Published online Jul 7, 2006. doi: 10.3748/wjg.v12.i25.4074
Revised: November 1, 2005
Accepted: November 10, 2005
Published online: July 7, 2006
AIM: To analyze the clinical characteristics of Chinese hereditary nonpolyposis colorectal cancer (HNPCC) families and to screen the germline mutations of human mismatch repair genes hMLH1 and hMSH2 in the probands.
METHODS: Thirty-one independent Chinese HNPCC families were collected in Zhejiang Province. All of them met Chinese HNPCC criteria. Clinical data about patient gender, site of colorectal cancer, age of onset, history of multiple colorectal cancer, associated extracolonic cancer were recorded. PCR and denaturing high performance liquid chromatography (DHPLC) were employed to screen the mutations. Sequencing analysis was used to find out the exact mutation site and characteristics of the samples showing abnormal DHPLC profiles.
RESULTS: One hundred and thirty-six malignant neoplasms were found in 107 patients including 14 multiple cancers.One hundred and six of the 136 neoplasms (77.9%) were diagnosed as colorectal cancer, with an average age of onset at 48.57 ± 29.00 years. Gastric cancer was the most common extracolonic cancer (10.3%) in these families. Twenty-three different sequence variations in hMLHl and hMSH2 genes were detected in these 17 families. Fifteen sequence variations were located in the exons, including 5 SNPs, 3 silent mutations, 3 missense mutations, 2 nonsense mutations and 2 frameshift mutations. The latter seven mutations seemed to be pathogenic.
CONCLUSION: Germline mutations of hMLH1 and hMSH2 genes are identified in about one-third HNPCC kindreds fulfilling Chinese HNPCC criteria. Chinese HNPCC families have some particular clinical characteristics, such as a left-sided predominance, less synchronous or metachronous colorectal cancer, and frequent occurrence of gastric cancer.
- Citation: Wang XL, Yuan Y, Zhang SZ, Cai SR, Huang YQ, Jiang Q, Zheng S. Clinical and genetic characteristics of Chinese hereditary nonpolyposis colorectal cancer families. World J Gastroenterol 2006; 12(25): 4074-4077
- URL: https://www.wjgnet.com/1007-9327/full/v12/i25/4074.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i25.4074
Hereditary nonpolyposis colorectal cancer (HNPCC) is an autosomal dominantly-inherited cancer susceptibility syndrome. It is estimated that HNPCC may account for 5%-10% of the total colorectal cancers (CRC) worldwide[1]. Clinically, HNPCC families are diagnosed based on the Amsterdam criteria. Although the Amsterdam criteria unify the diagnosis of HNPCC worldwide, they are too rigid to exclude extracolonic cancers associated with HNPCC. New criteria and guidelines have been proposed, such as the Japanese criteria, suspected HNPCC criteria. Based on the suspected HNPCC criteria and the specific characteristics of the tumor spectrum in Chinese population, the Chinese Hereditary Colorectal Cancer Collaboration has established the criteria for Chinese HNPCC families[2].
Six genes (hMSH2, hMLH1, hPMS1, hPMS2, hMSH6/GTBP and hMLH3) involved in DNA mismatch repair have been proven to be closely related with the development of HNPCC. hMLH1 and hMSH2 genes are thought to be the main genes responsible for HNPCC, because more than 90% detected germline mutations in HNPCC families are located in these two genes[3]. Denaturing high-performance liquid chromatography (DHPLC) is a mutation pre-screening method[4]. The major advantage of DHPLC is the low cost and the high speed of analysis. Therefore, in this study, we analyzed the clinical characteristics of our 31 Chinese HNPCC families registered in our cancer institute, and screened the germline mutations of hMLH1 and hMSH2 genes by DHPLC and DNA sequencing.
Thirty-one families involved in this study fulfilling the Chinese HNPCC criteria were collected in Zhejiang Province. Detailed familial and medical histories were obtained through an interview with the probands, a home visit to extended family members and an extensive review of medical records if available. Peripheral blood samples were collected from all participants after formal written consent was signed. Eligible HNPCC families were registered and family members were followed up intensively. All patients were reviewed by telephone or by outpatient visit at regular intervals. Data concerning sex, site of CRC, age of diagnosis, history of synchronous and/or metachronous CRC, instance of extracolonic cancers, and histopathology of tumors were documented and thoroughly verified.
