Candida albicans: A HUMAN FUNGAL PATHOGEN
Tremendous health care implications in terms of cost, morbidity and mortality are associated with fungemia worldwide[1-5]. The yeasts of the genus Candida are opportunistically invasive in individuals whose defense mechanisms are impaired. Pathogenic Candida species cause diseases ranging from superficial mycoses to fatal infections[6]. The incidence of candidiasis caused by different Candida species continues to increase in proportion to the growing number of immunocompromised hosts, cancer patients and organ transplant recipients. Other individuals also at risk include intensive care and postoperative patients, individuals with hematological malignancies, elderly patients, premature infants, patients under prolonged broad-spectrum antibiotic therapy and human immunodeficiency virus (HIV)-infected persons[7-10]. The infections caused by Candida spp. result in increased length of hospital stay and medical costs that constitute an important public health problem[11,12].
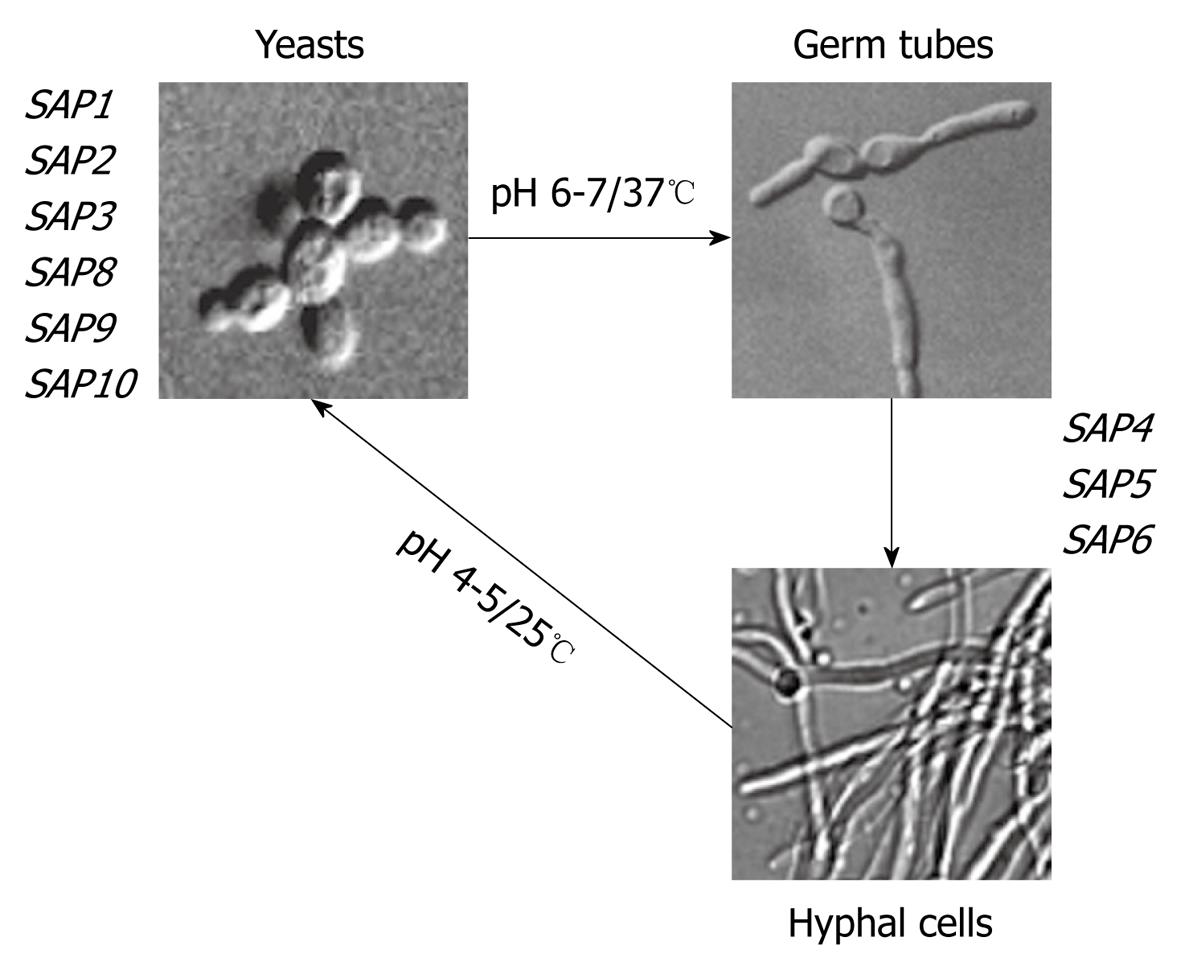
Candida albicans (C. albicans), a pleomorphic fungus (Figure 1), is one of the most common human commensals. It colonizes the mucocutaneous surfaces of the oral cavity, gastrointestinal tract and vagina[6]. Under certain host circumstances, C. albicans can proliferate in a saprophytic state and can become pathogenic. Given the high levels of morbidity and mortality associated with nosocomial candidiasis, the pathogenic adaptation of C. albicans has been the topic of extensive investigations[6]. Delayed therapy for invasive candidiasis contributes to a poor outcome. Unfortunately, traditional diagnostic techniques remain insensitive and slow. Whilst efficacious, antifungal prophylaxis is inefficient. As such, early antifungal strategies need to be targeted to maximize benefits and minimize adverse consequences; with no doubt, this remains the major challenge.
