Abstract
The peptide repertoire presented by classical as well as nonclassical MHC class I (MHC I) molecules is altered in the absence of the endoplasmic reticulum aminopeptidase associated with Ag processing (ERAAP). To characterize the extent of these changes, peptides from cells lacking ERAAP were eluted from the cell surface and analyzed by high-throughput mass spectrometry. We found that most peptides found in wild-type (WT) cells were retained in the absence of ERAAP. In contrast, a subset of “ERAAP-edited” peptides was lost in WT cells, and ERAAP-deficient cells presented a unique “unedited” repertoire. A substantial fraction of MHC-associated peptides from ERAAP-deficient cells contained N-terminal extensions and had a different molecular composition than did those from WT cells. We found that the number and immunogenicity of peptides associated with nonclassical MHC I was increased in the absence of ERAAP. Conversely, only peptides presented by classical MHC I were immunogenic in ERAAP-sufficient cells. Finally, MHC I peptides were also derived from different intracellular sources in ERAAP-deficient cells.
Introduction
Major histocompatibility complex class I (MHC I) molecules present peptides to CD8 T cells for recognition by their TCRs (1–3). CD8 T cells are tolerant to the “self” peptide–MHC I (pMHC I) complexes present at steady-state on the cell surface and are activated when infected or transformed cells present either foreign or “altered-self” peptides. An essential part of immune surveillance is the recognition and elimination of infected or transformed cells by cytotoxic CD8 T cells. However, inappropriate recognition of pMHC I complexes and consequent activation of CD8 T cells can result in autoimmune disease.
The generation and loading of peptides onto MHC class I are tightly regulated to prevent unintended activation of CD8 T cells (1–3). The failure of this regulation, as well as impairment of components of the Ag processing pathway, can lead to the presentation of novel immunogenic pMHC I complexes and possible autoimmunity (4). The endoplasmic reticulum aminopeptidase associated with Ag processing (ERAAP) (5) edits the N termini of peptides presented by MHC I molecules after transport into the ER by the TAP (2). Polymorphic variants of human ERAAP have been associated with a variety of autoimmune diseases (6–11), suggesting that ERAAP strongly influences the peptides presented by MHC I molecules. Indeed, changes in the pMHC I repertoire associated with ER aminopeptidase (ERAP)1 polymorphisms have been found (12).
The association of ERAAP with many different autoimmune diseases suggests that there may be differences in metabolic and cellular pathways in ERAAP-deficient cells that could predispose the host to autoimmunity. Additionally, using a mouse model for ERAAP deficiency (13), we have found that the loss of ERAAP results in significant alterations in the peptides presented by both classical, or MHC Ia, and nonclassical, or MHC Ib, molecules, resulting in robust CD8 T cell responses (14, 15). We therefore decided to analyze the MHC I–associated peptide repertoire in the absence of ERAAP using high-throughput peptidomics (16, 17) to determine both the molecular changes in peptide presentation and the systemic cellular changes that might occur as a result of ERAAP deficiency.
In this study, we present a global analysis of the peptides presented by MHC I molecules on the cell surface when ERAAP is absent. We found that ERAAP-deficient cells presented a larger number of peptides bound to nonpolymorphic MHC Ib molecules, and that both MHC Ia and MHC Ib molecules on these cells presented longer peptides. A substantial number of peptides were unaffected by the absence of ERAAP whereas another set of peptides was not detected in ERAAP-deficient cells, suggesting that ERAAP is essential for generating a distinct subset of peptides. Interestingly, a set of new unedited peptides was present on ERAAP-deficient cells but was not detected in wild-type cells. Additionally, a large fraction of the unedited peptides were Qa-1b associated and the CD8+ T cell response of WT mice to ERAAP-deficient cells was largely Qa-1b restricted.
This large dataset also allowed us to identify the general characteristics and sources of ERAAP-dependent MHC I–associated peptides (18, 19). We found that peptides presented on ERAAP-deficient cells were derived from a different constellation of proteins compared with WT cells. A peptide from the Fbxl19 gene was presented specifically by ERAAP-deficient cells and elicited a CTL response in WT mice. Because genome-wide association studies of psoriasis patients have linked both the Fbxl19 and ERAP1 genes to the disease (6, 7), the discovery of immunogenic Fbx19-derived peptides on ERAAP-deficient cells suggests a potential mechanism driving pathogenesis of psoriasis.
Materials and Methods
Mice
ERAAP−/− mice (B6.129.Arts1tm1Ucb) have been previously described. ERAAP−/−KbDb−/−, ERAAP−/−TAP−/−, ERAAP−/−β2-microglobulin (β2m)−/− and ERAAP−/−Qa-1b−/− mice were generated in our facility by crossing the respective knockout mice with ERAAP−/− mice. WT C57BL/6J and β2m−/− mice were either purchased from The Jackson Laboratory (Bar Harbor, ME) or bred in our facility. Qa-1b−/− mice were generated in the laboratory of Dr. Harvey Cantor (Harvard University) and were the gift of Dr. Edgar Engleman (Stanford University). Mice were housed and all procedures carried out with the approval of, and in accordance with, the Institutional Animal Care Guidelines of the University of California Berkeley and the University of Montreal.
MHC I peptide extraction and mass spectrometry analyses
MHC I peptides were isolated from bone marrow–derived dendritic cells (BMDCs) as described previously using mild acid elution (16). Two biological replicates (500 × 106 BMDCs per replicate) were prepared. MHC I peptides obtained after acid elution were separated using an off-line 1100 series binary liquid chromatography (LC) system (Agilent Technologies) to remove contaminating species. Peptides were loaded on a homemade strong cation exchange column (0.3 mm internal diameter × 50 mm length) packed with strong cation exchange bulk material (PolySulfoethyl A from PolyLC). Peptides were fractionated with a gradient of 0–25% B after 25 min, 25–60% B after 35 min (solvent A: 5 mM ammonium formate, 15% acetonitrile [pH 3.0]; solvent B: 2 M ammonium formate, 15% acetonitrile [pH 3.0]). MHC I peptides were collected in five consecutive fractions and brought to dryness using a SpeedVac. Peptide fractions were resuspended in 2% aqueous acetonitrile (0.2% formic acid) and analyzed by nano-LC–tandem mass spectrometry (LC-MS/MS) on an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). Full mass spectra were acquired with the Orbitrap analyzer operated at a resolving power of 60,000 (at m/z of 400), and collision-activated dissociation tandem mass spectra were acquired in data-dependent mode with the quadrupole linear ion trap analyzer. Mass calibration used an internal lock mass [protonated (Si(CH3)2O))6; m/z of 445.120025] and typically provided mass accuracy within 5 ppm for all nano–LC-MS/MS experiments.
