Abstract
Autophagy is a self-degradation system of cellular components through an autophagosomal-lysosomal pathway. Over the last 15 yr, yeast genetic screens led to the identification of a number of genes involved in the autophagic pathway. Most of these autophagy genes are present in higher eukaryotes and regulate autophagy process for cell survival and homeostasis. Significant progress has recently been made to better understand the molecular mechanisms of the autophagy machinery. Especially, autophagy process, including the regulation of autophagy induction through mTOR and the nucleation and elongation in autophagosome formation through class III phosphatidylinositol 3-kinase complex and ubiquitin-like conjugation systems, became evident. While many unanswered questions remain to be answered, here, we summarize the recent process of autophagy with emphasis on molecules and their protein complexes along with advanced molecular mechanisms that regulate the autophagy machinery.
Similar content being viewed by others
Introduction
Autophagy is an evolutionarily conserved and highly regulated lysosomal pathway that degrades macromolecules (e.g. proteins, glycogen, lipids and nucleotides) and organelles (Cuervo, 2004; Levine and Klionsky, 2004). Recent progress has demonstrated that autophagy plays an essential role in cellular development and differentiation (Levine and Klionsky, 2004) and its dysregulation is implicated in various diseases, including cancer, infectious disease, obesity, aging and neurodegenerative disorders such as Alzheimer's, Parkinson's and Huntington's diseases (Huang and Klionsky, 2007; Mizushima et al., 2008; Lee et al., 2012).
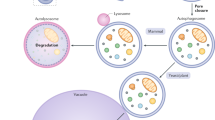
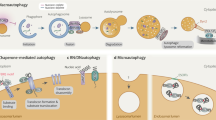
Depending on the delivery route of the cytoplasmic material to the lysosome, there are three major types of autophagy in eukaryotes; 1) chaperone-mediated autophagy (CMA), 2) microautophagy and 3) macroautophagy, hereafter referred to as autophagy (Klionsky, 2005). CMA allows the direct lysosomal import of unfolded, soluble proteins which contain a particular pentapeptide motif. In microautophagy, cytoplasmic material is directly engulfed into the lysosome at the surface of the lysosome by membrane rearrangement. Autophagy involves the sequestration of cytoplasm into a double-membrane cytosolic vesicle, referred to as an autophagosome that subsequently fuses with a lysosome to form an autolysosome for the degradation by lysosomal hydrolases (Klionsky and Emr, 2000). Autophagy is the major regulated-cellular pathway for degrading long-lived proteins and is the only known pathway for degrading cytoplasmic organelles (Yang and Klionsky, 2009). Autophagy consists of several sequential steps, which are induction, autophagosome formation, autophagosomelysosome fusion and degradation. Although autophagy has been extensively studied at the cellular level for more than four decades, its molecular mechanisms have just started to be elucidated in the past few years, mainly due to the application of yeast genetics. In this review, we summarize the molecular mechanisms of autophagy process, especially focusing on autophagosome formation.
The process of autophagy
While autophagy has been studied in mammals since the 1950's, yeast genetics has allowed us to understand this process at a molecular level. To date, 32 genes that are involved in autophagy have been identified in mammals (Table 1) and these have been termed as autophagy-related genes (Atg) (He and Klionsky, 2009). Among these, 16 genes (Atg 1-10, 12-14, 16 and 18) are required for all types of autophagy mentioned above (Suzuki and Ohsumi, 2007; Xie and Klionsky, 2007; Longatti and Tooze, 2009). These Atg proteins function at several physiologically continuous steps in autophagy and are generally classified into six groups. (1) The ULK1 kinase complex (ULK1-mAtg13-FIP200-Atg101) for the induction of autophagy, (2) Atg9 for recycling membrane, (3) class III phosphatidylinositol 3-kinase (PI3K) complex (Vps34-Beclin1-Vps15-mAtg14) for vesicle nucleation, (4) phosphatidylinositol 3-phosphate[PI(3)P]-binding Atg2-Atg18 complex (WIPI1/2 in mammals), (5) Atg12-Atg5-Atg16L conjugation system and (6) Atg8 conjugation system involving phosphatidylethanolamine (Atg8-PE) for membrane expansion (Mizushima, 2010).