Figure 1 Morphological stages of Candida albicans (C.
albicans) and differential expression of SAP genes. Both temperature and pH values of growth media directly influence the yeast into hyphal transition as well as the expression of SAP genes. In this sense, the expression of SAP1-SAP3 and SAP8-SAP10 has been detected in yeast cells, while SAP4-SAP6 has been associated with the filamentous forms.
Candidiasis may occur as a result of disturbed balance between host immunity and this opportunistic pathogen. This disorder is not only due to the immunological dysfunction of the host, but also to the ability of the fungus to adapt to new niches, dependent on the expression of infection-associated genes[13]. These genes and their products contribute to fungal pathogenicity and are described as virulence factors[14-17]. As a commensal and opportunistic pathogen, C. albicans possesses a range of determinants that contribute to survival, persistence and virulence. Among this repertoire of fitness and virulence attributes, the dimorphic transition, antigenic variability, ability to switch among different cell phenotypes, adhesion to inert and biological substrates, immunomodulation of host defense mechanisms and production of secreted hydrolytic enzymes, such as lipases and proteases, all stand out[14-17].
SECRETED ASPARTYL PROTEASES: THE MAJOR VIRULENCE FACTORS OF C. albicans
Aspartyl proteases constitute one of the four super families of proteolytic enzymes showing acidic optima pH for enzymatic activity, and are totally inhibited by pepstatin A, a hexapeptide produced by Streptomyces[18,19]. Secreted aspartyl proteases (Saps) are the major virulent factors produced by C. albicans[14]. Saps are encoded by a gene family with 10 distinct members (SAP1-SAP10) that are located on five different chromosomes. Alignments show that the products of SAP1, SAP2 and SAP3 are approximately 75% identical. Another distinct subgroup contains SAP4, SAP5 and SAP6 showing 90% identity to each other, which falls to 65% when compared with SAP1-SAP3. SAP8 is most similar to SAP1 (65%) and SAP9 and SAP10 have a C-terminal extension. SAP7 is the most diverged member of this family[20,21].
All 10 SAP genes of C. albicans encode preproenzymes approximately 60-200 amino acids longer than the mature enzyme, which are processed when transported via the classical secretory pathway. The pathway of protease synthesis starts in the nucleus, from where the newly synthesized messenger RNA (mRNA) is transferred to the cytoplasm and translated into the preproenzyme by ribosomes on the rough endoplasmic reticulum (RER). The N-terminal secretion signal of each Sap preproenzyme is cleaved by a signal peptidase in the RER[22], and the proenzyme transferred to the Golgi apparatus. Later in the trans Golgi network, the propeptide is removed by the subtilisin-like Kex2 serine protease, which cleaves a lysine-arginine sequence, in order to activate the protease[23]. This was confirmed for Sap2 by N-terminal sequencing[24] and its three dimensional structure[25]. However, alternative processing pathways for Saps must exist, because a Kex2 disruption mutant of C. albicans was shown to secrete aberrantly processed, but active Sap2[26]. The pro region probably acts as an intramolecular chaperone that is required for proper folding of the Sap protein in the RER[27]. Leaving the Golgi, the Saps are packaged into secretory vesicles and then transported to the plasma membrane and are either released into the extracellular environment (Sap1-8)[28] or remain attached to the cell membrane or cell wall via a glycosylphosphatidylinositol (GPI) anchor (Sap9 and Sap10)[21,29,30]. The mature enzyme in the range of 35-48 kDa contains sequence motifs typical for all aspartyl proteases, including the two conserved aspartate residues of the active site. Conserved cysteine residues are probably implicated in maintaining the three-dimensional structure of the enzyme[31].