MS/MS sequencing, hierarchical clustering, and identification of MHC I peptide-source proteins
Peptidomic data were analyzed using Xcalibur software, and peak lists were generated using Mascot Distiller (version 2.1.1, http://www.matrixscience.com). Database searches were performed against the International Protein Index mouse database (version 3.23 containing 51,536 sequences and 24,497,860 residues) using Mascot (version 2.2, http://www.matrixscience.com) with a mass precursor tolerance of 15 ppm and a fragment tolerance of ±0.5 Da. Searches were performed without enzyme specificity with variable modifications for oxidation (Met) and deamidation (Asn, Gln). Label-free quantitative proteomics was used to compare peptide abundance across sample sets as described previously (18). Briefly, peptide maps were aligned and clustered together to profile the abundance of Mascot-identified peptides using hierarchical clustering with criteria based on m/z and time tolerance (±0.02 m/z and ±1 min). This resulted in a list of nonredundant peptide clusters for both replicates of all samples. The list of all peptides and their source proteins is available from the Immune Epitope Database under accession no. 1000706 (http://www.iedb.org/SubID/1000706)
MHC I–specific peptides were determined by comparing peptides from WT and ERAAP-knockout (KO) eluates to the β2m-KO negative control. To keep as much as possible of the MHC I peptides (avoiding false negatives), a cutoff of 2-fold was used when comparing the intensity of peptides that were detected in both β2m replicates to the intensity detected in WT or in ERAAP-KO (16), with a permissive p value <0.1 because only two replicates were included in the analyses. Peptides that were detected only in one or none of the β2m replicates were also considered as MHC I–associated peptides.
To identify peptides specific to WT, specific to ERAAP-KO, or common to both conditions, we only selected those with at least a 5-fold change in abundance. Peptides that were below threshold were given an intensity value of 8000. WT-specific peptides were identified as those detected in both WT replicates and in either one or two replicates of ERAAP-KO (with a WT/ERAAP-KO fold change > 5 and p < 0.1), or not detected in either ERAAP-KO replicates. The same treatment was used to determine ERAAP-KO–specific peptides. Common peptides were those detected in both WT and both ERAAP-KO replicates, with a calculated fold change intensity of <5 (p > 0.1).
Finally, Qa-1–associated peptides were identified as those detected in both ERAAP-KO replicates and in either one or two replicates of Qa-1b–ERAAP–double KO (DKO) (with an ERAAP-KO/Qa-1–ERAAP–DKO fold change intensity > 5 and p < 0.1), or not detected in either Qa-1–ERAAP–DKO replicates.
Abs and peptides
Abs for flow cytometry were obtained from BD Biosciences (CD8α, CD4, B220, TCRβ, CD44, NK1.1, Qa-1b, CD11c, H2-Kb, H2-Db, I-Ab, and IFN-γ) and BioLegend (CD62L). All peptides were synthesized by Dr. David King (University of California Berkeley).
Analysis of pMHC binding and motifs
Peptide binding affinities for H2-Kb and H2-Db were determined using the NetMHC algorithm (20). Peptides between 8 and 14 residues in length were analyzed, and those with a predicted IC50 of ≤500 nM were classified as good binders. Peptides of nine residues and longer were analyzed using the Rankpep algorithm (21, 22) to determine Qa-2a binding peptides. All peptides with a core region that had a positive score were assigned as Qa-2a associated. Peptides with C-terminal extensions of the core region were excluded.
CTL and T cell activation assays
WT anti–ERAAP-KO CD8 T cells lines were generated by immunizing C57BL/6J mice once with 2 × 107 ERAAP−/− spleen cells i.p. ERAAP-KO anti-WT CD8 T cell lines were generated by immunizing ERAAP-KO mice once with 2 × 107 C57BL/6J spleen cells i.p. Spleens were harvested from immunized mice 10 d after immunization. Responder spleen cells (5 × 106) were restimulated in vitro with an equal number of irradiated stimulator spleen cells, which were the same genotype as used to immunize, with 20 U/ml recombinant human IL-2 (National Cancer Institute) per well in 24-well plates. T cells were used for IFN-γ assays 6 d after restimulation. To measure intracellular IFN-γ, CD8 T cell lines were harvested and stimulated for 5 h with CD4- and CD8-depleted LPS-blasted spleen APCs of the indicated genotypes. GolgiPlug (BD Biosciences) was added for the latter 4 h of the incubation period. Cells were then stained first with surface markers, or with tetramers followed by other surface markers, fixed, permeabilized, and stained for intracellular IFN-γ. For ex vivo analysis of IFN-γ production, spleen cells were harvested from immunized mice and plated in 96-well plates with 1 μM of the indicated peptides for 5 h in the presence of GolgiPlug. The cells were then stained as described.
Analysis of gene ontology terms and protein function keywords
The gene symbols for the source genes for WT-specific, ERAAP-KO–specific, or common peptides were annotated and analyzed using DAVID (http://david.abcc.ncifcrf.gov/) (23). The background for each set was set as the group of gene symbols in the Mouse Genomic Informatics database. Only the groups of gene symbols with corresponding annotations in DAVID were analyzed. The EASE threshold was set as 0.01, and gene ontology (GO) terms describing cellular components and protein function keywords (Swiss-Prot Protein Information Resource) preferentially enriched for each group were determined. Terms or keywords with a false discovery rate of >5% were excluded. GO terms and keywords that were associated uniquely with ERAAP-KO peptides, and not with WT or common peptides, were determined.
Results
Global changes in MHC I–associated peptides in ERAAP-deficient cells
Our earlier work has shown that unedited peptides are presented by the MHC Ia molecules H2-Kb and H2-Db in the absence of ERAAP (24). However, that analysis of immunoprecipitated MHC I molecules from lysed cells could not distinguish between cell surface pMHC I and intracellular intermediates. Furthermore, the absence of ERAAP results in conformational changes in both MHC Ia– and MHC Ib–associated peptides (14, 15), which may not be recognized by conventional Abs raised against WT MHC I molecules. To circumvent these concerns, we used mild acid treatment to potentially elute all peptides from the surface of WT (16), ERAAP-deficient (ERAAP-KO), and Qa-1b– and ERAAP–double deficient (Qa-1–ERAAP–DKO) BMDCs. Peptides were separated by LC and sequenced by MS/MS, as previously described (Table I, http://www.iedb.org/SubID/1000706) (16, 18). Peptides eluted from the surface of β2m-KO BMDCs were used to exclude MHC I–independent, nonspecific peptides.
| . | WT . | ERAAP-KO . | Common . |
|---|---|---|---|
| Total no. of peptides | 439 | 389 | 2367 |
| Range of lengths | 6–20 | 6–25 | 5–28 |
| Qa-1 peptides | 6 | 67 | 86 |
| Qa-2 peptides | 29 | 24 | 169 |
| . | WT . | ERAAP-KO . | Common . |
|---|---|---|---|
| Total no. of peptides | 439 | 389 | 2367 |
| Range of lengths | 6–20 | 6–25 | 5–28 |
| Qa-1 peptides | 6 | 67 | 86 |
| Qa-2 peptides | 29 | 24 | 169 |
The numbers and lengths of the peptides identified by high-throughput LC-MS are shown.