Regulation of autophagy induction through mTOR and ULK1 complexes
Under stress conditions such as amino acid starvation, autophagy is strongly induced in many types of cultured cells. The effects of individual amino acids differ in their abilities to regulate autophagy. Amino acids including Leu, Tyr, Phe, Gln, Pro, His, Trp, Met and Ala suppress autophagy in ex vivo perfused liver (Mortimore and Pösö, 1987). However, such profiles depend on cell types showing their different amino acid metabolisms in tissues. The questions on how cells sense amino acid concentration and physiological significance of autophagy regulation by amino acid starvation are not fully understood yet. Accumulated reports demonstrated that amino acid signaling pathways exist, which involve activation of serine/threonine kinase mammalian Target of rapamycin (mTOR) and the subsequent regulation of the class III PI3K. The mTOR is involved in the control of multiple cell processes in response to changes in nutrient conditions (Nobukuni et al., 2005). Especially, mTOR complex1 (mTORC1) requires Rag GTPase, Rheb and Vps34 for its activation and subsequent inhibition of autophagy in response to amino acids (Wullschleger et al., 2006; Sancak et al., 2010). Energy levels are primarily sensed by AMP-activated protein kinase (AMPK), a key factor for cellular energy homeostasis. In low energy states, AMPK is activated and the activated AMPK then inactivates mTORC1 through TSC1/TSC2 and Rheb protein (Gwinn et al., 2008). Thus, inactivation of mTORC1 is essential for the induction of autophagy and plays a central role in autophagy. In addition to amino acid signaling, hormones, growth factors and many other factors, including bcl-2 (Levine et al., 2008), reactive oxygen species (ROS) (Botti et al., 2006), calcium (Green and Wang, 2010), BNIP3 (Tracy et al., 2007), p19ARF (Sherr, 2006), DRAM (Crighton et al., 2007), calpain (Xia et al., 2010), TRAIL (Mills et al., 2004), FADD (Pyo et al., 2005) and myo-inositol-1,4,5-triphosphate (IP3) (Sarkar and Rubinsztein, 2006), have also been reported to regulate autophagy. But, not all autophagy signals are transduced through mTOR signaling. A recent study showed that small-molecule enhancers of the cytostatic effects of rapamycin (called SMERs) induce autophagy independently of mTOR (Sarkar et al., 2007).
The essential process of autophagy is conserved from yeast to mammals. A distinct difference between yeast and mammalian autophagy is the presence of transient pre-autophagosome (PAS) in yeast. A protein complex composed of Atg1 (serine/threonine kinase), Atg13 (scaffold protein), Atg17, Atg29 and Atg31 is required for the formation of PAS structure and functions in the initial step of autophagosome formation in yeast. Similarly, the ULK1 kinase complex consisting of ULK1 (Atg1), mAtg13 (Atg13), FIP200 (Atg17) and Atg101 exists in mammals. Unlike in yeast, however, the ULK1 kinase complex in mammal is likely to be stably formed for autophagosome formation regardless of nutrient conditions (Ganley et al., 2009; Mercer et al., 2009; Kuma and Mizushima, 2010).
Activities of the ULK1 kinase complex are regulated by mTOR, depending on nutrient conditions. Under growing and high-nutrient conditions, the active mTORC1 interacts with the ULK1 kinase complex (ULK1-mAtg13-FIP200-Atg101) and phosphorylates ULK1 and mAtg13, and thus inhibits the membrane targeting of the ULK1 kinase complex. During starvation condition, on the other hand, the inactivated mTORC1 dissociates from the ULK1 kinase complex and results in the ULK1 kinase complex free to phosphorylate components, such as mAtg13 and FIP200, in the ULK1 kinase complex, leading to autophagy induction (Mizushima, 2010) (Figure 1).
The Class III PI3K complex in autophagosome nucleation
Autophagosome formation process is composed of isolation membrane nucleation, elongation and completion steps. In mammals, the class III PI3K complex plays an essential role in isolation membrane nucleation during autophagy (Mariño and López-Otín, 2004), while the class I PI3K pathway is also involved in autophagy regulation through insulin signaling cascade to activate mTOR and PKB (Yang and Klionsky, 2009). The class III PI3K (Vps34) is associated with Beclin1 (Atg6) and p150, the homolog of Vps15 (phosphoinositide-3-kinase, regulatory subunit 4), to form the class III PI3K core complex. As the first step of autophagosome formation, autophagosome nucleation requires Beclin1. Mammalian Beclin1, which was identified as an interaction partner of Bcl-2 (Liang et al., 1998), associates with the class III PI3K core complex to generate PI(3)P (Funderburk et al., 2010). The interaction of Beclin1 with Vps34 is known to promote the catalytic activity of VPS34 and increase levels of PI(3)P, but is dispensable for the normal function of Vps34 in protein trafficking or recruitment of endocytic events (Wurmser et al., 1999; Zeng et al., 2005).
