Spatial Potential Energy Weighted Maximum Simplex Algorithm for Hyperspectral Endmember Extraction
Abstract
:1. Introduction
2. Relative Research Works
2.1. LMM
2.2. Space Energy
2.3. SENMAV
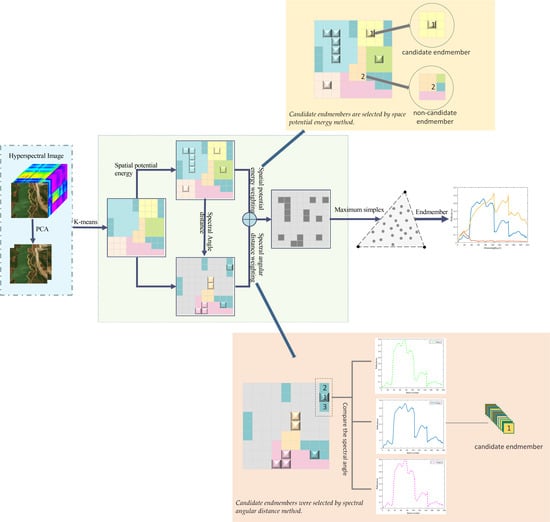
3. Spatial Potential Energy Weighted Maximum Simplex Algorithm
3.1. Space Potential Energy Weighting
3.2. Spectral Distance Weighting
3.3. SPEW Algorithm
4. Experimental Results and Analysis
4.1. Evaluation Criteria
4.2. Experimental Dataset
4.2.1. Synthetic Dataset
4.2.2. Hyperspectral Digital Imagery Collection Experiment (Hydice) Dataset
4.2.3. Samson Dataset
4.2.4. Jasper Ridge Dataset
4.2.5. Urban Dataset
4.2.6. Cuprite Dataset
4.3. Experimental Results
4.3.1. Performance of Locating Endmembers
4.3.2. Performance of Unmixing
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Bioucas-Dias, J.M.; Plaza, A.; Camps-Valls, G.; Scheunders, P.; Nasrabadi, N.M.; Chanussot, J. Hyperspectral remote sensing data analysis and future challenges. IEEE Geosci. Remote Sens. Mag. 2013, 1, 6–36. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Li, J.; Li, S.; Plaza, A. Generalized Morphological Component Analysis for Hyperspectral Unmixing. IEEE Trans. Geosci. Remote Sens. 2020, 58, 2817–2832. [Google Scholar] [CrossRef]
- Brown, A.J.; Hook, S.J.; Baldridge, A.M.; Crowley, J.K.; Bridges, N.T.; Thomson, B.J.; Marion, G.M.; Filho, C.R.D.S.; Bishop, J.L. Hydrothermal formation of Clay-Carbonate alteration assemblages in the Nili Fossae region of Mars. Earth Planet. Sci. Lett. 2010, 297, 174–182. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.I.; Wen, C.H.; Wu, C.C. Relationship exploration among PPI, ATGP and VCA via theoretical analysis. Int. J. Comput. Sci. Eng. 2013, 8, 361–367. [Google Scholar] [CrossRef]
- Ren, H.; Chang, C.-I. Automatic spectral target recognition in hyperspectral imagery. IEEE Trans. Aerosp. Electron. Syst. 2003, 39, 1232–1249. [Google Scholar] [CrossRef] [Green Version]
- Nascimento, J.M.; Dias, J.M. Vertex component analysis: A fast algorithm to unmix hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2005, 43, 898–910. [Google Scholar] [CrossRef] [Green Version]
- Winter, M.E. N-finder: An algorithm for fast autonomous spectral endmember determination in hyperspectral data. In Proceedings of the SPIE′s International Symposium on Optical Science, Engineering, and Instrumentation, Denver, CO, USA, 18–23 July 1999; International Society for Optics and Photonics: Bellingham, WA, USA, 1999; Volume 3753, pp. 266–276. [Google Scholar]
- Xiong, W.; Chang, C.-I.; Wu, C.-C.; Kalpakis, K.; Chen, H.M. Fast Algorithms to Implement N-FINDR for Hyperspectral Endmember Extraction. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2011, 4, 545–564. [Google Scholar] [CrossRef]
- Chang, C.-I.; Wu, C.-C.; Liu, W.-M.; Ouyang, Y.-C. A New Growing Method for Simplex-Based Endmember Extraction Algorithm. IEEE Trans. Geosci. Remote Sens. 2006, 44, 2804–2819. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, H.; Guo, X.; Xu, G. Endmember Extraction Using Minimum Volume and Information Constraint Nonnegative Matrix Factorization. IEEE Geosci. Remote Sens. Lett. 2019, 16, 1427–1431. [Google Scholar] [CrossRef]
- Miao, L.; Qi, H. Endmember Extraction From Highly Mixed Data Using Minimum Volume Constrained Nonnegative Matrix Factorization. IEEE Trans. Geosci. Remote Sens. 2007, 45, 765–777. [Google Scholar] [CrossRef]
