Abstract
The Mapping Nearby Galaxies at Apache Point Observatory (MaNGA) survey, one of three core programs of the fourth-generation Sloan Digital Sky Survey (SDSS-IV), is producing a massive, high-dimensional integral field spectroscopic data set. However, leveraging the MaNGA data set to address key questions about galaxy formation presents serious data-related challenges due to the combination of its spatially interconnected measurements and sheer volume. For each galaxy, the MaNGA pipelines produce relatively large data files to preserve the spatial correlations of the spectra and measurements, but this comes at the expense of storing the data set in coarse units or "chunks." This coarse chunking and the total volume of the data make it time-consuming to download and curate locally stored data. Thus, accessing, querying, visually exploring, and performing statistical analyses across the whole data set at a fine-grained scale is extremely challenging using just FITS files. To overcome these challenges, we have developed Marvin, a toolkit consisting of a Python package, Application Programming Interface, and web application utilizing a remote database. Marvin allows users to seamlessly work with MaNGA data by abstracting both remote and local (on-disk) interactions to behind-the-scenes data-handling functions. Combining this capability with additional processing and querying tools, users can create powerful Python workflows that are easy to import and share. Marvin's web application uses these tools to enable "point-and-click" examination of data cubes and derived maps, as well as search queries for all publicly released MaNGA galaxies. Marvin's robust and sustainable design minimizes maintenance, while facilitating user-contributed extensions such as high-level analysis code.
Export citation and abstract BibTeX RIS
1. Introduction
Large astronomy collaborations with dedicated facilities pursuing multiyear surveys are producing massive data sets at furious rates. The data sets from the current generation of surveys, such as the Sloan Digital Sky Survey (SDSS; York et al. 2000; Strauss et al. 2002), require more disk space than is available on personal computers and some moderate-sized institution-level servers. However, the next generation of surveys, such as the Large Synoptic Sky Survey (Ivezić et al. 2019) and the Square Kilometer Array (Braun et al. 2015), will create data sets that will be far too large for all but a few dedicated national-level facilities. The real power of these immense data sets comes from simultaneously leveraging multiple sources of information (e.g., at different wavelengths) about each object, so connecting the relevant data sources for a comprehensive analysis is critical. Since individual users cannot store the data locally and need to access portions of the data remotely, bandwidth is often the primary bottleneck. Speed increases in internet bandwidth have lagged behind those in computer processors (i.e., Moore's law; Moore 1965) by 10% (Nielsen 1998); the effect of this lag has compounded over decades, up to the present, to exacerbate the gap. Consequently, only a subset of the data can be transferred. However, selecting this subset often requires access to the whole data set, which requires remote operations, especially queries.
SDSS was one of the earliest and remains one of the strongest driving forces in astronomy pushing the philosophy of public data releases that make astronomy a leader in open science. Crucially, these data releases are served with robust data distribution systems and come thoroughly documented. These two often-overlooked aspects have lowered the entry barrier and enabled thousands of professional astronomers and many times more public users to take advantage of this powerful data set. Marvin extends this mission by providing code to facilitate data use by professional astronomers, scientists in other fields (e.g., physics, computer science, and statistics), data scientists, citizen scientists, educators, and students.
The current phase (2014–2020) of SDSS, SDSS-IV (Blanton et al. 2017), consists of three simultaneous surveys, including the Mapping Nearby Galaxies at Apache Point Observatory (MaNGA; Bundy et al. 2015) survey. Legacy SDSS (York et al. 2000) took spectra of only the central regions of galaxies (Strauss et al. 2002), whereas MaNGA takes hundreds of spectra per galaxy arranged in a hexagonal grid across the face of the galaxy (Drory et al. 2015), using the SDSS/BOSS spectrographs (Smee et al. 2013) on the SDSS telescope (Gunn et al. 2006). Typically, there are three dithered sets of three individual exposures offset from each other that are combined into a data cube (Law et al. 2016; Yan et al. 2016a, 2016b). Thus, each object is not represented by just a single central spectrum, but rather a well-sampled grid of spectra.
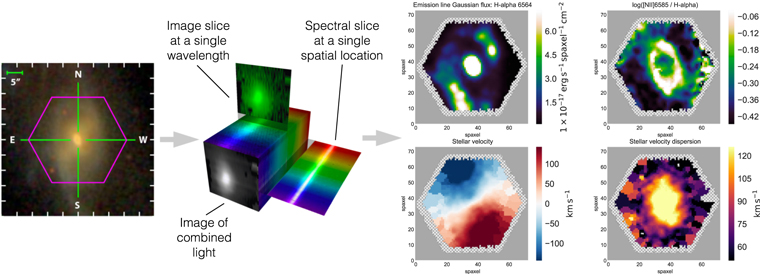
Figure 1 illustrates the format of the MaNGA data set. Each data cube consists of two spatial dimensions and one wavelength dimension. The one-dimensional spectrum at each spatial location can be interpreted in terms of measurements and physical parameters, yielding over 150 two-dimensional maps for each galaxy (Westfall et al. 2019), including gas emission lines, stellar absorption features, stellar surface density, star formation rate surface density, stellar velocities, and gas velocities. These maps can then be interpreted in terms of global properties of each galaxy: its mass in stars, its mass in dark matter, its total star formation rate, and other quantities. Marvin and the MaNGA maps for 4824 galaxies will be publicly released as part of Data Release 15 (Aguado et al. 2019).
Figure 1. Left: gri image of MaNGA 1-596678 with the IFU field of view shown in purple. Middle: IFU observations produce three-dimensional data cubes, with one spectral and two spatial dimensions (credit: Stephen Todd and Douglas Pierce-Price; IFS Wiki http://ifs.wikidot.com). Right: spectral analysis of individual spaxels produces hundreds of two-dimensional maps for each galaxy spanning a wide range of physical properties. The four example maps shown for 1-596678 are  flux (top left), log([N ii] λ6585/
flux (top left), log([N ii] λ6585/ ) flux (top right), stellar velocity (bottom left), and stellar velocity dispersion corrected for instrumental broadening (bottom right). The map plots were made using the Marvin code provided in the online documentation (https://sdss-marvin.readthedocs.io/en/stable/tutorials/plotting-tutorial.html).
) flux (top right), stellar velocity (bottom left), and stellar velocity dispersion corrected for instrumental broadening (bottom right). The map plots were made using the Marvin code provided in the online documentation (https://sdss-marvin.readthedocs.io/en/stable/tutorials/plotting-tutorial.html).
Download figure:
Standard image High-resolution imageIn addition to its complexity, MaNGA's data volume is significant. MaNGA will observe over 10,000 galaxies (Law et al. 2015; Wake et al. 2017), more than an order of magnitude larger than previous IFU surveys, such as the Atlas3D (Cappellari et al. 2011), DiskMass (Bershady et al. 2010), and CALIFA (Calar Alto Large Integral Field Area; Sánchez et al. 2012) surveys. All told, the final MaNGA data release will be 10 TB, or about 1 GB per galaxy in final summary data products, containing data cubes and row-stacked spectra in log and linear wavelength sampling, derived analysis maps, and model template data cubes. Individual data releases contain multiple analyses of each galaxy, each optimized for different science goals, resulting in multiple versions, e.g., different binning schemes, of the data cube and maps. The total volume for all of the MaNGA public data releases will be 35 TB owing to reanalyses of the same galaxies as the data pipelines improve. Because of these reanalyses, if a given scientific paper is to be replicable, easy access to previous data releases must also be provided.