Beclin1 plays an essential role in the initiation step of autophagy and is also involved in the pathogenesis of diseases such as pathogen infection, cancer and neurodegeneration (Levine and Kroemer, 2008). Despite the proposed roles, molecular function of Beclin1 is poorly understood. Some hints on the molecular function of Beclin1 are likely to be found in its many binding partners and several studies actually provide biological significance of these interactions. Depending on the proteins recruited by Beclin1, class III PI3K complexes differentially regulate the process of autophagosome formation (Proikas-Cezanne and Codogno, 2011). Various additional components of Beclin1 complex were recently identified. (1) Atg14L (the probable mammalian homologue of yeast Atg14) exists primarily in a Beclin1-Atg14L-Vps34-Vps15 complex that is essential for the formation of autophagosome (Itakura et al., 2008; Funderburk et al., 2010). (2) UV radiation resistance-associated gene (UVRAG) is present in a Beclin1-UVRAG-Vps34-Vps15 complex. Atg14L and UVRAG are located in the Beclin1-Vps34-Vps15 complex in a mutually exclusive manner (Liang et al., 2008). (3) Activating molecule in Beclin1-regulated autophagy (AMBRA1) is a positive regulator of the Beclin1-dependent autophagy and regulates developments of the nervous system (Fimia et al., 2007). (4) Bax-interacting factor 1 (Bif1) interacts with Beclin1 through UVRAG. Bif1 positively regulates autophagy and suppresses of tumorigenesis (Takahashi et al., 2007). Other additional proteins, including PTEN-induced putative kinase 1 (PINK1), neuronal isoform of protein-interaction, specifically with TC10 (nPIST) (Yue et al., 2002), IP3 receptor (IP3R) (Vicencio et al., 2009), the pancreatitis- associated protein, vacuole membrane protein 1 (VMP1) (Ropolo et al., 2007) and high mobility group box 1 (HMGB1) (Kang et al., 2010), have also been identified as Beclin1-interacting proteins.
In contrast to these positive regulators, there are negative regulators among Beclin1-interacting partners. (5) RUN domain- and cysteine-rich domain-containing Beclin1-interacting protein (Rubicon) negatively regulates autophagosome maturation by interacting with Beclin1 contrastively to Atg14L (Matsunaga et al., 2009). (6) Bcl2 and BclXL also bind to Beclin1 through their BH3 domain and inhibit autophagy by disrupting the interaction between Beclin1 and class III PI3K complex (Pattingre et al., 2005; He and Levine, 2010). Despite identification of numerous aforementioned molecules, it is not clear how class III PI3K complex regulates autophagosome nucleation. According to a recent study, one of Beclin1-interacting protein, Barkor/ Atg14(L), was suggested to directly bind to membrane composed of PI(3)P generated by PI3KC3 (Fan et al., 2011). By binding preferentially to the curved membranes incorporated with PI(3)P, Barkor may be capable of sensing and maintains membrane curvature to initiate autophagosomal membrane formation. However, the assembly of the class III PI3K complexes and how they act with other components in the class III PI3K complex need to be further characterized.
Ubiquitin-like protein conjugation systems in autophagosome expansion
The expansion of the isolation membrane is basically the simultaneous elongation and nucleation of little cistern. It is not known yet how the Atg12-Atg5 complex recruits additional membranes, but two ubiquitin-like protein conjugation systems are involved in the expansion of autophagosome membranes (Figure 2). The first ubiquitin-conjugation system is Atg12-Atg5-Atg16L which is essential for the formation of pre-autophagosomes. Atg12 is a 186-amino acid protein and is conjugated to Atg5 (Kuma et al., 2002). The carboxy-terminal glycine residue of Atg12 is activated by E1-like Atg7 through a high energy thioester bond in an ATP-dependent manner (Mizushima et al., 1998; Kim et al., 1999; Yuan et al., 1999; Tanida et al., 2001). Atg12 is then transferred to E2-like Atg10 (Shintani et al., 1999) and finally attached to lysine 149 of Atg5 via an isopeptide bond (Mizushima et al., 1998). The Atg12-Atg5 conjugate further interacts with Atg16L1 to form a ~350 kDa multimeric Atg12-Atg5-Atg16 protein complex through the homo-oligomerization of Atg16 (Mizushima et al., 1999) (Figure 2, left). Once autophagosome formation is completed, Atg proteins are released back to the cytoplasm by a yet uncharacterized mechanism.