The SAP genes are differentially expressed depending on the C. albicans strain and environmental conditions[32-36] (Figure 1). For instance, eight of these proteases (Saps1-8) are secreted into the extracellular environment, while Sap9 and Sap10 are membrane GPI-anchored proteins[37]. Saps1-3, which are expressed predominantly in yeast cells, have optimal activity within the pH range of 2.5-5.5, whereas Saps4-6 have greater activity at a higher pH range (3.0-7.0) and are mainly expressed in hyphal forms[38]. Increasing the pH above 7.0 causes irreversible alkaline denaturation that involves protein dimerization[39]. Denaturation also occurs rapidly at temperatures above 45°C[40].
Enzymatic studies of these isoenzymes have also revealed significant differences in substrate specificities[40,41], suggesting that the large set of isoenzymes provides C. albicans with a capacity to hydrolyze a variety of substrates under a wide range of conditions. In this context, Sap1, Sap2, Sap3 and Sap6 are known to cleave peptide bonds between larger hydrophobic amino acids, but preferred the P1 and P’1 sites; Sap1, Sap2 and Sap6 preferred phenylalanine, while Sap3 preferred leucine at the P1 site[41]. Positively charged amino acids are also accommodated, especially by Sap2 and Sap3. The specificities at P’1 are broader than at P1 for all four studied enzymes. Sap6 prefers alanine, whereas other Saps prefer tyrosine[41]. In contrast, Sap9 and Sap10 seem to hydrolyze very distinct substrates similar to fungal yapsins and kexins[30]. The observed differences in residue preferences among Saps may be utilized in the design of specific substrates and inhibitors.
Most of the biochemical studies used culture conditions that favored SAP2 expression[32,38] and accordingly mainly characterized Sap2. This isoenzyme acts mainly at acidic pH values (pH for optimal activity ranging from 3.2-3.5) and seemed to have broad substrate specificity[31]. Collectively, its broad capability in degrading different proteinaceous structures, such as serum proteins, extracellular matrix components, sialylated proteins and antimicrobial peptides, allow fungal cells to obliterate relevant protein barriers culminating in the dissemination and escape of the host immune response[16,37].
The presence of 10 SAP genes indicated that different proteases might also target a variety of host cells and tissues as they are differentially regulated and expressed under a variety of laboratory growth conditions and during C. albicans infectious processes. Curiously, the promoter regions of SAP genes are distinctive, which would not only allow the regulation of individual SAP genes by several transcriptional regulators but also the coordinated expression of SAP genes with other virulence attributes, including hyphal formation and phenotypic switching[16]. Detailed analysis of the in vitro expression of SAP genes revealed that the major protease gene expressed in C. albicans forming at 30-37°C in defined media containing protein (such as albumin and hemoglobin) as the sole nitrogen source, was SAP2[38]. Thus, the synthesis and secretion of Sap2 is regulated by exogenous protein via a positive feedback mechanism, in which the accumulation of peptides resulting from proteolysis of high-molecular mass proteins leads to the induction of SAP2 gene expression[38]. On the other hand, yeasts grown at 25°C showed levels of SAP8 transcripts and Sap8 protein increased, suggesting that the expression of this gene is temperature regulated[21]. SAP1 and SAP3 genes are differentially expressed in vitro when C. albicans undergoes phenotypic switching[32,42], a phenomenon whereby the fungus randomly switches its phenotype, especially in response to stress. In this context, SAP1 mRNA expression is associated with the white-opaque switch in strain WO-1. Log-phase opaque cells contained high levels of SAP1 mRNA that rapidly decreased upon conversion to the white-phase[43]. Expression of SAP9 and SAP10 appears to be independent of environmental conditions, in view of the fact that they are constitutively expressed in both yeast and hyphal forms[44,45]. SAP4-6 genes are almost exclusively expressed during hyphal formation at neutral pH[46]. Interestingly, antifungal agents (itraconazole, miconazole, flucytosine and caspofungin) with three different mechanisms of action similarly generate a rise in expression of SAP2 gene and activity of the secreted Sap2 gene product in C. albicans[47]. Similarly, exposure of C. albicans to subinhibitory concentrations of fluconazole in RPMI 1640 in the absence of serum led to up-regulation of the virulence-associated genes SAP4, SAP5 and SAP6 in hyphae and long pseudohyphae forms[48]. In addition, SAP genes are regulated by biofilm formation, because greater levels of SAP5, SAP6 and SAP9 mRNA transcripts were detected in biofilm rather than during planktonic growth[49].