- Qian, Y.; Jia, S.; Zhou, J.; Robles-Kelly, A. Hyperspectral unmixing via L1/2 sparsity-constrained nonnegative matrix fac-torization. IEEE Trans. Geosci. Remote Sens. 2011, 49, 4282–4297. [Google Scholar] [CrossRef] [Green Version]
- Parente, M.; Clark, J.T.; Brown, A.J.; Bishop, J. End-to-End Simulation and Analytical Model of Remote-Sensing Systems: Application to CRISM. IEEE Trans. Geosci. Remote Sens. 2010, 48, 3877–3888. [Google Scholar] [CrossRef]
- Mei, S.; He, M.; Wang, Z.; Feng, D. Spatial Purity Based Endmember Extraction for Spectral Mixture Analysis. IEEE Trans. Geosci. Remote Sens. 2010, 48, 3434–3445. [Google Scholar] [CrossRef]
- Li, H.; Zhang, L. A Hybrid Automatic Endmember Extraction Algorithm Based on a Local Window. IEEE Trans. Geosci. Remote Sens. 2011, 49, 4223–4238. [Google Scholar] [CrossRef]
- Xu, M.; Du, B.; Zhang, L. Spatial-Spectral Information Based Abundance-Constrained Endmember Extraction Methods. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2004–2015. [Google Scholar] [CrossRef]
- Madronero, M.T.; Velez-Reyes, M. Integrating Spatial Information in Unsupervised Unmixing of Hyperspectral Imagery Using Multiscale Representation. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 1985–1993. [Google Scholar] [CrossRef]
- Xu, M.; Zhang, L.; Du, B. An image-based endmember bundle extraction algorithm using both spatial and spectral in-formation. IEEE J. Sel. Topics Appl. Earth Observ. Remote Sens. 2015, 8, 2607–2617. [Google Scholar] [CrossRef]
- Wang, L.; Shi, C.; Diao, C.; Ji, W.; Yin, D. A survey of methods incorporating spatial information in image classification and spectral unmixing. Int. J. Remote Sens. 2016, 37, 3870–3910. [Google Scholar] [CrossRef]
- Shen, X.; Bao, W.; Qu, K. Clustering based spatial spectral preprocessing for hyperspectral unmxing. In Proceedings of the 4th International Conference on Communication and Information Processing—ICCIP’ 18, Qingdao, China, 2–4 November 2018; Association for Computing Machinery (ACM): New York, NY, USA; pp. 313–316. [Google Scholar]
- Shen, X.; Bao, W. A Spatial Energy And Spectral Purity Based Preprocessing Algorithm For Fast Hyperspectral Endmember Extraction. In Proceedings of the 2019 10th Workshop on Hyperspectral Imaging and Signal Processing: Evolution in Remote Sensing (WHISPERS), Amsterdam, The Netherlands, 24–26 September 2019; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA; pp. 1–5. [Google Scholar]
- Shen, X.; Bao, W. Hyperspectral Endmember Extraction Using Spatially Weighted Simplex Strategy. Remote Sens. 2019, 11, 2147. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Hua, W.; Liu, X.; Cui, Z.; Diao, D. Spatial–spectral preprocessing for spectral unmixing. Int. J. Remote Sens. 2018, 40, 1357–1373. [Google Scholar] [CrossRef]
- Plaza, A.; Martinez, P.; Perez, R.; Plaza, J. Spatial/spectral endmember extraction by multidimensional morphological operations. IEEE Trans. Geosci. Remote Sens. 2002, 40, 2025–2041. [Google Scholar] [CrossRef] [Green Version]
- Zortea, M.; Plaza, A. Spatial Preprocessing for Endmember Extraction. IEEE Trans. Geosci. Remote Sens. 2009, 47, 2679–2693. [Google Scholar] [CrossRef]
- Martín, G.; Plaza, A. Region-Based Spatial Preprocessing for Endmember Extraction and Spectral Unmixing. IEEE Geosci. Remote Sens. Lett. 2011, 8, 745–749. [Google Scholar] [CrossRef] [Green Version]