The second ubiquitin-like protein conjugation system is the modification of LC3 (a mammalian homolog of Atg8) by the phospholipid phosphatidylethanolamine (PE) (Ichimura et al., 2000), an essential process for the formation of autophagosomes (Figure 2, right). LC3 is cleaved by cysteine protease Atg4 and then conjugated with PE by Atg7 and Atg3, a second E2-like enzyme. This lipidated LC3-II then associates with newly forming autophagosome membranes. LC3-II remains on mature autophagosomes until its fusion with lysosomes (Burman and Ktistakis, 2010). The conversion of LC3 to LC3-II is thus well-known as a marker of autophagy-induction. However, the increase of LC3-II alone is not enough to show autophagy activation because the inhibition of LC3-II degradation in the lysosome by the impaired autophagy flux can also cause its accumulation.
While the origin of autophagic vacuoles remains disputable, several hypotheses have been proposed for the source of autophagosomal membrane during autophagosome formation. The first hypothesis is "de novo" formation of autophagosome by Atg9 reservoirs (Mari et al., 2010). In the second hypothesis, various organelles such as ER (Hayashi-Nishino et al., 2009), mitochondria (Hailey et al., 2010) and plasma membrane (Ravikumar et al., 2010) are used as an origin for the formation of the phagophore (Figure 1). Recently, cup-shaped structures called omegasome, a discrete region of the ER, was identified as a platform for autophagosome formation (Tooze and Yoshimori, 2010). The Atg5 complex, LC3 and ULK1 were shown to be recruited into the omegasome after starvation, and Atg5- and LC3-positive membranes seem to emerge from the omegasome. It was also observed that omegasomes form in close proximity to the Vps34-containing vesicles which may synthesize the PI(3)P. This hypothesis is also supported by a notion of a physical association between the ER and early autophagic membranes (Hayashi-Nishino et al., 2009).
Vesicle completion and lysosomal degradation
Autophagosome then fuses with lysosomes/vacuoles, which is an essential process for completion of the autophagy pathway. Sequestration of cytoplasm into a double-membrane cytosolic vesicle is followed by the fusion of the vesicle with a late endosome or lysosome to form an autophagolysosome (or autolysosome). Then, inner membrane of the autophagosome and autophagosome-containing cytoplasm-derived materials are degraded by lysosomal/vacuolar hydrolases inside the autophagosome. The molecular mechanisms underlying the transport and fusion of autophagosomes are just beginning to be understood, and through active investigations, several major events involved in the process have recently been clarified.
In yeast, the fusion of autophagosomes with the vacuole requires SNARE machinery and proteins such as the vacuolar syntaxin homologue Vam3 (Darsow et al., 1997), the SNAP-25 homologue Vam7 (Sato et al., 1998), the Rab family GTP-binding protein Ypt7 (Mayer et al., 1997) or the orthologue of the N-ethylmaleimide-sensitive fusion (NSF) protein, Sec18 (Ishihara et al., 2001). In mammalian cells, autophagosome maturation is a prior step for the fusion between autophagosomes and lysosomes. Like in yeast, the activity of monomeric GTPases such as Rab22 and Rab24 is required for autophagosome maturation (Petiot et al., 2000), and mammalian orthologues of SNARE protein family members and the NSF protein may also be involved in the maturation of autophagic vesicles.