Genomic DNA was extracted using the QIAamp DNA isolation kit (Qiagen, Valencia, CA) according to the manufacturer's recommendations. PCR was performed using 100 ng of genomic DNA as template. A 25 μL reaction mixture containing 10-20 pmol of each primer, 1.5 U of Taq DNA polymerase (Transgenomics, UK) with a final concentration of 2 mmol/L Mg2+ and 0.2 mmol/L of dNTPs was used. PCR conditions were as follows: an initial denaturing at 95°C for 5 min, followed by 40 cycles at 95°C for 30 s, at 55-60°C for 30 s, at 72°C for 40 s, and a final extension at 72°C for 10 min. A total of 35 sets of primers including 19 sets for hMLH1 gene and 16 sets for hMSH2 gene were used [5].
DHPLC analysis was carried out on an automated HPLC device equipped with a DNA separation column (WAVE: Transgenomic, San Jose, CA, USA) as previously described[6].
PCR products displaying abnormal DHPLC peak were purified with microconcentrator filters (Amicon, Beverly, MA) and sequenced with a Bioasia-1524-030/3730DNA sequencer. All mutations were sequenced in both directions.
A total of 31 kindreds meeting the Chinese HNPCC criteria or the Amsterdam criteria were studied. One hundred and thirty six malignant neoplasms were found in 107 patients (14 multiple cancers), including 106 CRCs, 14 gastric cancers, 3 esophageal cancers, 2 lung cancers, 2 cervical cancers, 2 leukemia, 1 breast cancer, 1 ovarian cancer, 1 oral cancer, 1 thyroid cancer, 1 hepatic cancer, 1 urinary cancer, 1 malignant histocytosis. CRC accounted for 77.9% (106/136) of the cancers. Nine multiple colorectal tumors and 5 colorectal tumors were associated with extracolonic cancer, accounting for 8.5% and 4.5% of the total CRCs, respectively. Of the 106 CRCs, 18 were located in the right-sided colon, 8 in the transverse colon, 15 in the left-sided colon, 25 in the rectum and 40 in unidentified site. Left-sided colon cancers constituted of 60.6% (40/66) of all CRCs. Individuals suffering from gastric cancer accounted for 10.3% (14/136) of total patients. The average age of malignant neoplasm onset in all the patients was 48.82 ± 13.32 years and the ratio of males to females was 1.7:1. The average age of CRC patients was 48.57 ± 29.00 years. All predispositions in kindreds were followed up.
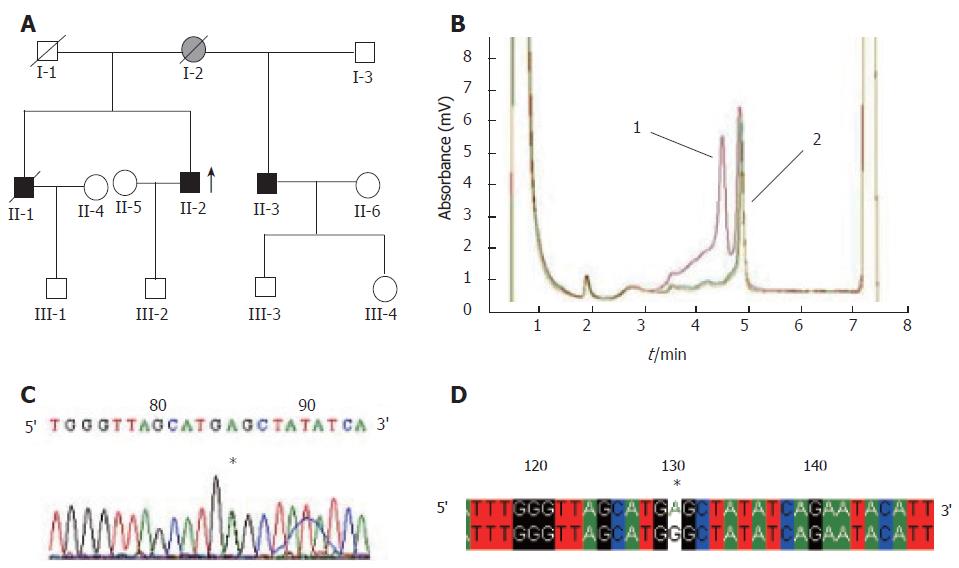
A total of 23 abnormal peak profiles were found in 31 probands from 31 families. Finally, 17 probands were identified with 23 base variations by sequencing analysis, of which 7 base substitutions were located in the hMLH1 gene (6 in the exons, 1 in the intron) while the other 16 base substitutions were located in the hMSH2 gene (9 in exons, 7 in introns). Of the fifteen base substitutions in coding sequences, five were proven to be polymorphisms (SNPs), while the other ten were proven to be mutations, including 3 missense mutations(samples H4, H11 and H12), 2 nonsense mutations(samples H20 and H27), 2 frameshift mutations(samples H24 and H29), and 3 silent mutations(samples H5, H16 and H28). The detailed information is shown in Table 1 and Figure 1.