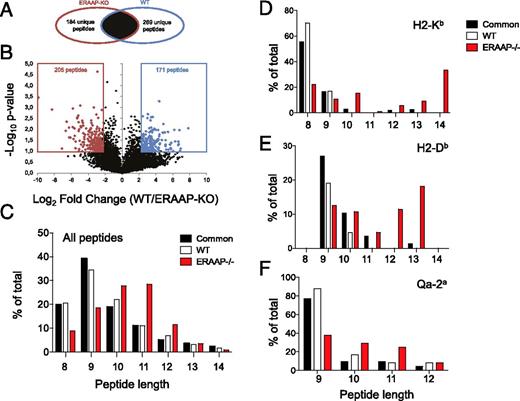
We defined peptides as specific to WT or ERAAP-deficient cells when they were unique (Fig. 1A) to either condition, or overexpressed (>5-fold, p < 0.1, Fig. 1B) in one sample versus another. Common peptides were defined as those found in both WT and ERAAP-deficient cells with a <5-fold difference in intensity between WT and ERAAP-deficient cells (Fig. 1B). We identified 2367 peptides common to both WT and ERAAP-deficient cells (common peptides), 440 specific to WT cells (WT-specific peptides; 269 unique [Fig. 1A] plus 171 overexpressed [Fig. 1B]) and 389 specific to ERAAP-deficient cells (ERAAP-KO–specific peptides; 184 unique [Fig. 1A] plus 205 overexpressed [Fig. 1B]; Table I, complete lists of peptides are available at http://www.iedb.org/SubID/1000706).
High-throughput MS detects unique longer peptides in ERAAP-deficient cells. (A) Venn diagrams showing the exclusive and common subsets of peptides found in ERAAP-KO or WT cells. (B) Volcano plot, showing that common peptides were segregated into ERAAP-KO–specific or WT-specific when they were significantly overexpressed (>5-fold, p < 0.1, two-tailed Student t test). The numbers indicate different peptides in each category. The average of two independent experiments is shown. (C) The proportion of peptides of distinct lengths either common to both or specific only to WT or ERAAP-deficient cells. (D and E) NetMHC was used to identify (D) H2-Kb– and (E) H2-Db–associated peptides with a predicted IC50 of 500 ≥nM. (F) RANKPEP was used to identify Qa-2a–associated peptides. The lengths of Qa-2a–associated peptides common to both WT and ERAAP-KO cells, or specific to WT or ERAAP-KO cells, were compared.
High-throughput MS detects unique longer peptides in ERAAP-deficient cells. (A) Venn diagrams showing the exclusive and common subsets of peptides found in ERAAP-KO or WT cells. (B) Volcano plot, showing that common peptides were segregated into ERAAP-KO–specific or WT-specific when they were significantly overexpressed (>5-fold, p < 0.1, two-tailed Student t test). The numbers indicate different peptides in each category. The average of two independent experiments is shown. (C) The proportion of peptides of distinct lengths either common to both or specific only to WT or ERAAP-deficient cells. (D and E) NetMHC was used to identify (D) H2-Kb– and (E) H2-Db–associated peptides with a predicted IC50 of 500 ≥nM. (F) RANKPEP was used to identify Qa-2a–associated peptides. The lengths of Qa-2a–associated peptides common to both WT and ERAAP-KO cells, or specific to WT or ERAAP-KO cells, were compared.
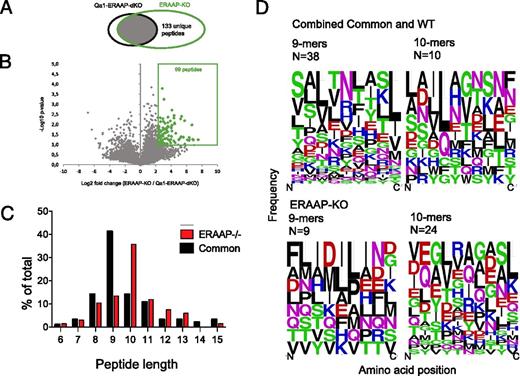
Algorithms for identifying Qa-1b–associated peptides are not available, and the peptide-binding motif is poorly defined. We therefore used a subtractive strategy to determine the sequences of Qa-1b–associated peptides from ERAAP-deficient cells. We compared the peptides isolated from Qa-1b– and ERAAP–double deficient BMDCs and compared the results to those from BMDCs deficient only in ERAAP. As described earlier, peptides that were detected in ERAAP-deficient cells but not in Qa-1b– and ERAAP–double deficient cells, or that were detected at intensities ≥5-fold in ERAAP-deficient cells (p < 0.1), were designated as ERAAP-KO–specific Qa-1b–associated peptides (Fig. 2, http://www.iedb.org/SubID/1000706). We found 232 Qa-1–associated peptides (133 unique and 99 overexpressed, Fig. 2A). Seventy-three of these peptides were detected in both replicates as associated with ERAAP-KO but only in single replicates as associated with WT. We could therefore assign them as unambiguously present only in ERAAP-KO, and we excluded these peptides from further analyses. Similarly, we identified WT-specific Qa-1b–associated peptides as those found only in, or overexpressed by, WT cells when compared with both ERAAP-KO and Qa-1–ERAAP–DKO cells. Common Qa-1b–associated peptides were those found in, or overexpressed by, both WT and ERAAP-deficient cells, when compared with Qa-1b–ERAAP–double deficient cells. We found 86 Qa-1b–associated peptides common to both WT and ERAAP-KO cells, 6 specific to WT cells, and 67 peptides specific to ERAAP-KO cells (http://www.iedb.org/SubID/1000706).
A subtractive approach defines Qa-1b–associated peptides in ERAAP-deficient cells. (A) Venn diagrams showing the subsets of Qa-1b–associated peptides found in Qa-1b plus ERAAP-KO cells versus Qa-1b–ERAAP–DKO cells. (B) Volcano plots showing that some peptides were significantly overexpressed in ERAAP-KO cells compared with Qa-1b–ERAAP–DKO cells (>5-fold, p < 0.1, two-tailed Student t test). The numbers indicate distinct peptides found in each category. The average of two independent experiments is shown. (C) The proportion of peptides of distinct lengths either specific to ERAAP-deficient cells or common to both ERAAP-KO and WT cells. (D) WebLogo plots showing the amino acid composition of 9- or 10-reside-long Qa-1b–associated peptides, either specific to ERAAP-deficient cells or common to both ERAAP-deficient and WT cells. Amino acids are represented by their single letter code, and the size of the letter is proportional to the frequency with which that amino acid occurs at the position indicated on the x-axis. The color of the letter indicates the chemical nature of the amino acid as follows: green, polar; black, hydrophobic; red, acidic; magenta/blue, basic. N indicates the numbers of peptides in each group.