- Martín, G.; Plaza, A. Spatial-Spectral Preprocessing Prior to Endmember Identification and Unmixing of Remotely Sensed Hyperspectral Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 5, 380–395. [Google Scholar] [CrossRef]
- Mei, S.; Zhang, G.; Li, J.; Zhang, Y.; Du, Q. Improving Spectral-Based Endmember Finding by Exploring Spatial Context for Hyperspectral Unmixing. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 3336–3349. [Google Scholar] [CrossRef]
- Shah, D.; Zaveri, T.; Trivedi, Y.N.; Plaza, A. Entropy-Based Convex Set Optimization for Spatial–Spectral Endmember Extraction From Hyperspectral Images. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 4200–4213. [Google Scholar] [CrossRef]
- Shen, X.; Bao, W.; Qu, K. Spatial-Spectral Hyperspectral Endmember Extraction Using a Spatial Energy Prior Constrained Maximum Simplex Volume Approach. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 1347–1361. [Google Scholar] [CrossRef]
- Tarabalka, Y.; Fauvel, M.; Chanussot, J.; Benediktsson, J.A. SVM- and MRF-Based Method for Accurate Classification of Hyperspectral Images. IEEE Geosci. Remote Sens. Lett. 2010, 7, 736–740. [Google Scholar] [CrossRef] [Green Version]
- Khodadadzadeh, M.; Li, J.; Plaza, A.; Ghassemian, H.; Bioucas-Dias, J.M.; Li, X. pectral-spatial classification of hyper-spectral data using local and global probabilities for mixed pixel characterization. IEEE Trans. Geosci. Remote Sens. 2014, 52, 6298–6314. [Google Scholar] [CrossRef]
- Xia, J.; Chanussot, J.; Du, P.; He, X. Spectral–Spatial Classification for Hyperspectral Data Using Rotation Forests With Local Feature Extraction and Markov Random Fields. IEEE Trans. Geosci. Remote Sens. 2014, 53, 2532–2546. [Google Scholar] [CrossRef]
- Eches, O.; Dobigeon, N.; Tourneret, J.-Y. Enhancing Hyperspectral Image Unmixing With Spatial Correlations. IEEE Trans. Geosci. Remote Sens. 2011, 49, 4239–4247. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Prasad, S.; Fowler, J.E. Hyperspectral Image Classification Using Gaussian Mixture Models and Markov Random Fields. IEEE Geosci. Remote Sens. Lett. 2014, 11, 153–157. [Google Scholar] [CrossRef] [Green Version]
- Du, Q.; Raksuntorn, N.; Younan, N.H.; King, R.L. End-member extraction for hyperspectral image analysis. Appl. Opt. 2008, 47, F77–F84. [Google Scholar] [CrossRef] [PubMed]
- Hartigan, J.A.; Wong, M.A. Algorithm AS 136: A K-Means Clustering Algorithm. J. R. Stat. Soc. Ser. (Appl. Stat.) 1979, 28, 100–108. [Google Scholar] [CrossRef]
- Chang, C.-I.; Chen, S.-Y.; Li, H.-C.; Chen, H.-M.; Wen, C.-H. Comparative Study and Analysis Among ATGP, VCA, and SGA for Finding Endmembers in Hyperspectral Imagery. IEEE J. Sel. Topics Appl. Earth Observ. Remote Sen. 2016, 9, 4280–4306. [Google Scholar] [CrossRef]










| Algorithms | VCA | ATGP | SGA | ECSO | SENMAV | SPEW |
|---|---|---|---|---|---|---|
| Soil | 0.0399 | 0.0219 | 0.0407 | 0.0404 | 0.0404 | 0.1224 |
| Tree | 0.0236 | 0.0404 | 0.0404 | 0.0713 | 0.0407 | 0.0407 |
| Water | 0.1504 | 1.0948 | 1.1483 | 0.1553 | 0.1296 | 0.0404 |
| Mean | 0.0713 | 0.3857 | 0.4098 | 0.0890 | 0.0702 | 0.0678 |
| RMSE | 0.3253 | 0.3359 | 0.3689 | 0.2733 | 0.3237 | 0.3233 |
| RE | 0.0014 | 0.2421 | 0.2429 | 0.0057 | 0.0028 | 0.0020 |
| Algorithms | VCA | ATGP | SGA | ECSO | SENMAV | SPEW |
|---|---|---|---|---|---|---|
| Soil | 0.1481 | 0.1069 | 0.1180 | 0.2030 | 0.2652 | 0.1233 |
| Tree | 0.2554 | 0.1159 | 0.1069 | 0.2540 | 0.1559 | 0.2543 |
| Water | 0.0901 | 0.1336 | 0.1336 | 0.1961 | 0.1069 | 0.1559 |
| