A Fluorescent Cell-Based System for Imaging Zika Virus Infection in Real-Time
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Plasmids and Transfections
2.3. Generation of Stable ZIKV-NLS-GFP Reporter Cell Lines
2.4. ZIKV and DENV-2 Infection
2.5. Microscopy
2.6. Live Cell Imaging and Analysis
2.7. Immunoblotting
2.8. Nuclear:Total GFP Quantification
2.9. Percent of ZIKV+ Cells Quantification
3. Results
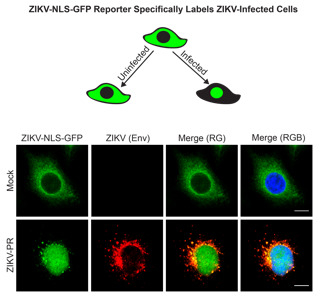
3.1. A Cleavable GFP Reporter to Identify ZIKV-Infected Cells
3.2. The Cleavable ZIKV-NLS-GFP Reporter Can Detect ZIKV Infection
3.3. Two Divergent Strains of ZIKV Can Both Cleave the ZIKV-NLS-GFP Reporter
3.4. Live Cell Imaging Determines Kinetics of Nuclear Localization of GFP Following ZIKV Infection
3.5. ZIKV Induces Cell Death Specifically in Infected Cells
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lazear, H.M.; Diamond, M.S. Zika virus: New clinical syndromes and its emergence in the western hemisphere. J. Virol. 2016, 90, 4864–4875. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.R.; Chen, T.H.; Hancock, W.T.; Powers, A.M.; Kool, J.L.; Lanciotti, R.S.; Pretrick, M.; Marfel, M.; Holzbauer, S.; Dubray, C.; et al. Zika virus outbreak on yap island, federated states of micronesia. N. Engl. J. Med. 2009, 360, 2536–2543. [Google Scholar] [CrossRef] [PubMed]
- Cao-Lormeau, V.M.; Blake, A.; Mons, S.; Lastere, S.; Roche, C.; Vanhomwegen, J.; Dub, T.; Baudouin, L.; Teissier, A.; Larre, P.; et al. Guillain-barre syndrome outbreak associated with Zika virus infection in French polynesia: A case-control study. Lancet 2016, 387, 1531–1539. [Google Scholar] [CrossRef]
- Parra, B.; Lizarazo, J.; Jimenez-Arango, J.A.; Zea-Vera, A.F.; Gonzalez-Manrique, G.; Vargas, J.; Angarita, J.A.; Zuniga, G.; Lopez-Gonzalez, R.; Beltran, C.L.; et al. Guillain-barre syndrome associated with Zika virus infection in Colombia. N. Engl. J. Med. 2016, 375, 1513–1523. [Google Scholar] [CrossRef] [PubMed]
- Adebanjo, T.; Godfred-Cato, S.; Viens, L.; Fischer, M.; Staples, J.E.; Kuhnert-Tallman, W.; Walke, H.; Oduyebo, T.; Polen, K.; Peacock, G.; et al. Update: Interim guidance for the diagnosis, evaluation, and management of infants with possible congenital Zika virus infection—United States, October 2017. MMWR Morb. Mortal. Wkly. Rep. 2017, 66, 1089–1099. [Google Scholar] [CrossRef] [PubMed]
- ECDC. Rapid Risk Assessment. Zika Virus Epidemic in the Americas: Potential Association with Microcephaly and Guillain-Barre Syndrome. Available online: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/zika-virus-americas-association-with-microcephaly-rapid-risk-assessment.pdf (accessed on 15 October 2017).
