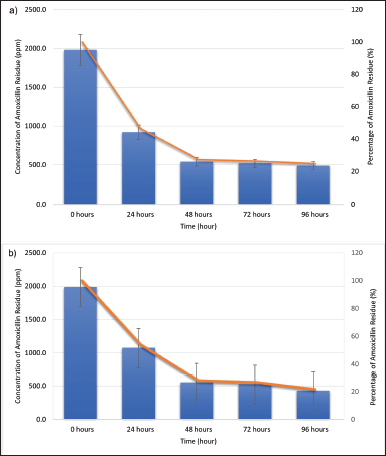 | Figure 3. Biodegradation profiles of amoxicillin (expressed as the concentration of amoxicillin residue in ppm (blue column) and percentage (%) of amoxicillin residue (brown line) after the biodegradation process at room temperature and 120 rpm. (a) Biodegradation profile by A. tamarii isolate 58. (b) Biodegradation profile by L. ramosa strain R. [Click here to view] |
According to a previous study, the β-lactamase of Aspergillus species can break down amoxicillin into amoxicillin acid by opening the β-lactam ring (Ji et al., 2021). Antibiotics become inactive when β-lactamase cleaves the amide linkage of the β-lactam ring (Tooke et al., 2019). The β-lactamase discovered on the surface of Aspergillus spp. initially breaks down the amoxicillin’s β-lactam ring before decarboxylating (Ji et al., 2021). The chemical bonds in β-lactam antibiotics are randomly broken by chemical and physical methods. Consequently, a variety of degradation mechanisms occur (Timm et al., 2019). The filamentous fungi can completely break down β-lactam antibiotics when they are metabolized by β-lactamase into fragments with low toxicity (Rodriguez et al., 2020).
An antibacterial assay was conducted to verify the biodeactivation of amoxicillin. Three bacteria, E. coli ATCC 25922, S. aureus ATCC 25923, and B. subtilis ATCC 19615, were used to test for resistance to amoxicillin. These bacteria were selected because they are susceptible to amoxicillin (Mohammad et al., 2018). After incubating the A. tamarii isolate 58 for 24 hours, the amoxicillin began to degrade (Fig. 4). The antimicrobial assay revealed the percentage of growth inhibition of E. coli ATCC 25922 (79.31%), followed by B. subtilis ATCC 19615 (83.37%) and S. aureus ATCC 25923 (91.66%). Following a 48-hour incubation period, S. aureus ATCC 25923 exhibits the greatest degree of degradation (58.33%), followed by B. subtilis ATCC 19615 (66.66%), and E. coli ATCC 25922 (68.97%). Furthermore, B. subtilis ATCC 19615 and S. aureus ATCC 25923 both displayed 33.33% after 72 hours, whereas E. coli ATCC 25922 showed 58.62% of inhibition. Bacillus subtilis ATCC 19615 reveals that the antibiotic completely degraded at 96 hours, while S. aureus ATCC 25923 demands 120 hours for amoxicillin to degrade with a 0% percentage of inhibition. Aspergillus tamarii’s aggressive growth characteristics allow it to break down amoxicillin more effectively. Figure 5 indicates the percentage of inhibition after 120 hours by L. ramosa strain R. The antimicrobial assay reveals the percentage of growth inhibition of E. coli (63.33%), followed by S. aureus ATCC 25923 (66.66%) and B. subtilis ATCC 19615 (71.43%). Escherichia coli ATCC 25922 and B. subtilis ATCC 19615 displayed roughly the same percentages of growth inhibition throughout the 48 hours, at 56.66% and 57.14%, respectively. The smallest inhibitory zone, shown on S. aureus ATCC 25923, indicates greater amoxicillin degradation (58.33%). Following B. subtilis ATCC 19615, S. aureus ATCC 25923 and E. coli ATCC 25922 exhibited the maximum amoxicillin degradation and lowest percentage of inhibition from 72 to 120 hours. Only B. subtilis ATCC 19615 exhibited 0% percentage of inhibition as opposed to the other two bacteria.