The importance of specific Sap isoenzymes for the pathogenicity of C. albicans has been investigated by comparing the virulence of mutants deleted for individual or multiple SAP genes with that of a wild-type control strain in different infection models. For example, mutants deleted for either SAP1, SAP2 or SAP3 were found to be less virulent in a rat model of Candida vaginitis, whereas mutants lacking SAP4-SAP6 did not exhibit a detectable virulence defect under these conditions[50]. In contrast, only the latter mutants showed reduced virulence in a murine model of Candida peritonitis, while deletion of SAP1, SAP2 or SAP3 had no significant effect in this infection model[51]. In another work, SAP7 gene transcript was detected in 60% of oral candidiasis patients as opposed to 25% of Candida carriers by means of real time-polymerase chain reaction[52]. In order to corroborate the relevance of Saps during human infection, several studies described the presence of Saps’ proteins in different anatomical sites. For example, high titers of anti-Sap antibodies were observed in the sera of candidiasis patients[53], and Sap antigens were detected in biopsies of oral epithelial lesions collected from HIV-infected patients[28] and in almost all organs of immunocompromised patients who had died of systemic C. albicans infections[54]. The importance of Saps in establishing systemic murine candidiasis was first demonstrated by the protective role of pepstatin A, an archetypal aspartyl protease inhibitor[55]. Due to multifaceted roles in both the physiological and pathological processes of C. albicans cells, Saps are targets for the development of future antifungal drugs.
ASPARTYL PROTEASE INHIBITORS: COMPOUNDS WITH ANTI-Candida PROPERTIES
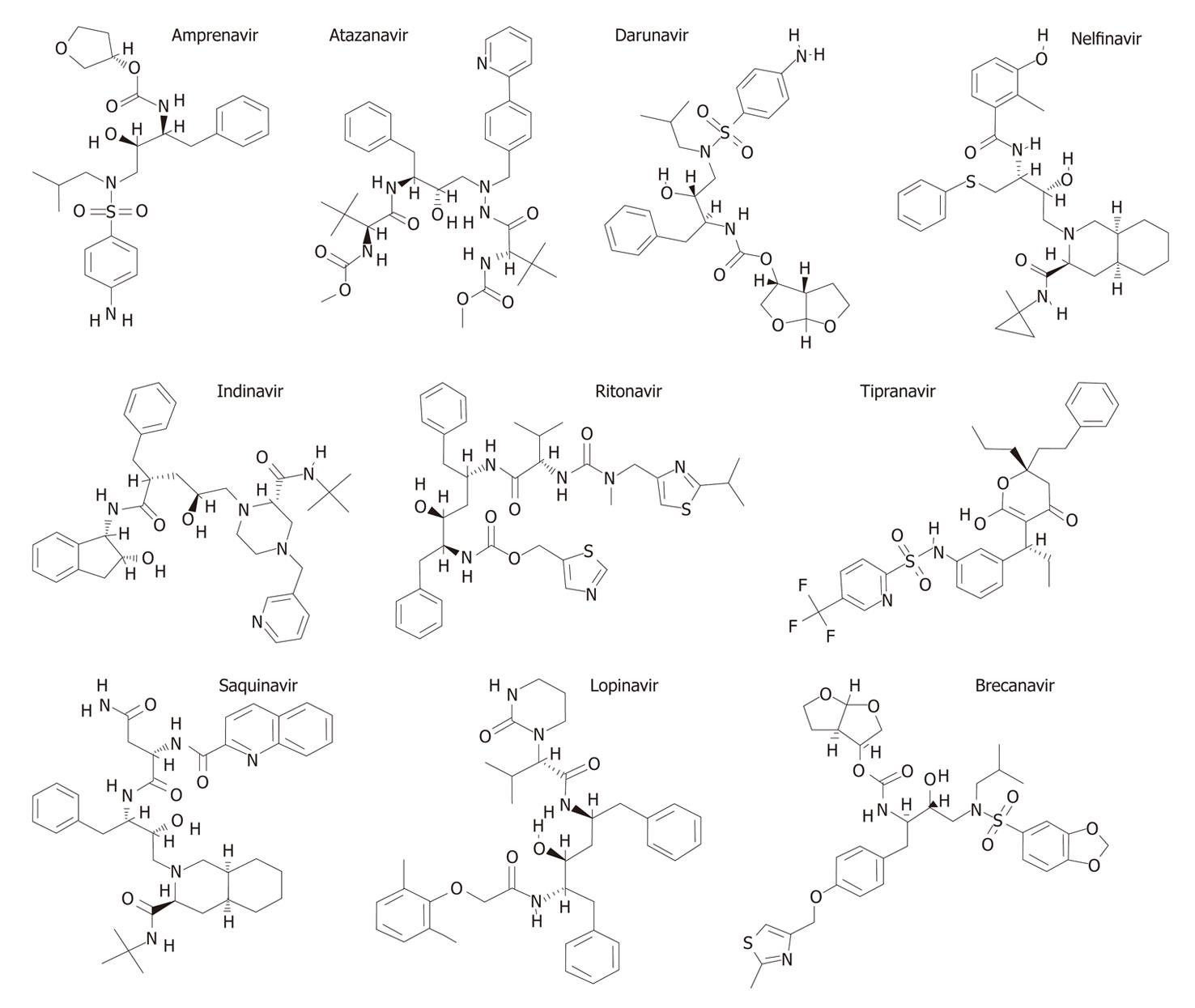
The last decade has witnessed an effervescence of research in the development of potent inhibitors of various aspartyl proteases due to their implications in several pathologies[56-58], including renin in hypertension[59], cathepsin D in breast cancer metastases[60], cathepsin E in the immune system[61], β-secretase in the Alzheimer’s disease[62], pepsin in various gastric disorders[63], plasmepsins in malaria[64], HIV protease in acquired immunodeficiency syndrome (AIDS)[63] and Saps in candidal infections[65]. In this context, the HIV protease inhibitors (PIs) were the first success in structure-based drug design[66]. There are at present ten PIs licensed for clinical use in the treatment of HIV infection: saquinavir, ritonavir, indinavir, nelfinavir, amprenavir (fosamprenavir), lopinavir, atazanavir, brecanavir, tipranavir and darunavir (Figure 2).