- Wu, K.Y.; Zuo, G.L.; Li, X.F.; Ye, Q.; Deng, Y.Q.; Huang, X.Y.; Cao, W.C.; Qin, C.F.; Luo, Z.G. Vertical transmission of Zika virus targeting the radial glial cells affects cortex development of offspring mice. Cell Res. 2016, 26, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Miner, J.J.; Cao, B.; Govero, J.; Smith, A.M.; Fernandez, E.; Cabrera, O.H.; Garber, C.; Noll, M.; Klein, R.S.; Noguchi, K.K.; et al. Zika virus infection during pregnancy in mice causes placental damage and fetal demise. Cell 2016, 165, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Cugola, F.R.; Fernandes, I.R.; Russo, F.B.; Freitas, B.C.; Dias, J.L.; Guimaraes, K.P.; Benazzato, C.; Almeida, N.; Pignatari, G.C.; Romero, S.; et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature 2016, 534, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xu, D.; Ye, Q.; Hong, S.; Jiang, Y.; Liu, X.; Zhang, N.; Shi, L.; Qin, C.F.; Xu, Z. Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell 2016, 19, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Huang, X.Y.; Liu, Z.Y.; Zhang, F.; Zhu, X.L.; Yu, J.Y.; Ji, X.; Xu, Y.P.; Li, G.; Li, C.; et al. A single mutation in the PRM protein of Zika virus contributes to fetal microcephaly. Science 2017, 358, 933–936. [Google Scholar] [CrossRef] [PubMed]
- Adams Waldorf, K.M.; Stencel-Baerenwald, J.E.; Kapur, R.P.; Studholme, C.; Boldenow, E.; Vornhagen, J.; Baldessari, A.; Dighe, M.K.; Thiel, J.; Merillat, S.; et al. Fetal brain lesions after subcutaneous inoculation of zika virus in a pregnant nonhuman primate. Nat. Med. 2016, 22, 1256–1259. [Google Scholar] [CrossRef] [PubMed]
- Lazear, H.M.; Govero, J.; Smith, A.M.; Platt, D.J.; Fernandez, E.; Miner, J.J.; Diamond, M.S. A mouse model of Zika virus pathogenesis. Cell Host Microbe. 2016, 19, 720–730. [Google Scholar] [CrossRef] [PubMed]
- Lossia, O.V.; Conway, M.J.; Tree, M.O.; Williams, R.J.; Goldthorpe, S.C.; Srinageshwar, B.; Dunbar, G.L.; Rossignol, J. Zika virus induces astrocyte differentiation in neural stem cells. J. Neurovirol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Garcez, P.P.; Loiola, E.C.; Madeiro da Costa, R.; Higa, L.M.; Trindade, P.; Delvecchio, R.; Nascimento, J.M.; Brindeiro, R.; Tanuri, A.; Rehen, S.K. Zika virus impairs growth in human neurospheres and brain organoids. Science 2016, 352, 816–818. [Google Scholar] [CrossRef] [PubMed]
- Ghouzzi, V.E.; Bianchi, F.T.; Molineris, I.; Mounce, B.C.; Berto, G.E.; Rak, M.; Lebon, S.; Aubry, L.; Tocco, C.; Gai, M.; et al. Zika virus elicits p53 activation and genotoxic stress in human neural progenitors similar to mutations involved in severe forms of genetic microcephaly and p53. Cell Death Dis. 2016, 7, e2440. [Google Scholar] [CrossRef] [PubMed]
- Souza, B.S.; Sampaio, G.L.; Pereira, C.S.; Campos, G.S.; Sardi, S.I.; Freitas, L.A.; Figueira, C.P.; Paredes, B.D.; Nonaka, C.K.; Azevedo, C.M.; et al. Zika virus infection induces mitosis abnormalities and apoptotic cell death of human neural progenitor cells. Sci. Rep. 2016, 6, 39775. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Hammack, C.; Ogden, S.C.; Wen, Z.; Qian, X.; Li, Y.; Yao, B.; Shin, J.; Zhang, F.; Lee, E.M.; et al. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell 2016, 18, 587–590. [Google Scholar] [CrossRef] [PubMed]
- Dang, J.; Tiwari, S.K.; Lichinchi, G.; Qin, Y.; Patil, V.S.; Eroshkin, A.M.; Rana, T.M. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell 2016, 19, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W.; Dorner, M.; Feulner, M.; Imanaka, N.; Murphy, M.Y.; Ploss, A.; Rice, C.M. Dengue reporter viruses reveal viral dynamics in interferon receptor-deficient mice and sensitivity to interferon effectors in vitro. Proc. Natl. Acad. Sci. USA 2012, 109, 14610–14615. [Google Scholar] [CrossRef] [PubMed]