Further complicating analysis of MaNGA data is its coarsely chunked storage across separate files for the spectra and derived property maps for each galaxy. Traditionally data are stored this way to optimize for an object-by-object catalog of files. These coarsely chunked data make querying on MaNGA's spatially resolved data quite difficult without extensive manual preparation of all files and tracking of correct cross-matches, so queries can only easily be done on global properties. Exploratory analysis and visualization are cumbersome with coarsely chunked data, which is compounded by the disconnected packaging of the spectra and maps. Finally, coarsely chunked data unnecessarily strain bandwidth and disk space resources because superfluous data need to be transferred and stored. These challenges encourage traditional object-by-object analyses instead of innovative ones that leverage the statistically significant sample size of MaNGA.
This paper presents software to address these challenges. Section 2 describes a typical science use case for MaNGA and how Marvin streamlines its implementation. Section 3 describes the initial prototype and its inherent limitations, the core design philosophy of Marvin, and the components involved. Section 4 describes the variety of client-based programmatic tools available in Marvin. Section 5 describes the front-facing web portion of Marvin, which serves as the exploratory portal of entry for new users. The server-side features and back-end capabilities are discussed in Section 6. In Section 7.1, we discuss the deployment and future directions for Marvin. We summarize Marvin in Section 8.
2. Workflow
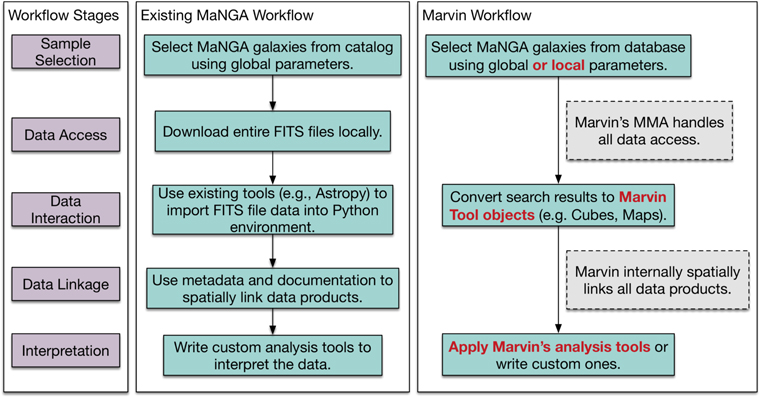
Figure 2 highlights an example workflow going through the stages of Sample Selection, Data Access, Data Interaction, Data Linkage, and Interpretation. Prior to Marvin, the workflow for an analysis of MaNGA data, as indicated in the middle panel, would typically consist of a user selecting galaxies based on global galaxy parameters, downloading all the data files for those galaxies locally, and then using existing tools (e.g., Astropy) to load the data into generic (i.e., not MaNGA-specific) FITS objects in a programming environment. To retrieve all relevant information for a target, the user must load, access, and construct the spatial links between data in separate files. Finally, users must write their own custom, often reinvented, analysis tools to visualize and interpret the data for their science.
Figure 2. Typical MaNGA workflow stages from sample selection through interpretation (left column) performed with existing tools (middle column) and Marvin (right column). At each stage, Marvin either enhances the existing capabilities (highlighted in red) or automatically handles tasks (gray boxes).
Download figure:
Standard image High-resolution imageThe Marvin framework streamlines the existing workflow as shown in the right panel of Figure 2. Marvin enhances existing workflow steps (shown as red text) and obviates workflow steps that require logistical overhead effort for data handling (shown in gray dashed boxes). While the methods involved in the existing workflow are functional, they contain the following problems, which Marvin redresses:
- 1.The first four tasks in the workflow, i.e., selecting a target sample, downloading and linking FITS files, are easy to describe but require moderate effort to implement, which is compounded when iterating over selection criteria during exploratory analyses. In contrast, Marvin provides functionality to handle these ubiquitous tasks. In particular, Marvin's front-end web interface enables rapid preliminary visual exploration without downloading data or writing code.
- 2.The selection of galaxies to analyze can only be done on global quantities, not the maps or the spectra. In contrast, search capabilities in Marvin are far more powerful, flexible, and detailed. Marvin can perform complex queries on the maps and spectra (e.g., search for galaxies with a high star formation rate surface density near the center).
- 3.Unnecessary data are inevitably downloaded, since only entire files (containing the entire data cube or hundreds of maps) are available for download, increasing bandwidth and disk resources. In contrast, Marvin adds substructure to the data so that only the explicitly requested data are downloaded (i.e., a single map or spectrum), minimizing bandwidth and local disk use, if desired.
- 4.Individuals must build their own tools to manage the download of data to the local server, which can be complicated to manage efficiently without substantial effort. Additionally, individuals must define a different set of tools for accessing remote data versus local files. These logistical issues pose a significant barrier to new users. In contrast, Marvin comes with such tools that automatically avoid multiple downloads of the same data. The same set of Marvin tools can be used in different hardware locations (i.e., with either local or remote access to the data) with only a single configuration change.
- 5.To compare map quantities and spectra, individuals must build their own tools to link spatial locations in the maps to spectra. In contrast, most of the detailed data access tools are built into Marvin and internally perform all necessary linkage in a standardized fashion.
- 6.Visualizing the data is cumbersome and time-consuming, as it requires that all data be local and relies on manual plotting scripts or the repetitive use of third-party tools. In contrast, Marvin's web interface and Python package provide visualization tools for fast iteration and exploratory analyses.
- 7.Individuals' analysis code remains siloed and is not reused. In contrast, Marvin includes some analysis code and serves as a foundation and repository for shared analysis code, which minimizes code duplication across researchers and projects.
Marvin is structured as a complete ecosystem such that the entire workflow can be performed in a single Python environment or program, but its modular design allows many aspects of Marvin to be used independently of each other. Data can be either accessed through Marvin's provided Tools or downloaded using Marvin but imported and analyzed with other tools. This flexibility makes Marvin a useful tool to a broad range of astronomers.
3. Core Design
3.1. The Marvin Prototype
To address the challenge of visually exploring MaNGA data, we developed a prototype version of Marvin that existed as a pure web application. The prototype displayed optical images, spectra, and property maps for individual galaxies. These visual displays, in conjunction with a basic annotation system, proved useful for quality assessment of an early version of the MaNGA pipelines. The prototype also featured a simple query system and provided links to download the FITS data files.
The design choices for the prototype enabled rapid development but ultimately limited its utility and sustainability. The images, spectra, and maps were static PNG files, which could not provide the interactive experience required for a complete visual exploration of the complex suite of available parameters. Queries could only be performed on global properties, not local (spatially resolved) ones. Data could only be accessed via large files that contained all of the spectra or property maps for a galaxy, making it impossible to retrieve just the spectrum or a single property of an individual spaxel. Because expanding the feature set of the prototype required creating new static files, the prototype was difficult to extend and time-consuming to maintain.