Figure 2 Aspartyl protease inhibitors used in the current human immunodeficiency virus (HIV) chemotherapy.
Clinical evidence of the beneficial use of HIV PIs
The introduction of HIV PIs has revolutionized the treatment of HIV disease. Their clinical use has been associated with a dramatic decline in opportunistic infections, including fungal infections, the number of deaths due to AIDS, the number of new cases of AIDS and the number of hospitalization days[67,68]. For instance, Palella et al[68] analyzed data from 1255 HIV-infected patients in the United States of America from January 1994 through June 1997 who had a CD4+ cell count lower than 100/mm3. Mortality from AIDS declined from 29.4 per 100 person-years in 1995 to 8.8 per 100 person-years in the second quarter of 1997. The incidence of major opportunistic infections also decreased, from 21.9 per 100 person-years in 1994 to 3.7 per 100 person-years by mid-1997[68].
HIV PIs rapidly and profoundly reduce the viral load, as indicated by a decline in the plasma HIV RNA concentrations within a few days after the start of treatment. Reductions in the viral load are paralleled by increases in CD4+ T lymphocyte counts[69] and stimulation in the survival and activation of neutrophils, monocytes and endothelial cells[70-73]. HIV PIs also exert unpredicted actions, which may have beneficial effects. For example, they inhibit inflammatory cytokine production and modulate antigen presentation and T-cell responses[73-75]. Consequently, HIV PIs have helped to dramatically reduce AIDS disease progression and mortality, prolonging survival of HIV-infected individuals[68,76,77].
Oropharyngeal candidiasis is one of the most common opportunistic infections in HIV and AIDS patients, being considered a clinical marker for disease progression[78,79]. Oropharyngeal candidiasis was highly prevalent at the beginning of the HIV/AIDS epidemic[80,81]. Findings from clinical trials with HIV-infected populations have shown a decrease in oral candidal infections as well as other oral manifestations of the HIV infection[82-84]. For instance, in a multinational cohort study involving 6941 HIV-positive individuals from Australia and ten European countries, when comparing the periods 1997-2001 and 1994-1996, there was a significant HIV PI-induced decrease in progression of candidiasis from 17.0% to 5.7%[85].