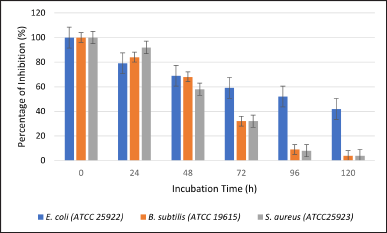 | Figure 4. Percentage of inhibition against E. coli (ATCC 25922), B. subtilis (ATCC 19615), and S. aureus (ATCC25923) with 0–120 hours amoxicillin exposure to A. tamarii isolate 58. [Click here to view] |
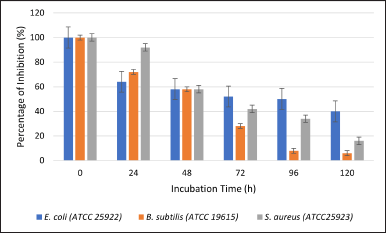 | Figure 5. Percentage of inhibition against E. coli (ATCC 25922), B. subtilis (ATCC 19615), and S. aureus (ATCC25923) with 0 to 120 hours amoxicillin exposure to L. ramosa strain R. [Click here to view] |
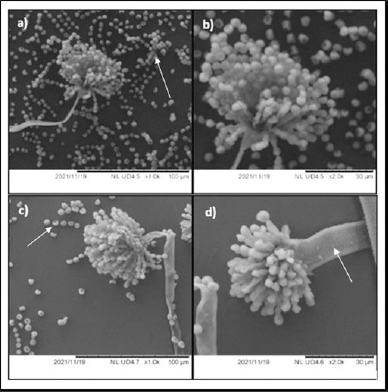 | Figure 6. SEM of A. tamarii isolate 58. (a) Untreated A. tamarii isolate 58 with 1,000× magnification shows numerous spores with a homogenous surface. (b) Untreated A. tamarii isolate 58 with 2,000× magnification. (c) Aspergillus tamarii isolate 58 treated with amoxicillin at 96 hours of incubation with 1,000× magnification shows a decline in spore count. (d) Aspergillus tamarii isolate 58 treated with amoxicillin at 96 hours of incubation with 2,000× magnification shows a shrunken hypha. [Click here to view] |
The β-lactam ring of amoxicillin can inhibit the cross-linkage between the linear peptidoglycan polymer chains of the bacterial cell wall. The enzymes produced by the fungi were able to hydrolyze or open the β-lactam ring and yield amoxicillin penicilloic acid, which contains a free carboxylic acid group and higher polarity than the parent molecule. This can be ascribed to the significant decrease in amoxicillin activity observed in the selected bacteria (Zhang et al., 2019).
Analyze the impact of amoxicillin using a SEM
Figure 6a and b depict the untreated A. tamarii isolate 58 with 1,000× and 2,000× magnification, respectively. The figures displayed numerous spores in homogenous size with smooth surfaces. However, Figure 6c and d depict the structure at 96 hours, after the impact of the antibiotic amoxicillin with 1,000× and 2,000× magnification, respectively, showed a significant decline in spore counts. A similar effect was also observed with L. ramosa strain R. The untreated L. ramosa strain R displayed numerous spores with smooth surfaces (Fig. 7a and b) and a marginal reduction after 96 hours of amoxicillin incubation (Fig. 7c and d).
Chandler (2017) stated that oxygen and water are necessary for the spore germination of fungi. PDB limits the amount of water that the fungi can exist in and affects spore formation with the presence of amoxicillin. In this study, the hyphae were shrinking rather than swelling, indicating that the hyphae were dehydrated (Figs. 6c and d and 7c and d). Despite having a strong cell wall, the fungi’s structure shrinks, and their spore germination is affected due to water scarcity. Thus, amoxicillin has an impact on the structure of fungi.
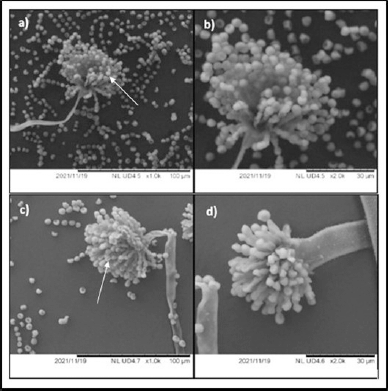 | Figure 7. SEM of L. ramosa strain R. (a) Untreated L. ramosa strain R with 1,000× magnification shows numerous spores with homogenous surfaces. (b) Untreated L. ramosa strain R with 2,000× magnification. (c) Lichtheimia ramosa strain R treated with amoxicillin at 96 hours of incubation with 1,000× magnification shows a decline in spore count. (d) Lichtheimia ramosa strain R treated with amoxicillin at 96 hours of incubation with 2,000× magnification shows a shrunken hypha. [Click here to view] |
CONCLUSION
In conclusion, morphological studies and molecular techniques successfully identified both fungi A. tamarii isolate 58 and L. ramosa strain R. The HPLC-DAD was used to successfully quantify the amount of amoxicillin residue after biodegradation for both fungi. After 96 hours, A. tamarii isolate 58 and L. ramosa strain R had residual amoxicillin levels of 51.0% and 42.0%, respectively. After 120 hours, the inhibition zone diameter for both fungi in E. coli ATCC 25922, B. subtilis ATCC 6633, and S. aureus ATCC 25923 decreased due to antimicrobial activity. The antibacterial effects of the amoxicillin were diminished after biodegradation by A. tamarii isolate 58 and L. ramosa strain R. Finally, SEM images showed a significant decline in spore counts from 0 to 96 hours.