Furthermore, none of the components in the prototype web application were usable in a command line form. Users were forced to reinvent the same visual and search tools if they wanted to use them programmatically. Such tools could serve as the basis for and be related to advanced programmatic analysis tools. Every user would end up developing similar tools but within different frameworks, such that each individual's analysis code would not be interoperable with that of other users.
These limitations of the prototype design failed to address any of the inherent challenges of the MaNGA data set. Thus, a complete redesign and refactor was required to fix these shortcomings, which led to a new design philosophy of Marvin.
3.2. Design Philosophy and Core Components
Marvin's design philosophy focuses on eliminating the overhead costs and limitations of accessing the large, coarsely chunked, and incompletely linked MaNGA data set. Solving these issues enables on-demand data access, interactive visual exploration, minimal downloads, spatially resolved queries, and statistical analyses at a spaxel level. Marvin provides a feature-rich framework that serves as the building blocks for user-developed analysis tools that can be contributed back into Marvin to maximize code reuse and accelerate scientific progress.
Marvin is a complete toolkit designed for overcoming the challenges of searching, accessing, and visualizing the MaNGA data. The core design is centered around a few main components:
- 1.A Multi-Modal Access (MMA) system that handles all data flow paths.
- 2.An Application Programming Interface (API) based on the Representational State Transfer (REST) architectural style that handles all communication between the client and server.
- 3.A Brain, a common core package that handles generic functionalities and abstracts common methods needed during data gathering.
- 4.A programmatic DataModel that simplifies handling of a large suite of parameters that may differ between data releases and formats.
Marvin combines and builds on top of these core pieces to provide the following additional tools:
- 1.A suite of interconnected Python tools, all based off a core Python tool with the MMA system built in, with two main tool types:
- I.Data Product Tool: wraps your data products and retrieves specific chunks of data (e.g., Cube or Maps in Section 4.1).
- II.Query Tool: performs SQL queries against the remote data, with a pseudo-natural language syntax parser to simplify the user input.
- 2.A Python Interaction class providing a uniform interface to the API, integrated into all the Tools.
- 3.A web application, built on top of the Tools, for quick data visualization and exploration.
These tools work with each other, allowing for multiple entry points into the data, making it easy for users of various domain expertise (i.e., from students to power users) to access the data using the same suite of tools.
3.3. Multi-modal Access
In the case of MaNGA, the amount of data produced (the final data release will be of order 10 TB) sits on the boundary of what a user can store and analyze locally with normal computing resources. Future surveys (e.g., the Large Synoptic Survey Telescope) will produce data sets many orders of magnitude larger than MaNGA's, thus requiring the development of new ways to access data.
One of Marvin's core design choices is that data access should be abstracted in a way that makes the origin of the data irrelevant to the final user. Marvin accomplishes this goal with an MMA system with a decision tree that defines what access mode to use and the code implementation that executes it. Below we describe the data access modes: opening local files, searching local databases, or making API calls to a remote web server. Each of these data formats carries a series of advantages and disadvantages, but Marvin's MMA allows users to leverage the advantages while minimizing the disadvantages.
Files (e.g., FITS) provide portable data that can be heavily compressed, and they are the current standard for astronomical data distribution. However, data access can be slow (especially from compressed files), and the data are usually stored in a way that requires a degree of familiarity with the data model. Moreover, doing searches and cross-analyses between multiple targets usually demands accessing a large number of files and keeping a significant amount of data in memory.
Relational databases solve some of these problems by storing the whole data set in an optimized and well-indexed way, which enables running complex queries efficiently and provides quicker data access in most situations. In this case, the main disadvantages are the large size of a monolithic database (comparable to downloading all of the uncompressed files that compose the data set) and the difficulty of learning how to access data, especially compared to access via files.
Finally, data can be stored in servers (either as files or in databases) and accessed remotely via an API call that returns only the subset of data requested in the call. APIs are convenient for the user since they obviate the need to download data files to a local computer and can be used to abstract the data model. Their main downsides are that the internet is required to access the data and that applications that require access to large amounts of data can be slow to run.
Marvin Tools (see Section 4) include implementations that allow loading data from files, from a database, or via a series of API calls. However, once the data have been loaded, the Tools behave the same and produce the same results regardless of the data origin.
Figure 3 shows the decision tree followed by each tool to decide from where to load data. If the MMA is being run in "local" mode and a target identifier is provided (a plate-IFU or mangaid, which define a unique observation or a single target, respectively; see Yan et al. 2016a), the code checks whether a database is available and, if so, loads the data using it. If a database cannot be found, the default path file corresponding to that identifier and data release (generated as described in Section 3.3.1) is used, if the file exists locally. Alternatively, a file path can be passed to the MMA, in which case that file will be used.
Figure 3. Decision tree for the MMA system. The MMA system operates in three possible modes: local, remote, and auto. See text for a detailed explanation.
Download figure:
Standard image High-resolution imageIn "remote" mode, an API call is done to a remote server with the target identifier and the data release as inputs; the remote server uses the same MMA in "local" mode to access the necessary data from a database containing the complete MaNGA data set and returns them.
The default mode for Marvin is "auto" mode, which tries to access the data in "local" mode first and will try in "remote" mode upon failure. This order prioritizes local over remote data access because the former is usually faster, while seamlessly transitioning to the latter if the data are not available locally.
In principle, it would also be possible to set up a system with a complete MaNGA database and use Marvin to access it locally. While setting up such a system would be nontrivial from a technical standpoint, there are situations in which it could be advantageous (e.g., in the case of an institution that wants to provide a local mirror of the MaNGA data set).
Figure 4 shows a high-level overview of the user interface in Marvin. The user has two main access points: the local Marvin client or the web browser interface. While the browser interface communicates directly with the Marvin server, the MMA operating on the client side decides whether to access data locally or remotely via API calls to the Marvin server. The Marvin server (following the MMA decision tree) first attempts to access data from a local database and will fall back to files when needed.
Figure 4. High-level user interface of Marvin and the MMA system, depicting the two major paths of user flow through the system, namely, via the browser, which communicates directly with the Marvin server, or via the Marvin client, which uses the MMA to decide on local or remote access.
Download figure:
Standard image High-resolution image3.3.1. Abstract Path Generation
A machine-aware approach to file locations requires generalizing the ability to generate full paths to these files and removing all traces of the base filesystem root directory. In this way, Marvin can be agnostic to whether it is installed on a user's laptop or an SDSS host server. This layer of functionality is provided by the publicly available sdss-access (Cherinka et al. 2018) and sdss-tree (Cherinka & Brownstein 2018) software packages. sdss-tree provides the local system environment variable setup, allowing tools to understand the relative locations of data, while sdss-access provides a convenient way of navigating local and remote file paths. Paths to files are defined in a template format, specified with a shortcut name, plus a series of keyword arguments that specify variables within the filenames. This enables users to specify a robust path to any file simply by adjusting the input variable parameters. These packages are designed around relative path definitions, allowing a user to replicate a full environment by changing the definition of the base path. With a single root environment variable set by the user, these packages automatically create a local filesystem structure that mimics the filesystem of the SDSS Science Archive Server hosted at the University of Utah on which the full MaNGA data archive is stored.