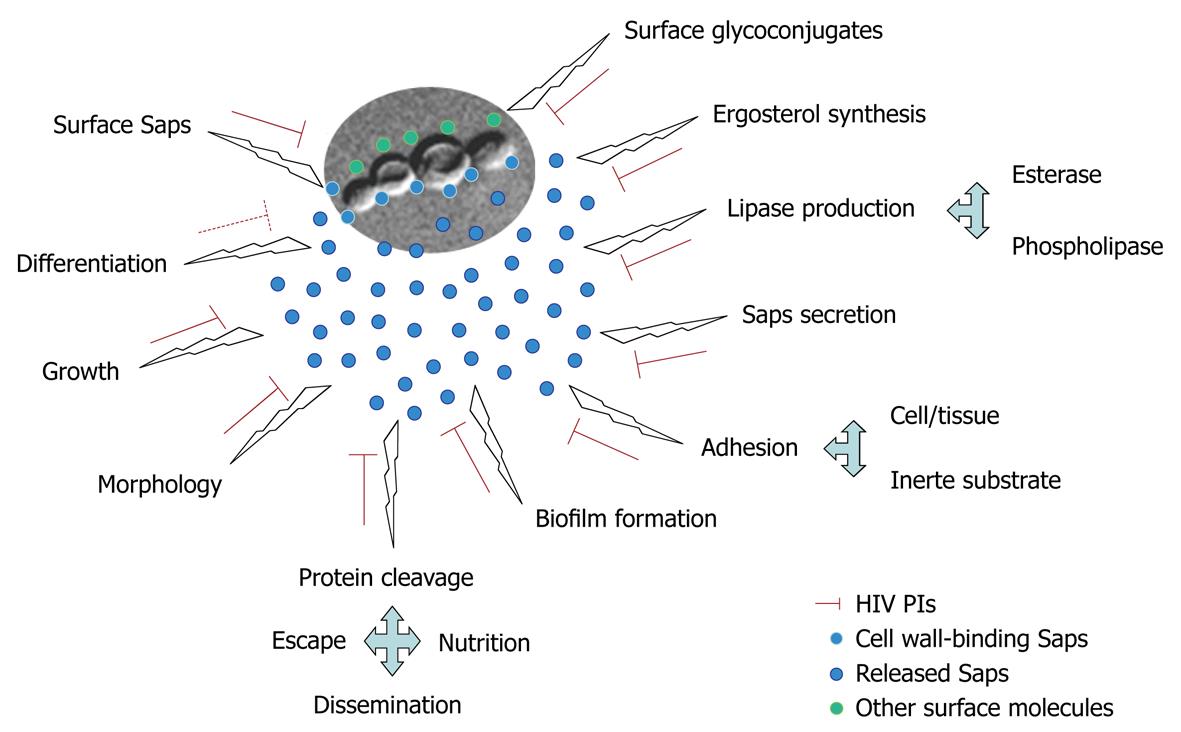
HIV PIs act directly on multiple relevant processes of C. albicans, reducing Candida carriage and recurrence of oropharyngeal candidiasis in HIV patients[86,87]. The attenuation of candidal infections in HIV-infected individuals in the highly active antiretroviral therapy (HAART) era might not solely have resulted from improved immunological status, but also as a result of direct inhibition of Candida Saps (Figure 3), which belong to the same family as HIV protease, by the HIV PIs contained in HAART[65].
Figure 3 Different roles performed by C.
albicans Saps in vital biological processes as well as during the different steps of interaction with host structures. Both physiological and pathological processes are inhibited (red lines) in different extensions, depending on HIV protease inhibitors (PIs), except for the yeast to mycelia transformation (differentiation) where conflicting results were observed (dotted red line).
Sap inhibition
The inhibition of Sap activity by HIV PIs was described for the first time in 1999 by distinct research groups. In this sense, saquinavir and indinavir restrained Sap activity when bovine serum albumin (BSA), hemoglobin and human stratum corneum were used as proteinaceous substrates[88]. In parallel, indinavir and ritonavir promoted a noticeable dose-dependent inhibition of Sap production and activity when evaluated both antigenically and as direct enzyme activity on BSA as substrate[89]. Similarly, Gruber et al[90] showed that indinavir promoted a dose-dependent decrease in cell wall-bound Sap antigen by flow cytometry analysis. Borg-von Zepelin et al[91] demonstrated that purified Sap1, Sap2 and Sap3 were inhibited by ritonavir, saquinavir, indinavir and nelfinavir in a concentration-dependent manner, in which ritonavir was by far the most effective inhibitor of these Sap isoenzymes. In contrast, only a slight inhibition of activity was detected for purified Sap4, Sap5 and Sap6. Recently, Braga-Silva et al[92] showed that amprenavir was found to be the most potent inhibitor of Sap2, which is the main pronounced virulence factor expressed during C. albicans infections, reducing the proteolytic activity by approximately 92%, while the other HIV PIs tested (ritonavir, saquinavir, indinavir, nelfinavir and lopinavir) inhibited around 45%-65% of the proteolytic activity. For that reason, amprenavir was selected to determine its dose response on Sap2, revealing that the inhibition increased from 85% to 100% as amprenavir concentration increased from 6.25 to 200 μmol/L[92].