Recent studies have identified new regulators of autophagosome maturation and degradation, including UVRAG (Liang et al., 2008), Rubicon (Matsunaga et al., 2009), presenillin-1 (Lee et al., 2010), valosin-containing protein (VCP) (Tresse et al., 2010) and syntaxin-5 SNARE complex proteins (Renna et al., 2011). In addition, the endosomal sorting complex required for transport (ESCRT), which had originally been identified in the recognition and sorting of ubiquitin-modified cargo proteins into multivesicular bodies (MVBs) (Rothman and Wieland, 1996), was recently found to play a role in autophagosome-lysosome fusion (Rusten et al., 2007). Furthermore, ESCRT machinery was shown to be required for phagophore closure (Raiborg and Stenmark, 2009), autophagosome fusion (Lee et al., 2007) and lysosome biogenesis (Raiborg et al., 2008). The degradation products, including macromolecules, are then exported to the cytosol for re-utilization by the cell. This process is poorly understood.
Conclusion
In the past decade there has been an extraordinary advance in our understanding of the molecular signaling involved in mammalian autophagy. Actually, genetic screens in yeast have identified numerous Atg genes that regulate autophagy process. In spite of those outcomes, many outstanding questions remain to be elucidated, including the origin of the membrane source for autophagosome formation, mechanism of phagophore expansion and autophagosome formation and regulation of ubiquitin-like conjugation system in autophagy process (Chen and Klionsky, 2011). Recently, two interesting approaches have been employed to identify new autophagy regulators: small molecules screening (Zhang et al., 2007; Farkas et al., 2009) and studies on structural information of Atg proteins. From our knowledge, autophagy is a major contributor to maintain cellular homeostasis and metabolism. It is also involved in the pathogenesis of human diseases. Thus, continued studies to identify key molecules regulating autophagy and a better understanding for the process at molecular level are required to be further proceeded.
Abbreviations
- AMPK:
-
AMP-activated protein kinase
- Atg:
-
Atg proteins
- CMA:
-
chaperone-mediated autophagy
- ESCRT:
-
endosomal sorting complex required for transport
- IP3:
-
myo-inositol-1,4,5-triphosphate
- mTOR:
-
mammalian Target of rapamycin
- mTORC1:
-
mTOR complex 1
- NSF:
-
orthologue of the N-ethylmaleimide-sensitive fusion
- PAS:
-
pre-autophagosome
- PE:
-
phosphatidylethanolamine
- PI(3)P:
-
phosphatidylinositol 3-phosphate
- PI3K:
-
phosphatidylinositol 3-kinase
- ROS:
-
reactive oxygen species
References
Botti J, Djavaheri-Mergny M, Pilatte Y, Codogno P . Autophagy signaling and the cogwheels of cancer . Autophagy 2006 ; 2 : 67 - 73
Burman C, Ktistakis NT . Autophagosome formation in mammalian cells . Semin Immunopathol 2010 ; 32 : 397 - 413
Chen Y, Klionsky DJ . The regulation of autophagy-unanswered questions . J Cell Sci 2011 ; 124 : 161 - 170
Crighton D, Wilkinson S, Ryan KM . DRAM links autophagy to p53 and programmed cell death . Autophagy 2007 ; 3 : 72 - 74
Cuervo AM . Autophagy: In sickness and in health . Trends Cell Biol 2004 ; 14 : 70 - 77
Darsow T, Rieder SE, Emr SD . A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole . J Cell Biol 1997 ; 138 : 517 - 529
Fan W, Nassiri A, Zhong Q . Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) . Proc Natl Acad Sci USA 2011 ; 108 : 7769 - 7774
Farkas T, Hoyer-Hansen M, Jäättelä M . Identification of novel autophagy regulators by a luciferase-based assay for the kinetics of autophagic flux . Autophagy 2009 ; 5 : 1018 - 1025
Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, Cecconi F . Ambra1 regulates autophagy and development of the nervous system . Nature 2007 ; 447 : 1121 - 1125
Funderburk SF, Wang QJ, Yue Z . The Beclin 1-VPS34 complex-at the crossroads of autophagy and beyond . Trends Cell Biol 2010 ; 20 : 355 - 362
Ganley IG, du Lam H, Wang J, Ding X, Chen S, Jiang X . ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy . J Biol Chem 2009 ; 284 : 12297 - 12305