| Family | Gene/exon | point of mutation | Peptide change | Mutation result | Significance | Reported by |
| H3 | hMLH1/12 | g.1151T > A,GTT-GAT | Val-Asp, V384D | SNP | Wang Y et al[7] | |
| H4 | hMLH1/2 | g.199G > A,GGG-AGG | Gly-Arg, G67R | missense mutation | pathological | Sasaki S et al[8] |
| H5 | hMSH2/1 | g.54C > G,GGC-GGG | Gly-Gly ,G18G | silent mutation | synonymous | |
| H6 | hMSH2/7 | g.1276 + 47T>A | intronic | uncertain | ||
| H7 | hMLH1/8 | g.637G > T,GTG-TTG | Val-Leu, V213L | SNP | ||
| H11 | hMLH1/15 | g.1668-20A > G | intronic | uncertain | ||
| hMSH2/12 | g.1886, A > G,CAA-CGA | Gln-Arg, Q629R | missense mutation | pathological | Kim JC et al[9] | |
| H12 | hMSH2/15 | g.2516, A > G,CAT-CGT | His-Arg, H839R | missense mutation | pathological | |
| H13 | hMSH2/10 | g.1661 + 12G > A | intronic | SNP | Scott RJ et al[10] | |
| H15 | hMSH2/1 | g.211+ 9C > G, | intronic | SNP | ||
| H16 | hMLH1/8 | g.649, C > G,CGC-TGC | Arg-Cys, R217C | SNP | Miyaki M et al[11] | |
| hMSH2/7 | g.1221, C > G,CTC-CTG | Leu-Leu, L407L | silent mutation | synonymous | ||
| hMSH2/13 | g.2006-6T > C | intronic | SNP | |||
| H20 | hMLH1/19 | g.2250, C > G,TAC-TAG | Tyr-X, Y750X | nonsense mutation | truncated peotein | Syngal S et al[12] |
| H21 | hMSH2/1 | g.23C > T, ACG-ATG | Thr-Met, T8M, | SNP | Nomura S et al[13] | |
| hMSH2/1 | g.211 + 9C > G, | intronic | SNP | |||
| H22 | hMSH2/1 | g.23C > G, ACG-ATG | Thr-Met, T8M | SNP | Nomura S et al[13] | |
| hMSH2/1 | g.211 + 9C > G, | intronic | SNP | |||
| H24 | hMSH2/11 | g.1664, delA, | Stopat odon556 | frameshift mutation | truncated protein | |
| hMSH2/11 | g.1662-2A > G | intronic | uncertain | |||
| H27 | hMSH2/11 | g.2292G > A, TGG-TGA | Trp-Stop, W764X | nonsense mutation | truncated protein | |
| H28 | hMSH2/5 | g.795T > C,GTT-GTC | Val-Val, V265V | silent mutation | synonymous | |
| H29 | hMLH1/14 | g.1591delGT | Stop at codon555 | frameshift mutation | truncated protein |
HNPCC is characterized by dominant right colon localization, early age of onset, high prevalence of synchronous and metachronous tumors, and certain extracolonic cancers[14]. In our study, patients with Chinese HNPCC developed CRC at an average age of 48.57 ± 29.00 years. As compared to the onset of sporadic colorectal malignancies at the age approximately 60 years, the onset of HNPCC was much earlier, similar to that in Western countries. In the present study, synchronous and metachronous colorectal cancers only accounted for 8.5% of all CRCs, lower than those in Western countries. The reason is still unclear. Left-sided colorectal cancers accounted for 60.6% (40/66) of all CRCs. This result might be due to the high incidence of left-sided colon cancer (including rectal cancer) in Chinese population. Gastric cancer was the most frequent extracolonic cancer in our 31 families, while endometrial cancer was the most common in Western countries. The reason might be due to the high incidence of gastric cancer in Chinese population. Liu et al[15] have reported similar results found in their cohort of Chinese HNPCC families. These characteristics might truly reflect the specific clinical phenotype of Chinese HNPCC.