- Shan, C.; Xie, X.; Muruato, A.E.; Rossi, S.L.; Roundy, C.M.; Azar, S.R.; Yang, Y.; Tesh, R.B.; Bourne, N.; Barrett, A.D.; et al. An infectious cDNA clone of Zika virus to study viral virulence, mosquito transmission, and antiviral inhibitors. Cell Host Microbe. 2016, 19, 891–900. [Google Scholar] [CrossRef] [PubMed]
- Gadea, G.; Bos, S.; Krejbich-Trotot, P.; Clain, E.; Viranaicken, W.; El-Kalamouni, C.; Mavingui, P.; Despres, P. A robust method for the rapid generation of recombinant Zika virus expressing the GFP reporter gene. Virology 2016, 497, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Tamura, T.; Fukuhara, T.; Uchida, T.; Ono, C.; Mori, H.; Sato, A.; Fauzyah, Y.; Okamoto, T.; Kurosu, T.; Setoh, Y.X.; et al. Characterization of recombinant flaviviridae viruses possessing a small reporter-tag. J. Virol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Blight, K.J.; McKeating, J.A.; Rice, C.M. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 2002, 76, 13001–13014. [Google Scholar] [CrossRef] [PubMed]
- Giard, D.J.; Aaronson, S.A.; Todaro, G.J.; Arnstein, P.; Kersey, J.H.; Dosik, H.; Parks, W.P. In vitro cultivation of human tumors: Establishment of cell lines derived from a series of solid tumors. J. Natl. Cancer Inst. 1973, 51, 1417–1423. [Google Scholar] [CrossRef] [PubMed]
- Medin, C.L.; Valois, S.; Patkar, C.G.; Rothman, A.L. A plasmid-based reporter system for live cell imaging of dengue virus infected cells. J. Virol. Methods 2015, 211, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Zennou, V.; Serguera, C.; Sarkis, C.; Colin, P.; Perret, E.; Mallet, J.; Charneau, P. The HIV-1 DNA flap stimulates HIV vector-mediated cell transduction in the brain. Nat. Biotechnol. 2001, 19, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Hirai, R.; Loo, Y.M.; Owen, D.; Johnson, C.L.; Sinha, S.C.; Akira, S.; Fujita, T.; Gale, M., Jr. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. USA 2007, 104, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Quicke, K.M.; Bowen, J.R.; Johnson, E.L.; McDonald, C.E.; Ma, H.; O'Neal, J.T.; Rajakumar, A.; Wrammert, J.; Rimawi, B.H.; Pulendran, B.; et al. Zika virus infects human placental macrophages. Cell Host. Microbe 2016, 20, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Le Sommer, C.; Barrows, N.J.; Bradrick, S.S.; Pearson, J.L.; Garcia-Blanco, M.A. G protein-coupled receptor kinase 2 promotes flaviviridae entry and replication. PLoS Negl. Trop. Dis. 2012, 6, e1820. [Google Scholar] [CrossRef] [PubMed]
- Jagannathan, S.; Nwosu, C.; Nicchitta, C.V. Analyzing mRNA localization to the endoplasmic reticulum via cell fractionation. Methods Mol. Biol. 2011, 714, 301–321. [Google Scholar] [CrossRef] [PubMed]
- Lerner, R.S.; Seiser, R.M.; Zheng, T.; Lager, P.J.; Reedy, M.C.; Keene, J.D.; Nicchitta, C.V. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA 2003, 9, 1123–1137. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.T.; Catanese, M.T.; Law, L.M.; Khetani, S.R.; Syder, A.J.; Ploss, A.; Oh, T.S.; Schoggins, J.W.; MacDonald, M.R.; Bhatia, S.N.; et al. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat. Biotechnol. 2010, 28, 167–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chambers, T.J.; Hahn, C.S.; Galler, R.; Rice, C.M. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 1990, 44, 649–688. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Vasudevan, S.G.; Lescar, J. The flavivirus NS2B-NS3 protease-helicase as a target for antiviral drug development. Antiviral Res. 2015, 118, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, E.; Prehn, S. The N-terminal region of the α-subunit of the trap complex has a conserved cluster of negative charges. FEBS Lett. 1994, 349, 324–326. [Google Scholar] [CrossRef]