Green DR, Wang R . Calcium and energy: making the cake and eating it too ? Cell 2010 ; 142 : 200 - 202
Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ . AMPK phosphorylation of raptor mediates a metabolic checkpoint . Mol Cell 2008 ; 30 : 214 - 226
Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J . Mitochondria supply membranes for autophagosome biogenesis during starvation . Cell 2010 ; 141 : 656 - 667
Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A . A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation . Nat Cell Biol 2009 ; 11 : 1433 - 1437
He C, Klionsky DJ . Regulation mechanisms and signaling pathways of autophagy . Annu Rev Genet 2009 ; 43 : 67 - 93
He C, Levine B . The Beclin 1 interactome . Curr Opin Cell Biol 2010 ; 22 : 140 - 149
Huang J, Klionsky DJ . Autophagy and human disease . Cell Cycle 2007 ; 6 : 1837 - 1849
Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A . Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion . Mol Biol Cell 2001 ; 12 : 3690 - 3702
Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N . A ubiquitin-like system mediates protein lipidation . Nature 2000 ; 408 : 488 - 492
Itakura E, Kishi C, Inoue K, Mizushima N . Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG . Mol Biol Cell 2008 ; 19 : 5360 - 5372
Kang R, Livesey KM, Zeh HJ, Loze MT, Tang D . HMGB1: a novel Beclin 1-binding protein active in autophagy . Autophagy 2010 ; 6 : 1209 - 1211
Kim J, Dalton VM, Eggerton KP, Scott SV, Klionsky DJ . Apg7p/Cvt2p is required for the Cvt, macroautophagy and peroxisome degradation pathway . Mol Biol Cell 1999 ; 10 : 1337 - 1351
Klionsky DJ . The molecular machinery of autophagy: unanswered questions . J Cell Sci 2005 ; 118 : 7 - 18
Klionsky DJ, Emr SD . Autophagy as a regulated pathway of cellular degradation . Science 2000 ; 290 : 1717 - 1721
Kuma A, Mizushima N . Physiological role of autophagy as an intracellular recycling system: with an emphasis on nutrient metabolism . Semin Cell Dev Biol 2010 ; 21 : 683 - 690
Kuma A, Mizuchima N, Ishihara N, Ohsumi Y . Formation of the approximately 350 kDa Apg12-Apg5-Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast . J Biol Chem 2002 ; 277 : 18619 - 18625
Lee H, Noh JY, Oh Y, Kim Y, Chang JW, Chung CW, Lee ST, Kim M, Ryu H, Jung YK . IRE1 plays an essential role in ER stress-mediated aggregation of mutant huntingtin via the inhibition of autophagy flux . Hum Mol Genet 2012 ; 21 : 101 - 114
Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB . ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration . Curr Biol 2007 ; 17 : 1561 - 1567
Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, Wolfe DM, Martinez-Vicente M, Massey AC, Sovak G, Uchiyama Y, Westaway D, Cuervo AM, Nixon RA . Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations . Cell 2010 ; 141 : 1146 - 1158
Levine B, Klionsky DJ . Development by self-digestion: Molecular mechanisms and biological functions of autophagy . Dev Cell 2004 ; 6 : 463 - 477
Levine B, Kroemer G . Autophagy in the pathogenesis of disease . Cell 2008 ; 132 : 27 - 42
Levine B, Sinha S, Kroemer G . Bcl-2 family members: dual regulators of apoptosis and autophagy . Autophagy 2008 ; 4 : 600 - 606
Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roverts EA, Vergne I, Deretic V, Feng P, Akazawa C . Beclin 1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocyticc trafficking . Nat Cell Biol 2008 ; 10 : 776 - 787
Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B . Protection against fatal Sindbis virus encephalitis by Beclin, a novel Bcl-2-interacting protein . J Virol 1998 ; 72 : 8586 - 8596
Longatti A, Tooze SA . Vesicular trafficking and autophagosome formation . Cell Death Differ 2009 ; 16 : 956 - 965
Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F . An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis . J Cell Biol 2010 ; 190 : 1005 - 1022
Mariño G, López-Otín C . Autophagy: molecular mechanisms, physiological functions and relevance in human pathology . Cell Mol Life Sci 2004 ; 61 : 1439 - 1454
Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T . Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages . Nat Cell Biol 2009 ; 11 : 385 - 396