For a given file, sdss-access has the ability to look up the full system path, generate the corresponding HTTP URL, and generate a remote access path for use with rsync. This flexibility allows Marvin to know precisely where to look for a given file locally and also quickly switch to a remote host when needed. sdss-access has the ability to download files from an SDSS server using multistream rsync, a technology derived from the SDSS Transfer Product (Weaver et al. 2015). This enables fast and robust file transfers, which are particularly helpful for speeding up downloads of many files. The hierarchy of files is created identically at the destination. As paths are added to the service, sdss-access eliminates redundant downloading by first checking for the existence of the file locally and only downloads files that do not currently exist.
3.4. Marvin's Brain
Marvin relies on many logistical tasks and functionalities for interacting with modern astronomical data interfaces that are not specific to MaNGA. Examples of logistical tasks are local file path management, submitting HTTP requests for accessing remote data, constructing SQL queries to access data from databases, and building web applications to serve data. These tasks incur logistical overhead costs on users and serve as barriers to data access. Since these tasks are not specific to MaNGA, they have been abstracted out of Marvin into a separate package called Marvin's Brain that provides a framework to automatically manage common data access logistics.
Currently, the key capability implemented in the Brain is a high-level wrapper, BrainInteraction, for handling HTTP requests to and from the API, which Marvin subclasses as Interaction (see Section 6.1). While BrainInteraction contains all of the generic functionality, Marvin's Interaction class customizes the configuration for MaNGA- and SDSS-specific needs, for example, adding MaNGA-specific input request parameter validation. Complementing the BrainInteraction class, the Brain also includes a generic authentication class used when accessing proprietary data via the API, generic utilities for streaming large volumes of compressed data, a generic dictionary class for the collection and lookup of all web and API routes from a given web application, and generic functions to process any incoming request data. As all of the Marvin Python Tools utilize the API for remote access, abstracting this functionality into a higher class provides a smooth entry point for each tool that is robust against API changes and reduces repeated necessary data handling. Additionally, the Brain also contains code for error logging, generic database functions (e.g., constructing network node graphs between database tables), efficient data compression, and a generic global configuration class, all of which Marvin either uses directly or subclasses for customization. While the Brain currently addresses the management of HTTP requests to an API, the goal is to expand its capabilities to additional common logistical overheads and to provide a template product for the development of future Marvin-like toolkits (see Section 7.3).
3.5. DataModel
As MaNGA is a long-running survey, the format of its DRP and DAP data product FITS files can change between data releases, also changing the MaNGA database content and structure. These changes can break analysis scripts or tools that access the underlying FITS files or database tables, creating inconsistencies that make reproducibility impossible. For example, a simple change is adding new properties, such as MPL-6 measuring 44 spectral indices not included in MPL-5. A more complicated case is the change in the default DAP binning scheme from MPL-6 (no spaxel binning) to MPL-7 (HYB10; different spaxel binnings for stellar kinematics and emission lines). Marvin programmatically implements the DRP and DAP data models by mapping the underlying FITS and database content into standardized DRPDataModel and DAPDataModel objects and classes. These classes remain constant in Marvin, acting as a translation layer between the underlying FITS file products, the equivalent database content, and the Marvin Python Tools. For example, the DRP FITS extensions containing "flux," "ivar," and a "mask," as well as its equivalent database table, are mapped to a single DataCube flux object. DAP FITS extensions containing "spectral indices" are mapped to Property and Channel objects. Through this standardized DataModel layer, data model changes introduced between releases are automatically propagated to the user-facing Data Product Tools simply by setting the MaNGA data release in a Marvin session. Thus, the Marvin Data Product Tools can be written independently of FITS files and database structure, delivering the same content through the same interface, regardless of data origin, which promotes the usage of consistent data versions and aids in scientific reproducibility for the users.
Ancillary benefits to the DataModel classes are the conveniences they provide for the user. Users can programmatically navigate the data model within the Data Product Tools, or as a standalone object, obviating the need to refer to online or published documentation of the MaNGA data model. The data model is simplified for users by utilizing FuzzyWuzzy, a fuzzy string matching algorithm, to fix incorrect but unambiguous user input (e.g., "gflux ha" maps to "emline gflux ha 6564"). Additionally, the documentation for the data model is automatically generated for online user reference.
4. Programmatic Tools
Marvin provides a programmatic interaction with the MaNGA data to enable rigorous and repeatable science-grade analyses beyond simply visualizing the data. These tools come in the form of a Python package that provides convenience classes and functions that simplify the processes of searching, accessing, downloading, and interacting with MaNGA data; selecting a sample; running user-defined analysis code; and producing publication quality figures. Marvin Tools are separated into two main categories: Data Product Tools and Query Tools. The Data Product Tools are object based and are constructed around classes that correspond to different levels of MaNGA data organization. The Query Tools are search based and are designed to provide the user the ability to remotely query the MaNGA galaxy data set and retrieve only the data they want. Marvin also provides a built-in data model, which describes the science deliverables for every data release of Marvin. Overall, these tools allow for easier access to the data without knowing much about the data model, by seamlessly connecting all the MaNGA data products, eliminating the need to micromanage a multitude of files. Figure 5 shows a visual guide to all our tools and highlights the interconnectivity between them. Tutorials with worked examples using the programmatic tools are available in the online documentation.16
Figure 5. Visual guide of the programmatic Tools, highlighting the complex interconnectivity between the tools. Green icons represent core tool classes, with orange ovals showing the connections between them. Pink icons are helper tools, and purple icons are endpoints for visually displaying data.
Download figure:
Standard image High-resolution image4.1. Galaxy Tools
These tools cover four main classes, Cube, RSS, Maps, and ModelCube, that are associated with the analogous Data Reduction Pipeline (DRP; Law et al. 2016) and Data Analysis Pipeline (DAP; Westfall et al. 2019) data products—namely, multidimensional data cubes, row-stacked spectra, derived analysis maps, and model cubes. The four main tools all inherit from a common core object, thus sharing much of their functionality and logic, such as the MMA. These tools are designed to do more than simply wrap and serve the underlying data and metadata contained in FITS files. Their goal is to streamline the users' interaction with that data and simplify common but often nontrivial tasks associated with handling the data. Via these tools, all data are delivered as Astropy Quantitys, with attached variance, mask, and any associated available properties. With Quantity variance and mask tracking, this enables robust and consistent arithmetic between any of the DAP Maps. Each tool has a built-in data model describing the format and content of the data it delivers. This data model also provides convenient top-level access to all properties available, with autocomplete navigation. Any given tool has convenient access to associated data products, as well as easy download capability for any data accessed remotely.
Features or functionalities that are common to multiple tools are designed as Python Mixin objects. These objects are designed as isolated pieces of code that can be "mixed in" with any other tool, giving that tool access to its parameters. Access to the NASA-Sloan Atlas (NSA) catalog (Blanton et al. 2011)17 and the DAP summary file, for instance, are implemented in this manner. Extracting spaxels within a specified aperture is a common functionality delivered to all tools as a Mixin.