LIMITATIONS AND FUTURE RESEARCH
Enzyme studies on amoxicillin degradation are limited by the absence of functional gene expression. Future studies are therefore, thus, needed to investigate the behavior of amoxicillin at lower concentrations and identify the specific enzyme needed to break down amoxicillin. Therefore, a method of safely decomposing amoxicillin in the environment can be developed.
ACKNOWLEDGMENT
The authors would like to thank Universiti Teknologi MARA and the Ministry of Higher Education Malaysia.
AUTHOR CONTRIBUTIONS
All authors contributed significantly to the work’s concept and design, data collection, analysis, and interpretation, as well as its drafting and critical revision for key intellectual content and the final approval of the published version. All authors have agreed to be accountable for all parts of the work and have participated to ensure that questions about the work’s accuracy or integrity are adequately investigated and resolved. All are entitled to be specified as authors according to the International Committee of Medical Journal Editors’ guidelines (ICMJE).
FINANCIAL SUPPORT
This research study was fully funded by FRGS/1/2018/WAB05/UITM/02/5, Universiti Teknologi MARA (UiTM), and the Ministry of Higher Education Malaysia.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVAL
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
This research article presents all the data that was generated and examined.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Ariffin ZZ, Mohammad NS, Saffian MF, Mohammad Ali NA. Antibacterial and ferric reducing ability activities of selected herbs essential oils. J Eng Appl Sci, 2017; 12:6934–8; DOI: 10.36478/jeasci.2017.6934.6938.
Blount BA, Lu X, Driessen MR, Jovicevic D, Sanchez MI, Ciurkot K, Zhao Y, Lauer S, McKiernan RM, Gowers GOF, Sweeney F, Fanfani V, Lobzaev E, Palacios-Flores K, Walker R, Hesketh A, Oliver SG, Cai Y, Stracquadanio G, Mitchell LA, Bader JS, Boeke JD, Ellis T. Synthetic yeast chromosome XI design enables extrachromosomal circular DNA formation on demand. bioRxiv, 2022; 2022-07.
Chandler D. Basic and applied research on entomopathogenic fungi. Microb Control Insect Mite Pests, 2017:69–89; https://DOI.org/10.1016/b978-0-12-803527-6.00005-6
de Andrade Silva CA, da Silva PGP, da SilvaGFA, Dantas DP, Leite RSR, Fonseca GG. Biotransformation of fruit residues via solid state bioprocess using Lichtheimia ramosa. SN Appl Sci, 2020; 2(5):1–10.
Guardado ALP, Belleville MP, Alanis MDJR, Saldivar RP, Sanchez-Marcano J. Effect of redox mediators in pharmaceuticals degradation by laccase: a comparative study. Process Biochem, 2019; 78:123–31.
Hamid AH, Ariffin ZZ. Bioremediation of amoxicillin, beta-lactam antibiotic by locally isolated soil fungi. Junior Sci Commun, 2020; 3:eISSN 2637–0689.
Homa M, Manikandan P, Szekeres A, Kiss N, Kocsubé S, Kredics L, Papp T. Characterization of Aspergillus tamarii strains from human keratomycoses: molecular identification, antifungal susceptibility patterns and cyclopiazonic acid producing abilities. Front Microbiol, 2019; 10; DOI:10.3389/fmicb.2019.02249.
Imade YN, Rukayat OK, Olubunmi TA, Busayo TA. Isolation of an emerging thermotolerant medically important fungus, Lichtheimia ramosa from the soil. Afr J Microbiol Res, 2020; 14(6):237–41; https://DOI.org/10.5897/ajmr2020.9358.