- Frumence, E.; Roche, M.; Krejbich-Trotot, P.; El-Kalamouni, C.; Nativel, B.; Rondeau, P.; Misse, D.; Gadea, G.; Viranaicken, W.; Despres, P. The south pacific epidemic strain of Zika virus replicates efficiently in human epithelial A549 cells leading to IFN-β production and apoptosis induction. Virology 2016, 493, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.C.; Abraham, R.; Shim, B.S.; Choe, H.; Page, D.T. Zika virus infection during the period of maximal brain growth causes microcephaly and corticospinal neuron apoptosis in wild type mice. Sci. Rep. 2016, 6, 34793. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Nguyen, H.N.; Song, M.M.; Hadiono, C.; Ogden, S.C.; Hammack, C.; Yao, B.; Hamersky, G.R.; Jacob, F.; Zhong, C.; et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 2016, 165, 1238–1254. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.Y.; Ames, H.M.; Tipton, A.; Vezina, G.; Liu, J.S.; Scafidi, J.; Torii, M.; Rodriguez, F.J.; du Plessis, A.; DeBiasi, R.L. Differential neuronal susceptibility and apoptosis in congenital Zika virus infection. Ann. Neurol. 2017, 82, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Simonin, Y.; Loustalot, F.; Desmetz, C.; Foulongne, V.; Constant, O.; Fournier-Wirth, C.; Leon, F.; Moles, J.P.; Goubaud, A.; Lemaitre, J.M.; et al. Zika virus strains potentially display different infectious profiles in human neural cells. EBioMedicine 2016, 12, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Retallack, H.; Di Lullo, E.; Arias, C.; Knopp, K.A.; Laurie, M.T.; Sandoval-Espinosa, C.; Mancia Leon, W.R.; Krencik, R.; Ullian, E.M.; Spatazza, J.; et al. Zika virus cell tropism in the developing human brain and inhibition by azithromycin. Proc. Natl. Acad. Sci. USA 2016, 113, 14408–14413. [Google Scholar] [CrossRef] [PubMed]
- Onorati, M.; Li, Z.; Liu, F.; Sousa, A.M.M.; Nakagawa, N.; Li, M.; Dell'Anno, M.T.; Gulden, F.O.; Pochareddy, S.; Tebbenkamp, A.T.N.; et al. Zika virus disrupts phospho-TBK1 localization and mitosis in human neuroepithelial stem cells and radial glia. Cell Rep. 2016, 16, 2576–2592. [Google Scholar] [CrossRef] [PubMed]
- Brault, J.B.; Khou, C.; Basset, J.; Coquand, L.; Fraisier, V.; Frenkiel, M.P.; Goud, B.; Manuguerra, J.C.; Pardigon, N.; Baffet, A.D. Comparative analysis between flaviviruses reveals specific neural stem cell tropism for zika virus in the mouse developing neocortex. EBioMedicine 2016, 10, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Humpel, C. Organotypic brain slice cultures: A review. Neuroscience 2015, 305, 86–98. [Google Scholar] [CrossRef] [PubMed]






| Primer Name | Primer Sequence |
|---|---|
| Primer 1 (734) | 5′-TTAAGCTTGCCACCATGGCGAATGAACTCGGATGGTTGGA-3′ |
| Primer 2 (735) | 5′-CATGGTGGCGAATTCCTCTCCCAGGGTCTCTCCTGTTC-3′ |
| Primer 3 (740) | 5′-TCTAGAGGATCCGGAAAGCTTGCCACCATGGCGAATGAA-3′ |
| Primer 4 (741) | 5′-TTTCTAGGTCTCGAGTTACTTGTACAGCTCGTCCATGCCG-3′ |
| Primer 5 (864) | 5′-ACGCTGGCTTGGTCAAGGCAGCTGGGGGTGGAACAGGAG-3′ |
| Primer 6 (865) | 5′-CTCCTGTTCCACCCCCAGCTGCCTTGACCAAGCCAGCGT-3′ |
| Primer 7 (880) | 5′-ATAAAGCGGCCGCTAGCTGGCCCCCTAGCGAAGT-3′ |
| Primer 8 (881) | 5′-AGCGGGTTTAAACTTATCTTTTCCCAGCGGCAAACT-3′ |
| Primer 9 (886) | 5′-ATTACCCAGCAGGAACTGCAGGATCTCCAATCCTA-3′ |
| Primer 10 (887) | 5′-TAGGATTGGAGATCCTGCAGTTCCTGCTGGGTAAT-3′ |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
McFadden, M.J.; Mitchell-Dick, A.; Vazquez, C.; Roder, A.E.; Labagnara, K.F.; McMahon, J.J.; Silver, D.L.; Horner, S.M. A Fluorescent Cell-Based System for Imaging Zika Virus Infection in Real-Time. Viruses 2018, 10, 95. https://doi.org/10.3390/v10020095
McFadden MJ, Mitchell-Dick A, Vazquez C, Roder AE, Labagnara KF, McMahon JJ, Silver DL, Horner SM. A Fluorescent Cell-Based System for Imaging Zika Virus Infection in Real-Time. Viruses. 2018; 10(2):95. https://doi.org/10.3390/v10020095
Chicago/Turabian StyleMcFadden, Michael J., Aaron Mitchell-Dick, Christine Vazquez, Allison E. Roder, Kevin F. Labagnara, John J. McMahon, Debra L. Silver, and Stacy M. Horner. 2018. "A Fluorescent Cell-Based System for Imaging Zika Virus Infection in Real-Time" Viruses 10, no. 2: 95. https://doi.org/10.3390/v10020095