There are additional tools that are not associated with a particular MaNGA data file but instead map to objects related to the MaNGA data. These tools behave in much the same way as the core tools. They utilize the MMA, allow for remote file downloading, and are seamlessly integrated with each other. The Plate tool corresponds to an observed SDSS plate used during MaNGA observations. This object provides a list of all of the Cubes observed on a given plate, along with additional metadata associated with the plate (exposure numbers, observation date, etc.). The Image tool provides interaction with the MaNGA IFU image cutout from SDSS multiband imaging. It allows for quick display of the IFU image, overplotting of the IFU hexagon, overplotting of the individual IFU bundle or sky fibers, or generating an entirely new image at a custom pixel scale. Additionally, a list of Image objects can be quickly generated and downloaded to the local client system. Image utilities also exist to quickly download a list of images in bulk using the streaming capability of sdss-access.
4.2. Subregion Tools
Marvin provides subregion galaxy tools, which are designed to access individual components within the main MaNGA data products. RSSFiber, Spaxel, and Bin provide access to the row-stacked spectra from individual fibers, data cube spaxels, or bins (for binned DAP data), respectively. These tools come with convenient plotting functions, as well as access to all the DRP and DAP properties associated with a given element. The DAP produces data products with different spectral binning schemes for different science cases: unbinned spectra (SPX), spectra binned to signal-to-noise ratio (S/N) ∼ 10 using the Voronoi binning algorithm (VOR10), and a hybrid binning scheme (HYB10), with spectra binned to S/N ∼ 10 for the stellar kinematics, but emission-line measurements are performed on the individual spaxels. The "HYB10" binning type for DAP products has complicated the underlying binning scheme of spaxels. The Spaxel and Bin tools make the binning much more straightforward. Each Spaxel property contains information about whether it is binned or not, hooking into the Bin tool when appropriate. The Bin tool displays only the relevant information for the underlying property and binning type, clearing up most of the obfuscation with accessing the "HYB10" binned files directly. From Bin, one can access all spaxels belonging to that bin, as well as generate masks for that bin.
4.3. Query Tools
Marvin provides tools for searching the MaNGA data set through an SQL-like interface, either via a web form or a Python class. The Marvin Query system uses a simplified SQL syntax (see Section 4.3.1) that focuses only on a filter condition using Boolean logic operators and a list of parameters to return. Not only does this simplify the syntax, but it also automatically performs the incredibly complex table joins required to extract data from the MaNGA database. Users can query the MaNGA sample on global galaxy properties, similar to searching through the DRP and DAP summary files. In the near future, users will be able to perform intragalaxy queries on individual spaxel measurements—a task that requires a database or loading all of the MaNGA spaxel data into RAM. Tutorials for querying with Marvin are available in the online documentation.18
4.3.1. Pseudo-natural Language Syntax
Figure 6 shows an example of a MaNGA query in (1) natural language, (2) full SQL syntax, and (3) simplified pseudo-natural language syntax. While the query is relatively easy to describe in natural language, the full SQL syntax (red panel in Figure 6) is immensely complicated to construct, even if the user already knows how to write SQL queries. SQL queries consist of three main parts: a select clause, a join clause, and a where clause. Constructing the select and join clauses requires detailed knowledge of the MaNGA database schema, table design, available columns, and the keys needed to join the tables. With the Marvin Query tool, rather than submitting the full SQL query, the user submits only a simplified where clause and an optional list of properties to return. The remainder of the query (the select and join clauses) is built dynamically behind the scenes, converted to raw SQL, and then submitted to the database. This allows the user to focus on the properties and their values in the selection criteria.
Figure 6. Example query on the MaNGA data set. The top describes the query in natural language syntax, i.e., how a user would describe it. The red panel shows the full SQL syntax needed to perform the same query on the MaNGA database. The green panel shows the corresponding pseudo-natural language syntax and its use in Marvin.
Download figure:
Standard image High-resolution imageMarvin uses SQLAlchemy to map Python "model" classes onto each of our database tables and columns. This provides the base ability to dynamically build and submit SQL queries in Python. With these model classes, Marvin constructs a singular lookup dictionary containing a mapping between a string parameter name, in the form of schema.table.column_name, and its Python counterpart. This provides an automatic way of looking up the database location for a given parameter name, effectively removing the select clause. Marvin uses networkx to map those model classes onto a network tree, which allows the construction of a proper SQL join clause given any two input parameters across all tables in all schema in the database. Finally, Marvin uses a customized version of sqlalchemy-boolean-search to simplify the where clause to a simple input string. This is a Boolean parser that takes a string Boolean filter condition, parses it, and converts to the proper SQLAlchemy filter object. The green panel in Figure 6 shows the pseudo-natural language equivalent of the desired query.
4.4. Utilities
Maskbit: MaNGA uses masks as a compact way to simultaneously convey information about the status of an object under many Boolean conditions. The MaNGA pipelines produce quality masks at each processing stage, which allow users to filter out specific types of undesirable data when performing science analyses. During target selection, MaNGA likewise creates targeting masks that encode the sample or program under which an object was selected to be targeted.
The Maskbit class is a general-purpose utility used by other Data Product Tools. It automatically loads the schema for a mask, which can be easily displayed for the user. It can then convert from the native integer value (e.g., 1025) to the list of bits set (e.g., [0, 10]) to the corresponding list of labels (e.g., ["NOCOV," "DONOTUSE"] for the MANGA_DRP3PIXMASK mask, which indicates that the spaxel has no coverage in the cube and should not be used for science). Users can create a mask by providing a list of labels instead of filtering bits. This class also enables searching on bits, which is particularly useful for target selection using the targeting masks.
Plotting Utilities: Marvin's plotting utilities enable users to quickly display images, spectra, and maps of individual MaNGA galaxies or galaxy subregions. The plotting utilities also can put galaxies or subregions in context via scatter and histogram plots of query results. As a general philosophy, Marvin's plotting utilities are designed to have smart defaults for quickly making useful visualizations while allowing for significant customization via standard Matplotlib methods, which is particularly important for displaying maps.
Querying is one of Marvin's most powerful features. Yet it is difficult, if not impossible, to discover trends in large tables of text produced from a query. To that end, Marvin includes utilities to make scatter and histogram plots of query results. Queries in Marvin can return results with anywhere from a few to millions of data points, so Marvin's scatter plot changes the underlying display technique depending on the number of data points (see Figure 7). Fewer than 1000 data points are shown individually (Figure 7(a)), 1000–500,000 data points are shown as a hex-binned density distribution (Figure 7(b)), and more than 500,000 data points are shown as a scatter density map (Figure 7(c)) that is responsive even with millions of data points. By default, scatter plots show marginal histograms with the mean and standard deviation. Users can also create histograms separately from a scatter plot and extract the data points in each bin.
Figure 7. Scatter-plotting capability from the Marvin Results Tool. Depending on the number of results, Marvin plots using a straight scatter plot, a hex-binned density distribution, or a scatter density map. (a) Matplotlib scatter plot for results with less than 1000 points. (b) Matplotlib hexbin plot for results with between 1000 and 500,000 points. (c) Matplotlib scatter density plot using https://github.com/astrofrog/mpl-scatter-density for results with more than 500,000 points.