A subtractive approach defines Qa-1b–associated peptides in ERAAP-deficient cells. (A) Venn diagrams showing the subsets of Qa-1b–associated peptides found in Qa-1b plus ERAAP-KO cells versus Qa-1b–ERAAP–DKO cells. (B) Volcano plots showing that some peptides were significantly overexpressed in ERAAP-KO cells compared with Qa-1b–ERAAP–DKO cells (>5-fold, p < 0.1, two-tailed Student t test). The numbers indicate distinct peptides found in each category. The average of two independent experiments is shown. (C) The proportion of peptides of distinct lengths either specific to ERAAP-deficient cells or common to both ERAAP-KO and WT cells. (D) WebLogo plots showing the amino acid composition of 9- or 10-reside-long Qa-1b–associated peptides, either specific to ERAAP-deficient cells or common to both ERAAP-deficient and WT cells. Amino acids are represented by their single letter code, and the size of the letter is proportional to the frequency with which that amino acid occurs at the position indicated on the x-axis. The color of the letter indicates the chemical nature of the amino acid as follows: green, polar; black, hydrophobic; red, acidic; magenta/blue, basic. N indicates the numbers of peptides in each group.
Longer peptides are presented on the cell surface in the absence of ERAAP
We analyzed the distribution of the lengths of ERAAP-KO– and WT-specific and common peptides and found that peptides eluted from ERAAP-deficient cells were longer than those specific to WT cells, or common to both (p = 0.0122, one-way ANOVA) (Fig. 1C). Because our analysis is specific to cell surface pMHC I complexes, and not to potential intracellular intermediates, we conclude that MHC I molecules present longer peptides on the cell surface in ERAAP-deficient cells.
We then analyzed specifically whether MHC Ia– or MHC Ib–associated peptides were longer in ERAAP-deficient cells. Non-Qa-1b–associated peptides were analyzed using the NetMHC algorithm (20) to predict their binding affinity to H2-Kb and H2-Db, the MHC Ia molecules expressed by the H-2b haplotype. NetMHC classifies peptides as strong MHC binders when their predicted IC50 is <50 nM, whereas weak binders correspond to those with IC50 of <500 nM. We therefore used an IC50 of 500 nM as the cutoff to classify peptides as H-2Kb or H-2Db binders (http://www.iedb.org/SubID/1000706). In this subset of peptides identified as good MHC binders, we found that most WT-specific and common peptides were of the canonical length, that is, eight residues for H2-Kb binders and nine residues for H2-Db binders (Fig. 1D, 1E). Strikingly, we found that a lesser proportion of the ERAAP-KO–specific good MHC binders were of the canonical length (Fig. 1D, 1E). Indeed, most ERAAP-KO–specific good MHC binders were longer than eight (for H2-Kb, Fig. 1D) or nine (for H2-Db, Fig. 1E) amino acid residues. These results show that ERAAP-deficient cells present longer peptides overall, and that these longer peptides were predicted to bind to MHC I with an affinity of <500 nM.
To identify peptides associated with Qa-2a, a nonclassical MHC I molecule, we analyzed all peptides nine residues in length or longer by the Rankpep algorithm (21, 22) (Fig. 1F, http://www.iedb.org/SubID/1000706, Supplemental Fig. 2). Rankpep defines a core region for Qa-2a binding, and it includes longer peptides that have either N- or C-terminal extensions to this core region. We excluded peptides with C-terminal extensions, because Qa-2a, similar to conventional MHC I molecules, is known to require a hydrophobic residue at the C terminus (25–27). Moreover, C-terminal extensions have been found to be deleterious for MHC I binding (28). We identified 169 Qa-2a–associated peptides common to both ERAAP-KO and WT cells, 29 specific to WT, and 24 specific to ERAAP-KO (Table I, http://www.iedb.org/SubID/1000706). WebLogo analysis of the amino acid composition of these predicted Qa-2–associated peptides confirmed the presence of the characteristic histidine at position 7 of the peptide (Supplemental Fig. 2). No clear motif emerged from an analysis of the N termini of these peptides (Supplemental Fig. 2). Strikingly, when we analyzed the lengths of these predicted Qa-2a–associated peptides, we found that ERAAP-KO–specific peptides were longer (Fig. 1F). These results suggest that the generation of the peptide repertoire presented by Qa-2a is also strongly influenced by ERAAP.
Qualitative and quantitative changes in Qa-1b–associated peptides in ERAAP-deficient cells
Relatively little is known about the peptide-binding preferences of the MHC Ib molecule Qa-1b. There have been very few Qa-1b–binding peptides definitively identified to serve as T cell ligands (29–33), and the numbers of endogenous peptide sequences are insufficient to derive general rules for the peptide-binding preferences of Qa-1b. When comparing peptides isolated from WT and ERAAP-deficient cells to those isolated from Qa-1b and ERAAP double-deficient cells, our subtractive strategy identified numerous endogenous peptides associated with Qa-1b (Fig. 2A, 2B, http://www.iedb.org/SubID/1000706).
Eighty-six peptides were found to be Qa-1b associated (i.e., absent in Qa-1–ERAAP–DKO cells) and common to Qa-1b plus WT and ERAAP-deficient cells. The present analysis cannot unambiguously assign WT-specific Qa-1b peptides, because Qa-1–KO ERAAP WT cells were not tested. However, 67 Qa-1b–associated peptides were identified from ERAAP-deficient cells, whereas only 6 Qa-1b–associated peptides were found in WT cells, and not ERAAP-deficient cells (Table I, Fig. 2A, 2B, http://www.iedb.org/SubID/1000706), suggesting that more peptides are presented by Qa-1b in the absence of ERAAP. Notably, we did identify the Qdm peptide (30), derived from the leader sequence of H2-Db, in WT cells, and to a lower extent in ERAAP-deficient cells in agreement with an earlier study (34). However, this peptide did not meet our filtering criteria for inclusion in our main analysis, because it was not found at amounts >5-fold higher in WT compared with ERAAP-KO (http://www.iedb.org/SubID/1000706).