In this study, we screened mutations of the hMLH1 and hMSH2 genes in 31 Chinese HNPCC families by DHPLC and PCR as well as DNA sequencing. Twenty-three sequence abnormalities were detected in 17 probands, 15 in exons and 8 in introns. Base variants identified were compared with those described in the human gene mutation database (HGMD) (http://www.uwcm.ac.uk/uwcm/mg/hgmd0.html). Seven of the 15 sequence abnormalities including 3 missense mutations (H4, H11, H12), 2 nonsense mutations (H20, H27), and 2 frameshift mutations (H24, H29) in coding regions of the hMLH1 and hMSH2 genes might probably affect the protein functions. Detailed information of these mutations is listed in Table 1. Among them, mutations of H4, H11 and H20 have been reported before[8,9,12], while the other four mutations of H12, H27, H24 and H29 are novel. The mutation rate was 22.6% (7/31). Nearly half of the mutations were missense mutations. Three (H5, H16, H28) of the fifteen sequence abnormalities in coding sequences are novel silent mutations in exons 1, 7, 5 of the hMSH2 gene, respectively. The last five (H3, H7, H16, H21 and H22) of the fifteen sequence abnormalities in coding sequences are known as SNPs[7,10,11,13].
The missense mutation detected in sample H4 located at codon 67 in exon 2 of the hMLH1 gene, has been reported as a pathological mutation[8]. The missense mutation detected in sample H11 located at codon 629 in exon 12 of the hMSH2 gene has also been reported[9]. In sample H12, a novel missense mutation at codon 839 in exon 15 of the hMSH2 gene was first discovered in our study, which is located in the Muts V structural motif (peptides 619-854) of the hMSH2 gene and leads to amino acid change from histidine to arginine. In order to determine the significance of this mutation, 100 blood samples from cancer-free donors were tested under the same conditions and none of the 100 normal samples harbored the same base change as sample H12, suggesting that this base change is pathologic.
The nonsense mutation detected in sample H20 altered nucleotide 2250 (genomic DNA) from C to G at codon 750 in exon 19 of the hMLH1 gene, resulting in a stop codon instead of tyrosine, forming a truncated protein of MutL losing 6 peptides from 751 residue to 756 residue. This nonsense mutation has been reported by Syngal et al [12]. The other nonsense mutation in sample H27 altered nucleotide 2292 (genomic DNA) from G to A at codon 764 in exon 11 of the hMSH2 gene, leading to the substitution of a stop codon TGA for tryptophan condon TGG, forming a truncated protein of MutC structural motif (peptides 619-854) losing 170 peptides from 765 residue to 934 residue, suggesting that these nonsense mutations may have a causative effect on cancer predisposition and further studies are needed to examine their co-segregation with the phenotype.
Two framshift mutations detected in samples H24 and H29 have not been reported previously. In sample H24, the base A deleted at codon 555 in exon 11 of the hMSH2 gene led to a stop codon at codon 556. While in sample H29, the GT deleted at codon 531 in exon 14 of the hMLH1 gene led to a stop codon at codon 555. Both mutations produced truncated proteins with insufficient function.
Of the 23 base variants, 8 were located in intronic regions. Five of these 8 variants have been previously reported and known as SNPs (http://www.ncbi.nlm.nih.gov/), the significance of the other 3 remains uncertain.
DHPLC is an elegant, new method for mutation detection. It detects heteroduplex formation in renaturated PCR fragments possessing heterozygous sequence variations, and has been applied successfully to mutation screening of various genes[3]. In the past few years, we have searched successfully for mutations in hMLH1 and hMSH2 genes in 29 HNPCC families and 34 suspected HNPCC families with PCR-SSCP and DNA sequencing methods[16] . However, SSCP is inefficient in detecting mutations and misses 20% of point and frameshift mutations. Though DNA sequencing is supposed to be a screening method with a high sensitivity, it is time consuming and expensive. It was reported that the sensitivity of DHPLC for screening mutations of the hMLH1 and hMSH2 genes can reach 97% without false negatives[6]. Furthermore, the system is highly automatic, the running time per sample averages only 7 min. Because of these advantages of DHPLC, it enables a broad screening of HNPCC families for human mismatch repair gene alterations in routine analysis.