Mayer A, Wickner W . Docking of yeast vacuoles is catalyzed by the Ras-like GTPase Ypt7p after symmetric priming by Sec18p (NSF) . J Cell Biol 1997 ; 136 : 307 - 317
Mercer CA, Kaliappan A, Dennis PB . A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy . Autophagy 2009 ; 5 : 649 - 662
Mills KR, Reginato M, Debnath J, Queenan B, Brugge JS . Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is required for induction of autophagy during lumen formation in vitro . Proc Natl Acad Sci USA 2004 ; 101 : 3438 - 3443
Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y . A protein conjugation system essential for autophagy . Nature 1998 ; 395 : 395 - 398
Mizushima N, Noda T, Ohsumi Y . Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway . EMBO J 1999 ; 18 : 3888 - 3896
Mizushima N, Levine B, Cuervo AM, Klionsky DJ . Autophagy fights disease through cellular self digestion . Nature 2008 ; 451 : 1069 - 1075
Mizushima N . The role of the Atg1/ULK1 complex in autophagy regulation . Curr Opin Cell Biol 2010 ; 22 : 132 - 139
Mortimore GE, Pösö AR . Intracellular protein catabolism and its control during nutrient deprivation and supply . Annu Rev Nutr 1987 ; 7 : 539 - 564
Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, Zwartkruis FJ, Thomas G . Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase . Proc Natl Acad Sci USA 2005 ; 102 : 14238 - 14243
Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B . Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy . Cell 2005 ; 122 : 927 - 936
Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P . Distinct classes of phosphatidylinositol 3'-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells . J Biol Chem 2000 ; 275 : 992 - 998
Proikas-Cezanne T, Codogno P . Beclin 1 or not Beclin 1 . Autophagy 2011 ; 7 : 671 - 672
Pyo JO, Jang MH, Kwon YK, Lee HJ, Jun JI, Woo HN, Cho DH, Choi B, Lee H, Kim JH, Mizushima N, Oshumi Y, Jung YK . Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death . J Biol Chem 2005 ; 280 : 20722 - 20729
Raiborg C, Malerod L, Pedersen NM, Stenmark H . Differential functions of Hrs and ESCRT proteins in endocytic membrane trafficking . Exp Cell Res 2008 ; 314 : 801 - 813
Raiborg C, Stenmark H . The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins . Nature 2009 ; 458 : 445 - 452
Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC . Plasma membrane contributes to the formation of pre-autophagosomal structures . Nat Cell Biol 2010 ; 12 : 747 - 757
Renna M, Schaffner C, Winslow AR, Menzies FM, Peden AA, Floto RA, Rubinsztein DC . Autophagic substrate clearance requires activity of the syntaxin-5 SNARE complex . J Cell Sci 2011 ; 124 : 469 - 482
Ropolo A, Grasso D, Pardo R, Sacchetti ML, Archange C, Lo Re A, Seux M, Nowak J, Gonzalez CD, Iovanna JL, Vaccaro MI . The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells . J Biol Chem 2007 ; 282 : 37124 - 37133
Rothman JE, Wieland FT . Protein sorting by transport vesicles . Science 1996 ; 272 : 227 - 234
Rusten TE, Stenmark H . How do ESCRT proteins control autophagy ? J Cell Sci 2009 ; 122 : 2179 - 2183
Rusten TE, Vaccari T, Lindmo K, Rodahl LM, Nezis IP, Sem-Jacobsen C, Wendler F, Vincent JP, Brech A, Bilder D . ESCRTs and Fab1 regulate distinct steps of autophagy . Curr Biol 2007 ; 17 : 1817 - 1825
Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM . Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids . Cell 2010 ; 141 : 290 - 303
Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, Maglathlin RL, Webster JA, Lewis TA, O'Kane CJ, Schreiber SL, Rubinsztein DC . Small molecules enhance autophagy and reduce toxicity in Huntington's disease models . Nat Chem Biol 2007 ; 3 : 331 - 338
Sarkar S, Rubinsztein DC . Inositol and IP3 levels regulate autophagy: biology and therapeutic speculations . Autophagy 2006 ; 2 : 132 - 134
Sato TK, Darsow T, Emr SD . Vam7p, a SNAP-25-like molecule, and Vam3p, a syntaxin homolog, function together in yeast vacuolar protein trafficking . Mol Cell Biol 1998 ; 18 : 5308 - 5319
Sherr CJ . Autophagy by ARF: a short story . Mol Cell 2006 ; 22 : 436 - 437