We identified Qa-1b–associated peptides ranging from 6 to 24 residues in length (Table I, http://www.iedb.org/SubID/1000706). Because there were only six WT-specific Qa-1b–associated peptides, three of which were eight residues long and three that were nine residues long, they were analyzed together with the common peptides, called combined common and WT peptides (Fig. 2C). Focusing on peptides between 6 and 15 residues in length, we found that most Qa-1b–associated peptides common to WT and ERAAP-deficient cells were nine residues long, in agreement with the canonical length of most MHC-associated peptides. However, consistent with MHC Ia– and Qa-2a–associated peptides (Fig. 1C–F), Qa-1b–associated peptides on the surface of ERAAP-deficient cells were also longer, with most of these peptides being 10 residues long (Fig. 2C).
We then analyzed the amino acid composition of the Qa-1b–associated peptides using WebLogo (Fig. 2D). The results are presented as frequency plots, where the size of the single letter amino acid symbol is in proportion to the frequency of its occurrence at that position. To determine a peptide-binding motif, we analyzed the complete sequences of all the nine-residue-long peptides from the combined WT and common peptide set, as these are the endogenous peptides of the canonical length found at steady-state (Fig. 2D, upper left panel). These peptides showed a strong preference for a hydrophobic residue (leucine, isoleucine, or valine) at the C terminus (P9) and a similar preference for a hydrophobic aliphatic residue (leucine, valine, or alanine) at P3 from the N terminus, and either asparagine or arginine at P5. In contrast to prior analyses of substituted Qdm peptides (35), we found no preference for a methionine at P2, suggesting that this position may not be an anchor position.
Remarkably, none of these preferences was retained in Qa-1b–associated peptides from ERAAP-deficient cells, or from common peptides 10 residues long (Fig. 2D, upper right and lower panels). Likewise, no strong amino acid preferences were evident at the C termini of 10-residue-long common peptides, or in the ERAAP-KO–specific peptides, although the common 10-residue-long peptides and ERAAP-KO–specific 9-mers retained a preference for a hydrophobic aliphatic residue at P3. Ten-residue-long ERAAP-KO–specific peptides showed a preference for either leucine or valine at P4, in common with 10-residue-long common peptides, which showed a preference for leucine at P4.
These findings suggest that Qa-1b has a permissive peptide-binding motif, with a preference for hydrophobic amino acids at P3 and the C terminus, and for canonical nine-residue-long peptides. Nevertheless, peptides longer than the canonical nine residues and peptides specific to ERAAP-KO cells are presented despite not conforming to this motif.
ERAAP edits a subset of peptides, generating a unique N terminus
We have previously established that the aminopeptidase ERAAP plays a crucial role in trimming the N termini of peptides that are eventually presented by MHC I (5). We wanted to determine whether the absence of ERAAP resulted in a global change in the nature of the N termini of MHC I–associated peptides. We therefore analyzed the amino acid composition of the N and C termini of the isolated peptides, using WebLogo (36) (Fig. 3, Supplemental Fig. 1). All the peptides were aligned at their C termini, and their sequences were analyzed from the C terminus to the −5 position (Supplemental Fig. 1). The analysis clearly showed that all the peptides (WT- and ERAAP-KO–specific, and common peptides) had the canonical hydrophobic C termini (Supplemental Fig. 1).
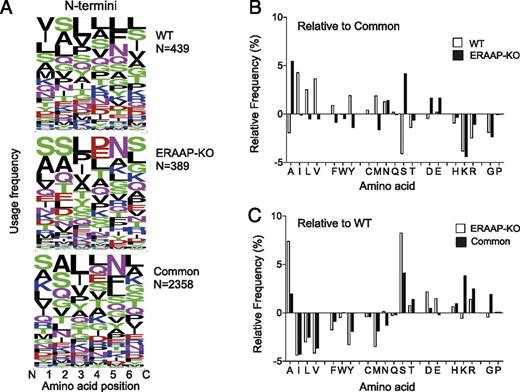
ERAAP editing results in a unique N-terminal peptide motif. (A) WebLogo plots showing the N-terminal amino acid composition of peptides either specific to WT and ERAAP-deficient cells or common to both. Amino acids are represented by their single letter symbol, and the size of the letter is proportional to the frequency with which that amino acid occurs at the position indicated on the x-axis. The color of the letter indicates the chemical nature of the amino acid as follows: green, polar; black, hydrophobic; red, acidic; magenta/blue, basic. N indicates the numbers of distinct peptides in the group. (B) The relative frequency of each amino acid at the N-terminal position of WT- and ERAAP-KO–specific peptides was calculated by subtracting the frequency of occurrence of the amino acid at that position in common peptides from the frequency of occurrence in WT- or ERAAP-KO–specific peptides. (C) Frequency of individual amino acids at the N terminus of common or ERAAP-KO–specific peptides relative to WT-specific peptides.
ERAAP editing results in a unique N-terminal peptide motif. (A) WebLogo plots showing the N-terminal amino acid composition of peptides either specific to WT and ERAAP-deficient cells or common to both. Amino acids are represented by their single letter symbol, and the size of the letter is proportional to the frequency with which that amino acid occurs at the position indicated on the x-axis. The color of the letter indicates the chemical nature of the amino acid as follows: green, polar; black, hydrophobic; red, acidic; magenta/blue, basic. N indicates the numbers of distinct peptides in the group. (B) The relative frequency of each amino acid at the N-terminal position of WT- and ERAAP-KO–specific peptides was calculated by subtracting the frequency of occurrence of the amino acid at that position in common peptides from the frequency of occurrence in WT- or ERAAP-KO–specific peptides. (C) Frequency of individual amino acids at the N terminus of common or ERAAP-KO–specific peptides relative to WT-specific peptides.
We then aligned all the peptides at their N termini and analyzed the composition of the first six amino acids (Fig. 3A). Intriguingly, we found that ERAAP-KO–specific peptides and the common peptides appeared to have a similar distribution of amino acids at their N termini (Fig. 3A, center and lower panels). Strikingly, WT-specific peptides had a different amino acid composition from either common or ERAAP-KO–specific peptides (Fig. 3A, upper panel). WT peptides appeared to have a preference for hydrophobic residues at five of the six N-terminal positions we analyzed. This distinctive preference for hydrophobic residues in the N-terminal profile is absent in common peptides and in those from ERAAP-deficient cells.
This is especially evident at the N-terminal residue, which would be generated by ERAAP cleavage (Fig. 3B, 3C). The relative frequencies of each amino acid at the N terminus were determined by subtracting the frequency of occurrence in common peptides from WT- or ERAAP-KO–specific peptides (Fig. 3B). WT-specific peptides have Ile, Leu, or Val more frequently at this position, whereas ERAAP-KO peptides have Ala and Ser. Conversely, WT peptides have Ser and Ala much less frequently than do common peptides (Fig. 3B). To visualize this trend more clearly, the relative frequencies of N-terminal amino acids in common and ERAAP-KO–specific peptides were determined relative to WT-specific peptides (Fig. 3C). The pattern of usage of individual amino acids by ERAAP-KO–specific and common peptides relative to WT is highly similar, with less frequent occurrences of hydrophobic residues Ile, Leu, and Val, and more frequent occurrence of Ala and Ser.