Download figure:
Standard image High-resolution imageAnalysis Tools: At the time of publication, we have prioritized development of aspects of Marvin required for interfacing with the MaNGA data over providing downstream analysis tools. However, Marvin is ideally suited to serve as a foundation for analysis tools that extend its functionality to additional processing steps. One such analysis tool that has already been developed is a tool to classify different regions of a galaxy according to classical emission-line ratios.
As discussed by, e.g., Baldwin et al. (1981, hereafter BPT), the nebular permitted ( ,
,  ) and forbidden emission-line transitions (e.g., [O ii]
) and forbidden emission-line transitions (e.g., [O ii]  , [O iii]
, [O iii]  , [N ii]
, [N ii]  ) are commonly strong and easy to detect in galaxies that contain significant quantities of gas. Since these transitions have different ionization potentials, their relative strengths encode information about both the metallicity of the gas and the hardness of the radiation field emitted by the source of ionizing photons. As such, easily measured line ratios such as [O iii]/
) are commonly strong and easy to detect in galaxies that contain significant quantities of gas. Since these transitions have different ionization potentials, their relative strengths encode information about both the metallicity of the gas and the hardness of the radiation field emitted by the source of ionizing photons. As such, easily measured line ratios such as [O iii]/ and [N ii]/
and [N ii]/ can be used to discriminate between H ii regions produced by thermal (i.e., star formation) and nonthermal processes (e.g., shocks and active galactic nuclei [AGNs]).
can be used to discriminate between H ii regions produced by thermal (i.e., star formation) and nonthermal processes (e.g., shocks and active galactic nuclei [AGNs]).
Marvin's BPT tool returns masks in which individual spaxels have been classified as "star-forming," "Seyfert," or "LINER-like" line ratios, such that a user can then plot diagnostic diagrams, categorial maps of the classifications, or maps filtered by these classifications (for instance, plotting the  flux for star-forming regions). Such analyses have revealed significant clues as to the physical origins of the ionizing photons, indicating, for example, that in many sources observed to have LINER-like line ratios in SDSS single-fiber spectroscopy the gas is spatially extended and likely ionized by hot evolved stars rather than a central AGN (Belfiore et al. 2016).
flux for star-forming regions). Such analyses have revealed significant clues as to the physical origins of the ionizing photons, indicating, for example, that in many sources observed to have LINER-like line ratios in SDSS single-fiber spectroscopy the gas is spatially extended and likely ionized by hot evolved stars rather than a central AGN (Belfiore et al. 2016).
4.5. Contributed Code
While the core of SDSS data releases centers around its base projects' science deliverables, smaller teams frequently contribute added value to its core deliverables with additional science products. These value-added data products or catalogs (VACS) are derived data products based on the core deliverables that are vetted, hosted, and released by SDSS in order to maximize the impact of SDSS data sets. To increase the visibility of MaNGA VACs, Marvin has hooks to allow users to contribute small pieces of code that plug their VACs into the overall system, immediately connecting their VAC into the larger suite of Marvin Tools and MaNGA Data Products. Each contributed code piece is well documented, adheres to the overall standards set by SDSS,19 and contains the proper software credit for the user.
The core design principles of Marvin are to perform most of the legwork for the users, making access as easy as possible, while allowing users to contribute their own code to help expand Marvin's functionalities. For VACs, contributors create a new class defining their VAC based on a predefined base class that provides unique target identifiers and automatic file retrieval methods needed to extract specific data from files. Contributors simply define the name of their VAC, the unique file path parameters, and a single method returning the data content. Contributors do not need to implement access to the core data products, which is already handled by Marvin.
More generally, the Marvin code is structured to ease contributions of drop-in utility or analysis methods that add functionality to Marvin. These functions can manipulate or extract data from existing Marvin Tools, perform some analysis, or return a plot or data. The BPT tool from the previous section serves as an example of such a drop-in function that easily wraps the existing Tools. Users are encouraged to contribute Marvin-based analysis code back into the project so that others can take advantage of their efforts. We have adopted the BSD 3-Clause open-source software license to facilitate and encourage contributions.
5. Marvin Web
The web, or browser-based information gathering, is often the first entry point for any user new to a field. Poor web design (e.g., cluttered content, complex interfaces) can quickly discourage users from interacting with the delivered content. Marvin provides a web front end that aims to be as intuitive and streamlined as possible, with a focus on quick visual exploration of the MaNGA data set, leaving more rigorous analysis to the programmatic Marvin Tools or the user's own scripts. This minimal but interactive interface encourages users to quickly engage with MaNGA data and, when ready, seamlessly transition into more advanced environments. Our web component is built using Flask, a Python-based, lightweight, micro web framework. Flask allows for quick deployment of a web application with minimal effort. It contains its own built-in web server for small-scale deployment, or can easily be integrated into more advanced web servers for production deployment. It has built-in hooks for modularity and extensibility and employs a templating system for writing front-end code like HTML or Javascript in a modular way.
MarvinWeb currently provides the following features:
- 1.a Galaxy page, for detailed information and interaction with individual galaxies in MaNGA;
- 2.a Query page, for searching the MaNGA data set using the simplified SQL pseudo-natural language syntax described in Section 4.3.1;
- 3.a Plate page, containing all MaNGA galaxies observed on a given SDSS plate; and
- 4.an Image Roulette page that randomly samples images of MaNGA galaxies, useful for browsing the wealth of variety in the 10,000-galaxy sample.
The Galaxy page (see Figure 8) provides dynamic, interactive, point-and-click views of individual galaxies to explore the output from the MaNGA DRP and DAP, namely, spectra and map properties, along with galaxy information from the NSA catalog. In contrast to the prototype, this page is completely interactive, with more galaxy metadata. These interactive features are deployed using a variety of third-party Javascript libraries: Dygraphs for the spectral viewer, Highcharts for the map and scatter plot viewers, OpenLayers for the interactive optical image display, and D3 for the box-and-whisker plots.
Figure 8. Marvin web Galaxy page, highlighting the interactive point-and-click feature. Users can dynamically interact with individual galaxy spaxels and maps. Clicking anywhere within the optical image, or the DAP maps, retrieves the spectrum at that spaxel location. The interactive spectrum displays both the flux (green) and model fit (blue) for the selected spaxel. Marvin displays three maps by default: the  emission line flux, the stellar velocity, and the D4000 spectral index. Drop-down menus provide additional maps to display or maps of different binning schemes.
emission line flux, the stellar velocity, and the D4000 spectral index. Drop-down menus provide additional maps to display or maps of different binning schemes.
Download figure:
Standard image High-resolution imageThe Query page provides the entry point for quickly searching through the MaNGA data set. In contrast to the prototype, this page provides search capability for all properties in the DRP and DAP summary files, with minimal impact on the design interface of the page. The search capability will soon be extended to the entire suite of MaNGA parameters. It is built on top of the Marvin Query tool, providing a single simple interface, for both the web and tool, that one needs to become familiar with. In addition, the query system can be easily extended for both web and client users at the same time. Performing a query produces a navigable table of results, with each row linking to the individual galaxy. One can optionally switch to a postage stamp view of all galaxies returned in the query subset.