Shintani T, Mizushima N, Ogawa Y, Matsuura A, Noda T, Ohsumi Y . Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast . EMBO J 1999 ; 18 : 5234 - 5241
Suzuki K, Ohsumi Y . Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae . FEBS Lett 2007 ; 581 : 2156 - 2161
Takahashi Y, Coppola D, Matsushita N, Cualing H, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ . Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis . Nat Cell Biol 2007 ; 9 : 1142 - 1151
Tanida I, Tanida-Miyake E, Ueno T, Kominami E . The human homolog of Saccharomyces cerevisiae Apg7p is a protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3 . J Biol Chem 2001 ; 276 : 1701 - 1706
Tooze SA, Yoshimori T . The origin of the autophagosomal membrane . Nat Cell Biol 2010 ; 12 : 831 - 835
Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF . BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy . Mol Cell Biol 2007 ; 27 : 6229 - 6242
Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, Yao TP, Dantuma NP, Taylor JP . VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD . Autophagy 2010 ; 6 : 217 - 227
Vicencio JM, Ortiz C, Criollo A, Jones AW, Kepp O, Galluzzi L, Joza N, Vitale I, Morselli E, Tailler M, Castedo M, Maiuri MC, Molgó J, Szabadkai G, Lavandero S, Kroemer G . The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1 . Cell Death Differ 2009 ; 16 : 1006 - 1017
Wullschleger S, Loewith R, Hall MN . TOR signaling in growth and metabolism . Cell 2006 ; 124 : 471 - 484
Wurmser AE, Gary JD, Emr SD . Phosphoinositide 3-kinases and their FYVE domain-containing effectors as regulators of vacuolar/lysosomal membrane trafficking pathways . J Biol Chem 1999 ; 274 : 9129 - 9132
Yang Z, Klionsky DJ . An overview of the molecular mechanism of autophagy . Curr Top Microbiol Immunol 2009 ; 335 : 1 - 32
Yuan W, Stromhaug PE, Dunn WA . Glucose-induced microautophagy of peroxysomes in Pichia pastoris requires a unique E1-like protein . Mol Biol Cell 1999 ; 10 : 1353 - 1366
Yue Z, Horton A, Bravin M, DeJager PL, Selimi F, Heintz N . A novel protein complex linking the delta 2 glutamate receptor and autophagy: implications for neurodegeneration in lurcher mice . Neuron 2002 ; 35 : 921 - 933
Xia HG, Zhang L, Chen G, Zhang T, Liu J, Jin M, Ma X, Ma D, Yuan J . Control of basal autophagy by calpain1 mediated cleavage of ATG5 . Autophagy 2010 ; 6 : 61 - 66
Xie Z, Klionsky DJ . Autophagosome formation: core machinery and adaptations . Nat Cell Biol 2007 ; 9 : 1102 - 1109
Zeng X, Overmeyer JH, Maltese WA . Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking . J Cell Sci 2006 ; 119 : 259 - 270
Zhang L, Yu J, Pan H, Hu P, Hao Y, Cai W, Zhu H, Yu AD, Xie X, Ma D . Small molecule regulators of autophagy identified by an image-based high-throughput screen . Proc Natl Acad Sci USA 2007 ; 104 : 19023 - 19028
Acknowledgements
The author thanks Yoo SM and Jung SM for preparation of the figures in this manuscript. Dr. Pyo JO and Nah J were partly supported by the Brain Korea-21 program. This work was supported by the grants of Global Research Laboratory program [K21004000002-11A0500-00210] and BAERI program (2011-0006314, Jung YK) and by the grant from Basic Research for Woman Scientists (Pyo JO) of the Korea Research Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Pyo, JO., Nah, J. & Jung, YK. Molecules and their functions in autophagy. Exp Mol Med 44, 73–80 (2012). https://doi.org/10.3858/emm.2012.44.2.029
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3858/emm.2012.44.2.029
Keywords
This article is cited by
-
Effects and mechanisms of Salmonella plasmid virulence gene spv on host-regulated cell death
Current Microbiology (2024)
-
DAP1 regulates osteoblast autophagy via the ATG16L1–LC3 axis in Graves’ disease-induced osteoporosis
Journal of Orthopaedic Surgery and Research (2023)
-
Resolvin D1 Induces mTOR-independent and ATG5-dependent Autophagy in BV-2 Microglial Cells
Current Medical Science (2023)
-
Autophagy Balances Neuroinflammation in Alzheimer’s Disease
Cellular and Molecular Neurobiology (2023)
-
Polymorphisms in autophagy genes are genetic susceptibility factors in glioblastoma development
BMC Cancer (2022)