The strong similarity in amino acid preferences between the N termini of the common and ERAAP-KO–specific peptides suggests that these peptides were generated in a manner that did not require ERAAP. We conclude that these peptides represent the repertoire of peptides unedited by ERAAP.
Differential immunogenicity of pMHC Ia versus pMHC Ib complexes due to ERAAP activity
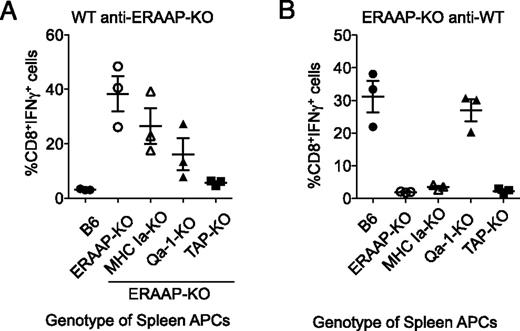
Given the striking changes in both MHC Ia– and MHC Ib–associated peptides in the absence of ERAAP, we wanted to test the relative immunogenicity of pMHC Ia versus pMHC Ib expressed by WT and ERAAP-KO cells. ERAAP-deficient mice were immunized with spleen cells from WT mice, and responding ERAAP-KO anti-WT T cells expanded by restimulation in vitro with irradiated WT splenic APCs (Fig. 4A). IFN-γ production by these CD8 T cell lines was measured in response to self (ERAAP-KO), WT, MHC Ia–deficient (H2-Kb–H2-Db–DKO), Qa-1b–deficient APCs (Qa-1–KO), and APCs lacking peptide transport into the ER (TAP-KO). Conversely, WT mice were immunized with ERAAP-deficient cells, and responding WT anti–ERAAP-KO CD8 T cells were tested for their responses to self (WT) APCs and ERAAP-deficient, ERAAP and MHC Ia–deficient, and Qa-1b– and TAP-deficient cells (Fig. 4B).
Differences in specificity of WT anti–ERAAP-KO and ERAAP-KO anti-WT immune responses. (A) WT B6 mice were immunized with ERAAP-KO spleen cells or (B) ERAAP-KO mice were immunized with WT spleen cells. Spleens from immunized mice were harvested after 10 d and restimulated in vitro with the same APCs used for immunization. The resulting CD8 T cell responses to the indicated APCs were measured as intracellular IFN-γ production. Forward and side scatter were used to exclude dead and granular cells, and the cells in the resulting lymphocyte gate were B220−CD4−CD8α+TCRβ+. Each point represents a different mouse responding to splenic APCs of the indicated genotype. The data are presented as the mean of five mice, with the bars representing SEM. When analyzed by nonparametric one-way ANOVA (Kruskal–Wallis with a Dunn posttest), the data are statistically significant, with p = 0.0157 and p = 0.0169 for (A) and (B), respectively.
Differences in specificity of WT anti–ERAAP-KO and ERAAP-KO anti-WT immune responses. (A) WT B6 mice were immunized with ERAAP-KO spleen cells or (B) ERAAP-KO mice were immunized with WT spleen cells. Spleens from immunized mice were harvested after 10 d and restimulated in vitro with the same APCs used for immunization. The resulting CD8 T cell responses to the indicated APCs were measured as intracellular IFN-γ production. Forward and side scatter were used to exclude dead and granular cells, and the cells in the resulting lymphocyte gate were B220−CD4−CD8α+TCRβ+. Each point represents a different mouse responding to splenic APCs of the indicated genotype. The data are presented as the mean of five mice, with the bars representing SEM. When analyzed by nonparametric one-way ANOVA (Kruskal–Wallis with a Dunn posttest), the data are statistically significant, with p = 0.0157 and p = 0.0169 for (A) and (B), respectively.
As previously reported (14, 15), we found that WT anti–ERAAP-KO CD8 T cells recognized both MHC Ia–sufficient and MHC Ia–deficient ERAAP-deficient APCs (Fig. 4B). WT anti–ERAAP-KO CD8 T cells required conventional peptide transport because they did not respond to TAP-deficient ERAAP-deficient APCs. WT anti–ERAAP-KO T cells also showed reduced responses to Qa-1b–deficient ERAAP-deficient APCs, suggesting that a large proportion of WT CD8 T cells respond to immunogenic peptide– Qa-1b complexes (Fig. 4A). In contrast, we found that CD8 T cells in ERAAP-deficient mice mounted a vigorous response against WT APCs (Fig. 4B), but did not respond to MHC Ia–deficient and TAP-deficient APCs, indicating that they did not recognize peptides presented by MHC Ib molecules on WT cells as foreign. The response of ERAAP-KO–anti-WT CTLs to Qa-1b–deficient APCs was also unimpaired (Fig. 4B), suggesting that WT APCs did not present immunogenic peptide– Qa-1b complexes to ERAAP-deficient CD8 T cells.
These results show that despite clear global changes in the pMHC Ib and pMHC Ia complexes presented by ERAAP-deficient cells, the relative immunogenicity of these changes is distinct: WT mice respond to immunogenic pMHC Ia and pMHC Ib complexes on ERAAP-deficient cells, but ERAAP-deficient mice respond exclusively to immunogenic pMHC Ia complexes.
MHC I peptides arise from different protein sources in ERAAP-deficient cells
We have previously shown that the pMHC I repertoire on the cell surface is a direct representation of the transcriptional and physiological state of the cell (18, 19). The sources of cell surface pMHC I change with the cellular state; for example, transformed cells present peptides encoded by genes characteristic of neoplastic transformation (16), whereas cells treated with rapamycin present peptides encoded by genes involved in mammalian target of rapamycin signaling (19). Furthermore, a substantial proportion of MHC I peptides are cell type specific (18). To test whether the absence of ERAAP led to the processing and presentation of peptides encoded by unique subsets of genes, we analyzed the source genes of MHC I–associated peptides from WT or ERAAP-deficient cells.
We used the DAVID database (23) to analyze lists of gene symbols for source genes for MHC I–associated peptides specific to WT or ERAAP-deficient cells, or common to both. We used a highly restrictive EASE threshold of 0.01 to filter our results and only counted results that had a false discovery rate of <5%. We focused our analysis on two areas: GO terms that described cellular components, and protein function (Swiss-Prot Protein Information Resource) keywords.