Ji J, Gao T, Salama ES, El-Dalatony MM, Peng L, Gong Y, Li X. Using Aspergillus niger whole-cell biocatalyst mycelial aerobic granular sludge to treat pharmaceutical wastewater containing β-lactam antibiotics. Chem Eng J, 2021; 412:128665.
Kamaruddin FF, Musa M, Mahat MM, Ariffin SHZ, Safian MF, Ariffin ZZ. Biodegradation of doxycycline hyclate by local Purpureocillium lilacinum strain PlHN17 and Trichoderma asperellum isolate Tullur: monitoring by antimicrobial activity. Jordan J Biol Sci, 2022; 15(3):457–61.
Kraemer SA, Ramachandran A, Perron GG. Antibiotic pollution in the environment: from microbial ecology to public policy. Microorganisms, 2019; 7(6):180.
Midorikawa GEO, Correa CL, Noronha EF, Filho EXF, Togawa RC, Costa MMDC, Silva-Junior O, Grynberg P, Miller RNG. Analysis of the transcriptome in Aspergillus tamarii during enzymatic degradation of sugarcane bagasse. Front Bioeng Biotechnol, 2018; 6; DOI: 10.3389/fbioe.2018.00123
Mohammad NS, Safian MF, Zainal Ariffin SH, Ariffin ZZ. Biotransformation of nitrofurans antibiotics by Aspergillus species-residual antibacterial activity. Malays J Biochem Mol Biol, 2018; 21(2):28–33.
Mukhopadhyay D, Khan N, Kamal N, Varjani S, Singh S, Sindhu R, Bhargava PC. Degradation of β-lactam antibiotic ampicillin using sustainable microbial peroxide producing cell system. Bioresour Technol, 2022; 361:127605.
Muthulakshmi C, Gomathi D, Kumar DG. Production, purification and characterization of protease by Aspergillus flavus under solid state fermentation. Jordan J Biol Sci, 2011; 147(621):1–12.
Rodriguez DC, Ahammad ZS, Penuela GA, Graham DW. Effect of β-lactamases associated to the resistance of Β-lactam antibiotics on the treatment of wastewater. J Environ Chem Eng, 2020; 8(1):102247.
Senwan MS, Safian MF, Noor ZM, Ariffin ZZ. Isolation and characterisation of filamentous fungi from animal agricultural farm soil. Pertanika J Sci Technol, 2017; 25:19–28.
Shanmugavel M, Vasantharaj S, Yazhmozhi A, Bhavsar P, Aswin P, Felshia C, Gnanamani A. A study on pectinases from Aspergillus tamarii: toward greener approach for cotton bioscouring and phytopigments processing. Biocatal Agric Biotechnol, 2018; 15:295–303; DOI: 10.1016/j.bcab.2018.06.013.
Timm A, Borowska E, Majewsky M, Merel S, Zwiener C, Bräse S, Horn H. Photolysis of four β?lactam antibiotics under simulated environmental conditions: degradation, transformation products and antibacterial activity. Sci Total Environ, 2019; 651:1605–12.
Tooke CL, Hinchliffe P, Bragginton EC, Colenso CK, Hirvonen VH, Takebayashi Y, Spencer J. β-Lactamases and β-lactamase inhibitors in the 21st century. J Mol Biol, 2019; 431(18):3472–500.
Venkitasamy C, Zhao L, Zhang R, Pan Z. Pomegranate. Integr Process Technol Food Agric By-Prod, 2019:181–216; DOI: 10.1016/b978-0-12-814138-0.00008-3
Wang P, Shen C, Cong Q, Xu K, Lu J. Enzyme-catalyzed biodegradation of penicillin fermentation residues by β-lactamase OtLac from Ochrobactrum tritici. Microb Cell Fact, 2021; 20:117; https://doi.org/10.1186/s12934-021-01606-2
Zhang Y, Xiao Y, Zhong Y, Lim TT. Comparison of amoxicillin photodegradation in the UV/H2O2 and UV/persulfate systems: reaction kinetics, degradation pathways, and antibacterial activity. Chem Eng J, 2019; 372:420–8.
Zulkifle NT, Abd Khalil K, Safian MF, Saat MN, Ariffin ZZ. Optimization of nitrofurazone degradation by local Aspergillus tamarii KX610719. Asia Pac J Mol Biol Biotechnol, 2022; 30(1):51–61.