While the input structure to the Marvin Query tool is simplified greatly from the underlying full SQL statement, the syntax can still be complicated to learn. Some users may find it cumbersome, delivering confusion instead of intuition. The Query page also includes an interface for dynamically constructing an SQL statement in a guided manner. This interface provides a series of parameter drop-downs that, in conjunction with operators and values, can be used to build conditions, and combined together with Boolean operands. A web video tutorial20 is available highlighting general usage, with more information available in the online documentation.
Because the web components are built on top of the Tools, all the galaxy and query features can be mapped to an underlying equivalent Marvin tool command. This allows users to re-create what they experience in the Web with the Marvin Tools locally on their system. On each page we provide feature-specific code snippets that indicate the equivalent commands for viewing galaxy maps or spaxels, or querying the data set. These snippets can be copied and pasted directly into the local iPython terminal.
For the back end, Flask provides the basis for the framework as well. It can be run in a "debug" mode for rapid development or be served in a production environment. For production deployment, Marvin is run using the NGINX web server, with uWSGI acting as the gateway interface between the Flask web-app and NGINX. Flask provides the basic framework on which the Marvin API and the web-facing front end are built. Our back-end Flask routes are built using the same suite of Marvin Tools available to the user on the client side. In this manner, we can build a single tool for the user while also using it to provide the same content directly over the API or integrated into the front end as a web feature. The server-side Marvin uses the same MMA system to determine data location, pulling first from a local database hosted at Utah and then from the files located on the Science Archive Server (SAS) filesystem.
6. Marvin Back End
6.1. REST-like API
To provide remote data access, Marvin employs a REST-like web API, which defines a set of rules for remote data acquisition through HTTP request methods (i.e., GET and POST). The API handles all requests and responses between the user and server. There are three ways to interface with the Marvin API: directly through HTTP (low level), with a Python helper class (mid level), or via Marvin Tools (high level).
The lowest access level provides direct HTTP access to the API routes. Our API routes use the underlying Marvin Tools and provide remote access to the most commonly desired features of the MaNGA data set. A list of the available routes, as well as what data they provide, can be found in the online documentation.21
While an experienced user can directly use the HTTP routes to retrieve data, not everyone is familiar with how to handle HTTP requests and responses. The middle layer wraps the direct API calls into a Python Interaction class. The Interaction class is a customized MaNGA subclass of the Brains BrainInteraction (see Section 3.4), which utilizes the requests package to handle GET/POST exception handling, set default request parameters, check the response, and provide convenience methods for parsing return data into Python data types.
At the highest level, the Interaction class is built into the core Marvin Tools (and any Tools customized from them), providing remote access ability to all Tools. Marvin contains a lookup dictionary for resolving API URL shortcuts into their full route names. This dictionary allows each Tool to understand its required remote call and provides robustness against server-side API route changes. In this access level, the user takes a hands-off approach to API requests. The Tool determines when to access data locally or remotely. If remote data access is required, it performs the API request without user input and shapes the data properly upon receiving the response. When using the API, the Tools employ a lazy-loading approach to remote data to minimize server load. The API returns the minimum amount of information needed to satisfy the user's request. Additional information requested through the Tools is acquired through additional API calls.
6.2. Database
All MaNGA data are stored in a PostgreSQL relational database. The database is the bedrock data storage component of Marvin. It provides the basis for interactive visualization in the web, spatially resolved queries, and selective data retrieval. For each release of MaNGA, we store the metadata and raw spectral output from the DRP and DAP pipelines. We currently store data from MaNGA internal data releases, referred to as MaNGA Product Launches (MPLs) 4–8, and the public data release DR15. The MaNGA database currently houses all galaxies across MPLs 4–8, totaling 12 TB in size. With galaxies from earlier MPLs/data releases being rereduced and reanalyzed in each MPL/data release, this results in a cumulative 26,932 galaxy reductions/analyses. MPL-8 alone contains 6779 galaxies contributing 3 TB to the database. The database contains three main schema: mangadatadb, which contains the DRP output; mangadapdb, which contains the DAP output; and mangasampledb, which contains information on MaNGA targets and the NSA catalog. In addition, there is an auxiliary schema for miscellaneous data and a history schema that stores user and query metrics. The main schema designs can be found in the online documentation.22
6.3. SAS Filesystem
The SAS filesystem is the data warehouse for SDSS. Hosted at the University of Utah and mirrored at the National Energy Research Scientific Computing Center, the SAS includes all of the raw and reduced SDSS data, including the intermediate and final data products from each survey's data reduction or analysis pipelines and value-added catalogs (∼1000 TB). The SAS serves both the collaboration (through a private gateway) and the public. The filesystem structure is organized hierarchically by survey to facilitate easy navigation. Both the software and data products are under version control, and the versions are explicitly included in the file paths. The explicit versioning in the file paths allows for consistent access and rapid deployment for internal and public data releases. All software and data products are frozen on a schedule set by the SDSS collaboration and tagged to maintain a self-consistent, reliable, and robust data system. These immutable tags and frozen reduction versions ensure the reproducibility of high-quality products. By developing Marvin to work on top of this structure, we can consistently deliver an archive-quality data product to the community, mitigating concerns about underlying intermittent data changes.
6.4. Authentication and Access
Marvin provides access to MaNGA data for both the SDSS collaboration and the public astronomy community. The SDSS collaboration provides data access rights for a proprietary period, so Marvin has collaboration-only and public access modes. The collaboration-only access mode provides access to the private gateway for both internal data releases and public data releases. In contrast, the public access mode provides access to the public gateway for only the public data releases. Approved SDSS collaboration members must authenticate with Marvin before access is granted to the private gateway. For the Web, Marvin uses the Flask extension Flask-Login, which uses session-based cookies to handle all login and authentication. For the API, Marvin uses Flask-JWT-Extended, which authenticates via JSON Web Tokens. After supplying their credentials in a Unix standard netrc file, the user is allowed to log in and receive a valid token. The token is inserted into every API request and authenticated on the back end.
7. Deployment and Future Directions
7.1. Deployment
We tag and deploy versions of Marvin that can be installed with the pip23 package manager, installed as a conda virtual environment,24 or cloned with git from the GitHub25 repository. We encourage open-source software development by adopting a BSD-3 license and hosting all code on Github. The documentation for every version of Marvin is built with the Sphinx package and available on Read the Docs,26 including the latest version pushed to the "master" branch of the GitHub repository. The tagged versions of Marvin have a digital object identifier (DOI) and are hosted on Zenodo.27
Marvin uses several packages and services for testing—pytest for Python code; the built-in Flask test server for web back-end and API routes; and a combination of Selenium, Pytest-Flask, and BrowserStack for web front-end interactions. The fraction of code covered by tests is tracked using Coveralls.28 Our full test suite is automated with Travis-CI, a continuous integration system. Finally, we use Sentry,29 an open-source error tracking client, to monitor and fix crashes in real time.