We looked for the GO terms and keywords that were preferentially enriched in source genes for common, WT-specific, and ERAAP-KO–specific peptides compared with the background set of all the genes in the Mouse Genome Informatics database. The complete lists of GO terms and keywords we found preferentially enriched in the three sets are listed in Supplemental Table I. Some GO terms and keywords appear to be generally characteristic of pMHC I presented on the surface of BMDCs; for example, 44–51% peptides in all three sets derived from phosphoproteins (Supplemental Table I). Because phosphorylation is often a prerequisite for ubiquitylation and targeting for degradation (37), this enrichment could reflect the nature of MHC I–associated peptides in general. We then filtered the list of GO terms and keywords uniquely associated with either the WT or ERAAP-KO set against the GO terms and functional keywords associated with common peptides to generate a list of GO terms and keywords specifically associated with peptides derived from ERAAP-deficient cells (Table I).
The analysis of GO terms showed that a fraction of MHC I peptides from ERAAP-deficient cells were preferentially derived from proteins from the Golgi (Table II). These observations are consistent with the idea that MHC I peptides can be processed in other compartments when ER processing is compromised (38). The pool of optimally trimmed MHC I peptides is reduced in ERAAP-deficient cells, as we, and others, have shown. As a consequence, pMHC I complexes that exit the ER in ERAAP-deficient cells are less stable and might be more susceptible to exchange with peptides of higher affinity, generated in post-ER cell compartments.
| Analysis Parameters . | Enriched in ERAAP-KO . |
|---|---|
| GO terms–cellular components | Golgi apparatus part, protein–DNA complex, nucleosome |
| Swiss-Prot Protein Information Resource keywords | WD repeat, leucine-rich repeat, methylation |
| Analysis Parameters . | Enriched in ERAAP-KO . |
|---|---|
| GO terms–cellular components | Golgi apparatus part, protein–DNA complex, nucleosome |
| Swiss-Prot Protein Information Resource keywords | WD repeat, leucine-rich repeat, methylation |
GO terms and protein function keywords that were uniquely associated with ERAAP-KO–specific peptides are shown.
Analyzing the protein function keywords associated with ERAAP-KO–specific peptides showed that the keywords “wd repeat,” “methylation,” and “leucine-rich repeat (LRR)” were specifically enriched (Table II). Of these keywords, only LRR was entirely unique to ERAAP-KO–specific peptides, because the other keywords were found in the other lists, albeit without reaching statistical significance (Supplemental Table I). We noticed that one of the peptides presented specifically in ERAAP-deficient cells was derived from the LRR-containing protein Fbxl19 (Supplemental Table I). Strikingly, the human FBXL19 gene has been associated with psoriasis in genome-wide association studies, which have also linked ERAP1, the human homolog of ERAAP, to psoriasis (6, 7).
To determine whether the preferential enrichment of the Fbxl19-derived DPSVHLLTA (DA9) peptide in our analyses was actually immunogenic, WT B6 mice were immunized with ERAAP-KO spleen cells and immune responses were measured by intracellular IFN-γ production in response to synthetic peptides directly ex vivo (Fig. 5). Remarkably, B6 anti–ERAAP-KO-immunized mice mounted robust responses to the DA9 peptide ex vivo. CD8 T cells from immunized, but not naive mice, downregulated l-selectin (CD62L), a marker of activation, and produced IFN-γ when restimulated ex vivo with the DA9 peptide (Fig. 5). As a positive control, we detected responses in the same mice against the FL9 peptide derived from the Fam49b protein, as shown previously (15). Naive mice, or immunized mice restimulated without peptide, did not produce IFN-γ (Fig. 5). Thus, among the many different peptides presented by ERAAP-KO cells, the DA9 peptide appears to induce an immunodominant T cell response. These results demonstrate a clear molecular link between presentation of an Fbxl19-derived peptide and the absence of ERAAP function in autoimmune disease.
The Fbxl19-encoded DA9 peptide is presented by ERAAP-KO cells to WT CD8 T cells. WT B6 mice were immunized with ERAAP-KO spleen cells, and immune responses to the DA9 and FL9 peptides were measured directly ex vivo by intracellular IFN-γ staining. Forward and side scatter were used to exclude dead and granular cells, and the cells in the resulting lymphocyte gate were B220−CD4−CD8α+TCRβ+. (A) Representative flow cytometry plots show CD8 T cells from immunized, but not naïve, mice downregulate l-selectin (CD62L) and produce IFN-γ when restimulated with either the DA9 or FL9 synthetic peptides. (B) Fraction of CD62LloIFN-γ+ cells in naive WT mice (open) or immunized with ERAAP-KO cells (closed). Each point represents an individual animal. Results from eight mice from two pooled experiments are shown, with the bars representing SEM. When analyzed by nonparametric one-way ANOVA (Kruskal–Wallis with a Dunn posttest), the data are statistically significant, with p = 0006.
The Fbxl19-encoded DA9 peptide is presented by ERAAP-KO cells to WT CD8 T cells. WT B6 mice were immunized with ERAAP-KO spleen cells, and immune responses to the DA9 and FL9 peptides were measured directly ex vivo by intracellular IFN-γ staining. Forward and side scatter were used to exclude dead and granular cells, and the cells in the resulting lymphocyte gate were B220−CD4−CD8α+TCRβ+. (A) Representative flow cytometry plots show CD8 T cells from immunized, but not naïve, mice downregulate l-selectin (CD62L) and produce IFN-γ when restimulated with either the DA9 or FL9 synthetic peptides. (B) Fraction of CD62LloIFN-γ+ cells in naive WT mice (open) or immunized with ERAAP-KO cells (closed). Each point represents an individual animal. Results from eight mice from two pooled experiments are shown, with the bars representing SEM. When analyzed by nonparametric one-way ANOVA (Kruskal–Wallis with a Dunn posttest), the data are statistically significant, with p = 0006.
Discussion
We used a combination of high-throughput MS and bioinformatics to analyze the changes in the pMHC I repertoire that occur when N-terminal peptide trimming by ERAAP is lost. The differences between the WT and ERAAP-deficient pMHC I repertoire are extensive and diverse and encompass both the highly polymorphic MHC Ia molecules and the nonpolymorphic MHC Ib molecules. The flexibility of our method, which isolates all cell surface MHC I–associated peptides regardless of whether they can be recovered by immunoprecipitation, allowed us to analyze common peptides found in both WT and ERAAP-deficient cells and those found specifically in WT or ERAAP-deficient cells. It is clear that MHC Ia– and MHC Ib–associated peptides specific to ERAAP-deficient cells are longer, compared with either the common or WT-specific peptides. This analysis provides insights into the role of N-terminal trimming in shaping the peptide repertoire presented by both MHC Ia and MHC Ib molecules.