Growth arrest
C. albicans cells were obliged to produce Saps in liquid medium containing only high-molecular-mass proteins in order to obtain peptides and amino acids for their growth and development, however, this was unnecessary in complex medium because of a sufficient supply of nutrients[32-34]. Indinavir and ritonavir markedly inhibited the growth of C. albicans when cultured in yeast carbon base medium supplemented with BSA as the sole nitrogen source. As expected, in nitrogen-unlimited complex media, where SAP expression is not required for growth, HIV PIs did not arrest C. albicans development[89]. Similarly, amprenavir impaired yeast growth in a concentration-dependent manner when 103 cells were used; conversely, treatment of 104 yeast cells with up to 200 μmol/L did not modify the growth behavior of C. albicans, showing a typical cell- and drug-concentration inhibition mechanism[92].
Ultrastructure alterations
By means of scanning electron microscopy analyses, Braga-Silva et al[92] reported alterations at the morphological level after treatment of C. albicans yeast cells with different concentrations of amprenavir. The normal morphology of yeasts, which varies from spherical to oval cells, was compared with the ultrastructure of amprenavir-treated cells which showed alterations in the shape and cell surface sculpturing, such as invaginations at the surface. Interestingly, the surface of normal yeast cells had numerous protrusions. In contrast, yeasts treated with amprenavir at 200 μmol/L had a smooth surface, whereas treatment with 100 μmol/L showed much smaller quantities of attached material, which might represent remnants of the former surface irregular layer[92].
Fungal-abiotic materials binding impediment
Candidal adhesion to any oral substrate is the first and essential stage in the formation of a pathogenic fungal biofilm, which is in turn a prerequisite for the microorganism to ingress into the human host. In general, yeast cells are known to have a remarkable potential to adhere to host surfaces, such as teeth or mucosa, and to artificial, non-biological surfaces, such as implanted dental devices[93]. Tsang et al[94] reported the inhibitory action of HIV PIs on the adhesion of C. albicans to acrylic materials, which are common components of oral appliances. In that study, C. albicans cells were pretreated with different concentrations of saquinavir, ritonavir or indinavir and then allowed to adhere on acrylic strips, which had been pretreated with pooled human saliva. Adhesion was reduced by approximately 50% at drug concentrations of 100, 100 and 20 μmol/L for saquinavir, ritonavir and indinavir, respectively. In another study, amprenavir was able to block the adhesion of C. albicans to polystyrene substrate, reducing the ability of fungal cells to form a biofilm[92].
Inhibition of fungal adhesion to host cells
Adhesion to host cells in various target tissues is widely accepted as a first step in the pathogenesis of candidiasis[95]. In this regard, Borg-von Zepelin et al[91] demonstrated that ritonavir and saquinavir inhibited the adhesion of C. albicans to epithelial cells of the Vero lineage, while indinavir had no inhibitory effect. In a similar manner, Bektić et al[96] showed that ritonavir, indinavir and saquinavir attenuated the adherence properties of C. albicans to the human vaginal epithelial cancer cell line HeLa S3; however, no effect on phagocytosis by polymorphonuclear leukocytes was observed. On the other hand, the adhesion process between C. albicans and human umbilical vein endothelial cells (HUVEC) was not reduced by ritonavir, indinavir or saquinavir, suggesting that inhibition of C. albicans adhesiveness by HIV PIs is clearly epithelial cell-specific[97].
Differentiation interference
It is well known that yeast-germ tube-hyphae transformation is a hallmark event during the pathogenesis of C. albicans[98]. C. albicans treated with or without indinavir retained their ability to form hyphae. However, the hyphal elongation of indinavir-treated cells was delayed[90]. Surprisingly, tipranavir, a non-peptidic HIV PI, triggered the mycelial transition in C. albicans yeasts[99]. Curiously, tipranavir impaired the in vitro growth of Cryptococcus neoformans, but not that of C. albicans. Tipranavir-treated C. neoformans, but not C. albicans, was more susceptible to killing by human neutrophils. These data indicate that tipranavir could act in multiple ways by diversifying its effects on various opportunistic/pathogenic fungi[99]. In an unpublished result from our laboratory, we showed that lopinavir efficiently blocked the yeast to germ tube transformation when C. albicans cells were incubated for 3 h at 37°C in the presence of fetal bovine serum, a condition that induces the fungal transition.