7.2. Ongoing SciServer Integration
While Marvin works either as a local analysis package or for browser-based visual exploration, it still requires local package installation for local analysis and focuses on single-user software usage. Local package installation can often interfere with custom user environments, while single-user usage and analysis limits the ability for collaborative science. However, an advantage of Marvin is that it can be deployed either as a client service or in a server mode distributing content, into existing archive systems or in different environments. To enable collaborative and remote scientific analysis, we are in the process of integrating Marvin into the SciServer platform. SciServer30 is a fully integrated cyber-infrastructure system encompassing related, integrated tools and services to enable researchers to cope with and collaborate around scientific big data.
SciServer integration will enable users to utilize the access and analysis capabilities of Marvin without having a local installation. Collaborative science will be enabled through remote, persistent Jupyter notebooks that can be shared among multiple users. Analysis can be offloaded to the SciServer system, removing the need for any data to be hosted locally. Additionally, through the SciServer Compute system, the Marvin Query tool will be expanded to include asynchronous query capability, allowing intensive queries to be submitted as jobs, similar to the existing SDSS CasJobs system, freeing up the user's local terminal.
7.3. Future Brain Generalization
Marvin is a toolkit specifically designed to serve MaNGA users, but key aspects of it could be generalized to form a template product that would serve as a starting point for a toolkit for other astronomy data sets. Section 2 describes a typical workflow with the MaNGA data set; however, this workflow is not unique to MaNGA and is similar to workflows with other astronomical data sets. As described in Section 3.4, there are many common logistical challenges and overheads when dealing with data access and delivery in astronomy, with no suite of tools providing out-of-the-box capabilities to address these challenges. Marvin's Brain currently contains some common important functionality necessary for a template product that can facilitate a more streamlined data workflow. Our goal is to further abstract out Marvin's building blocks (e.g., MMA system) into the Brain to create a complete framework for a data distribution system, providing seamless connections between web components, APIs, and programmatic Python tools. Such a template product would link underlying the Python packages (e.g., requests, Flask, SQLAlchemy) often used to solve logistical challenges but that do not provide usable solutions by themselves. This product, when given a file and a database presentation of that file, will provide base classes to provide a connected environment surrounding that file, with local file and remote API access, programmatic tools, a remote query system, and a web front end to the data with a minimum display view. Creating a functional application would simply involve building a new Python package based off the Brain, subclassing its base classes, and adding functionality and details necessary for the particular application. Such a product would be particularly interesting for future IFU data sets, as they are the most natural extension of Marvin, reusing the majority of its programmatic tools; however, the design of the Brain inherently makes it applicable to generic data sets within astronomy.
8. Summary
We have presented the first public release of the Marvin software, a toolkit for streamlining users' workflow (Section 2) on the SDSS-IV MaNGA data set. A novel aspect of Marvin is the MMA system (Section 3.3) that automatically switches between local files and a remote database to retrieve MaNGA data. The MMA is tightly integrated into a suite of Python Tools, enabling intuitive access into the MaNGA data products (Section 4.1), as well as remote querying of the entire MaNGA data set with a simplified query syntax (Section 4.3). Marvin provides a web front end (Section 5) for quick visual exploration of MaNGA data. We have adopted modern coding best practices for long-term open-source software sustainability (Section 7). Marvin is being integrated into the SciServer platform to enable collaborative and remote analysis (Section 7.2). Finally, we plan to generalize the Marvin framework (Section 7.3) to be easily adaptable for other IFU, astronomical, and scientific data sets.
We would like to acknowledge the MaNGA team for supporting this project, helping shape its design, and providing critical feedback and testing during various shakedown phases. We would like to thank Demitri Muna for his help with Flask and database services. We give thanks to 8485-1901 for its noble sacrifice as our guinea pig.
Funding for the Sloan Digital Sky Survey IV has been provided by the Alfred P. Sloan Foundation, the U.S. Department of Energy Office of Science, and the Participating Institutions. SDSS-IV acknowledges support and resources from the Center for High-Performance Computing at the University of Utah. The SDSS website is www.sdss.org.
SDSS-IV is managed by the Astrophysical Research Consortium for the Participating Institutions of the SDSS Collaboration, including the Brazilian Participation Group, the Carnegie Institution for Science, Carnegie Mellon University, the Chilean Participation Group, the French Participation Group, Harvard-Smithsonian Center for Astrophysics, Instituto de Astrofísica de Canarias, Johns Hopkins University, Kavli Institute for the Physics and Mathematics of the Universe (IPMU)/University of Tokyo, the Korean Participation Group, Lawrence Berkeley National Laboratory, Leibniz Institut für Astrophysik Potsdam (AIP), Max-Planck-Institut für Astronomie (MPIA Heidelberg), Max-Planck-Institut für Astrophysik (MPA Garching), Max-Planck-Institut für Extraterrestrische Physik (MPE), National Astronomical Observatories of China, New Mexico State University, New York University, University of Notre Dame, Observatário Nacional/MCTI, The Ohio State University, Pennsylvania State University, Shanghai Astronomical Observatory, United Kingdom Participation Group, Universidad Nacional Autónoma de México, University of Arizona, University of Colorado Boulder, University of Oxford, University of Portsmouth, University of Utah, University of Virginia, University of Washington, University of Wisconsin, Vanderbilt University, and Yale University.
Software: Anaconda (https://anaconda.org/anaconda/python), Astropy (Astropy Collaboration et al. 2013; The Astropy Collaboration et al. 2018, http://www.astropy.org), Bootstrap (https://getbootstrap.com), Browserstack (https://www.browserstack.com) brain (https://github.com/sdss/marvin_brain), Coveralls (https://coveralls.io/), D3 (https://d3js.org), DyGraphs (http://dygraphs.com), FITS (Pence et al. 2010), Flask (http://flask.pocoo.org), Flask-Login (https://flask-login.readthedocs.io), Flask-JWT-Extended (https://flask-jwt-extended.readthedocs.io), fuzzywuzzy (https://github.com/seatgeek/fuzzywuzzy), git (https://git-scm.com), Highcharts (https://www.highcharts.com), Jinja2 (http://jinja.pocoo.org/docs), JQuery (https://jquery.com), Jupyter (Kluyver et al. 2016, http://jupyter.org), Matplotlib (Hunter 2007, https://doi.org/10.5281/zenodo.61948), networkx (https://networkx.github.io), Nginx (https://www.nginx.com), OpenLayers (https://openlayers.org), pip (https://pypi.org/project/pip), Postgres (https://www.postgresql.org), pytest (https://docs.pytest.org/), Read the Docs (https://readthedocs.org/), requests (http://docs.python-requests.org), rsync (https://rsync.samba.org), sdss-access (https://doi.org/10.5281/zenodo.1410704), sdss-tree (https://doi.org/10.5281/zenodo.1410706), Selenium (https://www.seleniumhq.org), Sphinx (http://www.sphinx-doc.org), SQLAlchemy (https://www.sqlalchemy.org), sqlalchemy-boolean-search (https://github.com/sdss/sqlalchemy-boolean-search), Travis-CI (https://travis-ci.org/), uwsgi (https://uwsgi-docs.readthedocs.io).
Footnotes
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
pip install sdss-marvin.
- 24
- 25
- 26
- 27
- 28
- 29
- 30