ERAAP-KO–specific peptides have the canonical hydrophobic C termini, yet they are not simply N-terminally extended precursors of WT-specific peptides. Instead, ERAAP-KO–specific peptides showed an N-terminal pattern of amino acid usage strikingly similar to that of peptides common to both strains. The N termini of WT-specific peptides, however, showed a clearly distinct pattern of amino acids, with a preference for hydrophobic amino acids at five of six positions. This observation is especially striking at P1, the first residue of the peptide, and the one generated as a direct result of ERAAP cleavage. These results suggest that ERAAP generates a specific subset of peptides with a preference for certain N-terminal residues, consistent with the work of Schatz et al. (39) in which it was shown that peptides with serine at the N terminus are much less susceptible to endoplasmic aminopeptidase cleavage. When ERAAP is absent, this subset is no longer edited and is lost. The bulk of MHC I–associated peptides thus have the ERAAP unedited N-termini similar to those observed in the common set of peptides. These observations suggest that ERAAP plays a role in processing a subset of, but not all, peptides presented by MHC I.
Our analyses provided several interesting insights into the global MHC Ib–associated peptide repertoire. We found that similar to the peptides associated with the classical MHC Ia molecules, both Qa-1b– and Qa-2a–associated peptides were longer in ERAAP-deficient cells. Furthermore, some Qa-2a peptides found exclusively in WT cells are lost in ERAAP-deficient cells, and other novel peptides are associated with Qa-2a in ERAAP-deficient cells. These results suggest that ERAAP is required for trimming peptides for presentation by Qa-2a, in a manner similar to its effect on classical MHC Ia molecules.
We observed a striking increase in the number of Qa-1b–associated peptides specific to ERAAP-deficient cells. We found a total of 153 (67 ERAAP-KO–specific plus 86 common) peptides associated with Qa-1b in ERAAP-deficient cells, compared with a total of 92 in WT cells (6 found only in WT plus 86 common). Interestingly, these peptides had a wide distribution of lengths and showed no clear consensus motif. Instead, Qa-1b–binding peptides were enriched for a hydrophobic residue at the C terminus, with no other obvious anchor positions, except a preference for a hydrophobic residue at P3 and a polar residue at P5. Even these preferences are lost in 10-mer peptides and in peptides from ERAAP-KO cells. Recent work from Chen et al. (40) found a distinct peptide motif in peptides eluted from Qa-1b–H-2Db hybrid molecules transduced into human HeLa cells, a motif that we did not observe in this data set. It is possible that differences in peptide isolation methods as well as differential affinity of human β2m (in HeLa cells) and native mouse β2m could account for these differences.
Strikingly, Qa-1b was the only MHC I molecule that was associated with a larger number of peptides in the absence of ERAAP. This distinction can perhaps account for why WT CD8 T cells were predominantly specific for the peptide–Qa-1b repertoire presented by ERAAP-deficient cells, whereas CD8 T cells in ERAAP-deficient mice responded only to pMHC Ia complexes on WT APCs. Although we cannot formally exclude Qa-2a or other pMHC Ib at this time, the greatly increased numbers of unedited Qa-1b–associated peptides in ERAAP-deficient cells suggests that altered presentation by Qa-1b is an important determinant of the immunogenicity of ERAAP-deficient cells. It appears that Qa-1b occupies a unique niche, presenting a greater number of peptides under conditions of defective Ag processing, such as in the absence of TAP and ERAAP (15, 41). These results strongly suggest that presentation of a wide array of peptides by Qa-1b serves as a warning for defective Ag processing machinery in the ER.
We have shown before that MHC I–associated peptides are a faithful representation of the cellular state (16, 18, 19). We used GO terms to determine that peptides derived from Golgi-associated genes were specifically enriched in ERAAP-deficient cells. pMHC I complexes are less stable in ERAAP-deficient cells (13). This instability might make it possible for peptide exchange and loading to occur in post–ER compartments, such as the Golgi, leading to a shift in the sources of peptides. Indeed, studies have shown that MHC I molecules can be loaded in post–ER compartments (42, 43).
Another notable change we observed was a specific increase in peptides derived from LRR-containing proteins in ERAAP-deficient cells. Strikingly, the DA9 peptide, presented exclusively by ERAAP-deficient cells, was derived from the LRR-containing protein Fbxl19. The human FBXL19 gene and ERAP1 (the human ortholog of ERAAP) have both been associated with psoriasis in genome-wide association studies (6, 7). We found that the Fbxl19-derived DA9 peptide was not only presented by ERAAP-deficient cells, but was capable of inducing a CD8+ T cell response in WT mice. Psoriasis is a primarily CD8 T cell–mediated autoimmune disease, although the Ags associated with pathogenic T cells remain unknown (44). Whereas the significance of the FBXL19- specific CD8 T cells to psoriasis remains to be determined, our observations provide a potential molecular mechanism for ERAAP-associated autoimmunity.
In summary, we have shown that the loss of ERAAP leads to shifts in the nature and lengths of peptides presented by MHC I molecules on the cell surface. In the absence of ERAAP, MHC Ia and Ib molecules present longer peptides, whereas a distinct set of ERAAP-edited peptides presented in WT cells is lost. Strikingly, we also find changes in the antigenic precursors from which MHC I–associated peptides are derived, suggesting that mechanistic and metabolic changes may occur as a consequence of ERAAP deficiency. We also identified a peptide from an autoimmunity-associated gene that is presented only in the absence of ERAAP. We suggest that proteomic analyses of MHC I–associated peptides could be useful not only in identifying potential antigenic targets, but also in investigating metabolism and disease-related phenomena.
Acknowledgements
We acknowledge Federico Gonzalez, Caroline Côté, and the staff of the proteomic core facility at the Institute for Research in Immunology and Cancer for technical assistance and discussions. We thank David King for peptide synthesis.
Footnotes
This work was supported by Canadian Cancer Society Research Institute Grant 701564 (to C.P. and P.T.) and by grants from the National Institutes of Health (to N.S.). N.A.N. was supported in part by the Irvington Institute fellowship program of the Cancer Research Institute. D.A.d.V. was supported by a doctoral research award from the Fonds de Recherche Québec Santé. C.P. and P.T. hold Canada Research Chairs in Immunobiology and in Proteomics and Bioanalytical Spectrometry, respectively.
The online version of this article contains supplemental material.
Abbreviations used in this article:
- BMDC
bone marrow–derived dendritic cell
- DKO
double KO
- ER
endoplasmic reticulum
- ERAAP
endoplasmic reticulum aminopeptidase associated with Ag processing
- ERAP
ER aminopeptidase
- GO
gene ontology
- KO
knockout
- LC
liquid chromatography
- LC-MS/MS
LC–tandem mass spectrometry
- LRR
leucine-rich repeat
- β2m
β2-microglobulin
- MHC I
MHC class I
- MS
mass spectrometry
- pMHC
peptide–MHC
- WT
wild-type.
References
Disclosures
The authors have no financial conflicts of interest.