Down modulation of unrelated virulence factors
In C. albicans, tipranavir induced a significant reduction in both protease and phospholipase production (which catalyze the hydrolysis of proteins and phospholipids, respectively, that are major components of all cell membranes, thereby also facilitating fungal invasion)[99]. In addition, the treatment of yeast cells with amprenavir drastically reduced the expression of surface mannose- and sialic acid-rich glycoconjugates (which are carbohydrates directly linked to the yeast-adhesive processes) and inhibited esterase activity (an enzyme that hydrolyzes monoacylglycerol with a long chain fatty acid, aiding host membrane disruption), ergosterol content (which controls the fungal membrane fluidity), and biofilm formation (which is a common mechanism utilized by this fungus to survive hostile environments, such as the presence of antifungal agents) in C. albicans[92].
Inhibition of experimental candidiasis infection
Borg et al[100] detected Sap antigens on the surface of adhering blastoconidia and invading filamentous cells of C. albicans during the infection of human buccal mucosa by immuno-scanning electron microscopy. In addition, pepstatin A revealed an 89% reduction in the mucosal adherence of C. albicans[100]. Korting et al[88] described the first evidence for a direct protective role of saquinavir during experimental oral candidiasis. The authors showed that without saquinavir, at 12 h after C. albicans infection, a powerful epithelial lesion was observed with vacuolation, edema and detachment of all epithelial layers, and C. albicans was able to invade the epithelium. All the morphological alterations were robustly reduced when saquinavir was added at a concentration of 0.3 μmol/L[61]. The exciting work of Cassone et al[89] reported that indinavir and ritonavir were able to promote a therapeutic effect in an experimental model of vaginal candidiasis (estrogen-dependent rat vaginitis), with an efficacy comparable to that of fluconazole, a recognized anti-candidal agent.
Synergistic action
Combination therapy has arisen to diminish the fungal resistance phenomenon. The combination of HIV PIs and classical antimycotic drugs could help to enhance the therapeutic treatment of mycoses. In this context, the combination of antimycotic agents and saquinavir both at sub-inhibitory concentrations was effectively demonstrated against strains of C. albicans isolated from HIV-seropositive patients. The results described by Casolari et al[101] showed that the interaction between saquinavir and amphotericin B, 5-fluorocytosine, miconazole or fluconazole did not result in antagonism, and fluconazole acted in a more synergistic way. The advantage of the synergic effect of this combination was to attenuate the resistance and accompanying toxic phenomena, particularly and most importantly, in the event of long-term therapy as a result of the use of low dosages of both HIV PIs and anti-fungal drugs.
CONCLUSION
Nowadays, C. albicans is thought to be the major fungal pathogen in humans. Severe Candida infections are a serious problem, especially in individuals whose immune defense mechanisms have been weakened[102]. C. albicans has developed an extensive repertoire of putative virulence mechanisms that allows successful colonization and infection of the host under suitable predisposing conditions[13,103]. Efficient anti-Candida drugs need to be capable of blocking at least some of these virulence attributes. With this fact in mind, several beneficial effects of HIV PIs against C. albicans have been reported in the last 10 years. In this scenario, several studies described the inhibitory effects of HIV PIs on crucial physiological processes as well as on relevant steps in C. albicans-host interplays (Figure 3). However, it is also important to note that the inhibitory effects of HIV PIs both in in vitro and in vivo experimental models were observed at concentrations (μmol/L range), much higher than those needed for HIV protease inhibition (nmol/L range). This probably reflects a much lower affinity of these drugs for Sap than that for HIV protease[65,76]. Another explanation is that in contrast to the very small and structurally simplified HIV protease, Saps are larger and more complex[104,105]. They possess a relatively large active site which might be responsible for the broader substrate specificity and also their susceptibilities to distinct aspartyl PIs[105]. Nevertheless, the above concentrations may be achieved under current HAART regimens both in the blood[76] and (at least for indinavir) in human saliva, as shown by Hugen et al[106]. In future, derivatives of HIV PIs, being more specific for the C. albicans Saps, may form an alternative in the treatment of mucosal candidiasis insensitive to currently available anti-fungals. In this context, the crystal structure of Sap2 complexed with pepstatin A has been known since 1993[107], whereas the crystal structure of Sap3 and its complex with pepstatin A was first presented in 2007[108]. The secondary structures of Sap2 and Sap3 as well as Sap1 and Sap5 were recently described[109]. These data could help in the development of novel and more effective anti-C. albicans compounds. Furthermore, future research into the synergistic combinations of inhibitors targeting Saps, used at sub-inhibitory concentrations in order to obtain a reduction in their toxic effects, will offer the most exciting prospects[110-112].