S- Editor Wang J L- Editor Wang XL E- Editor Liu Y
| 1. | Lynch HT, Smyrk T. Hereditary nonpolyposis colorectal cancer (Lynch syndrome). An updated review. Cancer. 1996;78:1149-1167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 5] [Reference Citation Analysis (0)] |
| 2. | Yuan Y, Huang YQ, Cai SR, Song YM, Zheng S, Zhang SZ. Genetic characterization of Chinese hereditary non-polyposis colorectal cancer by DHPLC and multiplex PCR. Jpn J Clin Oncol. 2004;34:660-666. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 3. | Kurzawski G, Safranow K, Suchy J, Chlubek D, Scott RJ, Lubiński J. Mutation analysis of MLH1 and MSH2 genes performed by denaturing high-performance liquid chromatography. J Biochem Biophys Methods. 2002;51:89-100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 37] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 4. | Xiao W, Oefner PJ. Denaturing high-performance liquid chromatography: A review. Hum Mutat. 2001;17:439-474. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 510] [Cited by in F6Publishing: 536] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 5. | Weber TK, Conlon W, Petrelli NJ, Rodriguez-Bigas M, Keitz B, Pazik J, Farrell C, O'Malley L, Oshalim M, Abdo M. Genomic DNA-based hMSH2 and hMLH1 mutation screening in 32 Eastern United States hereditary nonpolyposis colorectal cancer pedigrees. Cancer Res. 1997;57:3798-3803. [PubMed] [Cited in This Article: ] |
| 6. | Holinski-Feder E, Müller-Koch Y, Friedl W, Moeslein G, Keller G, Plaschke J, Ballhausen W, Gross M, Baldwin-Jedele K, Jungck M. DHPLC mutation analysis of the hereditary nonpolyposis colon cancer (HNPCC) genes hMLH1 and hMSH2. J Biochem Biophys Methods. 2001;47:21-32. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 71] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 7. | Wang Y, Friedl W, Lamberti C, Nöthen MM, Kruse R, Propping P. A novel missense mutation in the DNA mismatch repair gene hMLH1 present among East Asians but not among Europeans. Hum Hered. 1998;48:87-91. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 20] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 8. | Sasaki S, Tokino T, Miyatsu T, Muto T, Nakamura Y. Mutational analysis of the hMLH1 gene using an automated two-dimensional DNA typing system. Hum Mutat. 1997;9:164-171. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 9. | Kim JC, Kim HC, Roh SA, Koo KH, Lee DH, Yu CS, Lee JH, Kim TW, Lee HL, Beck NE. hMLH1 and hMSH2 mutations in families with familial clustering of gastric cancer and hereditary non-polyposis colorectal cancer. Cancer Detect Prev. 2001;25:503-510. [PubMed] [Cited in This Article: ] |
| 10. | Scott RJ, McPhillips M, Meldrum CJ, Fitzgerald PE, Adams K, Spigelman AD, du Sart D, Tucker K, Kirk J. Hereditary nonpolyposis colorectal cancer in 95 families: differences and similarities between mutation-positive and mutation-negative kindreds. Am J Hum Genet. 2001;68:118-127. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 135] [Cited by in F6Publishing: 151] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 11. | Miyaki M, Konishi M, Muraoka M, Kikuchi-Yanoshita R, Tanaka K, Iwama T, Mori T, Koike M, Ushio K, Chiba M. Germ line mutations of hMSH2 and hMLH1 genes in Japanese families with hereditary nonpolyposis colorectal cancer (HNPCC): usefulness of DNA analysis for screening and diagnosis of HNPCC patients. J Mol Med (Berl). 1995;73:515-520. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 36] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 12. | Syngal S, Fox EA, Li C, Dovidio M, Eng C, Kolodner RD, Garber JE. Interpretation of genetic test results for hereditary nonpolyposis colorectal cancer: implications for clinical predisposition testing. JAMA. 1999;282:247-253. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 92] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 13. | Nomura S, Sugano K, Kashiwabara H, Taniguchi T, Fukayama N, Fujita S, Akasu T, Moriya Y, Ohhigashi S, Kakizoe T. Enhanced detection of deleterious and other germline mutations of hMSH2 and hMLH1 in Japanese hereditary nonpolyposis colorectal cancer kindreds. Biochem Biophys Res Commun. 2000;271:120-129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 37] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 14. | Velayos FS, Lee SH, Qiu H, Dykes S, Yiu R, Terdiman JP, Garcia-Aguilar J. The mechanism of microsatellite instability is different in synchronous and metachronous colorectal cancer. J Gastrointest Surg. 2005;9:329-335. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 15. | Liu SR, Zhao B, Wang ZJ, Wan YL, Huang YT. Clinical features and mismatch repair gene mutation screening in Chinese patients with hereditary nonpolyposis colorectal carcinoma. World J Gastroenterol. 2004;10:2647-2651. [PubMed] [Cited in This Article: ] |
| 16. | Yuan Y, Zheng S. [Mutations of hMLH1 and hMSH2 genes in suspected hereditary nonpolyposis colorectal cancer]. Zhonghua Yi Xue Za Zhi. 1999;79:346-348. [PubMed] [Cited in This Article: ] |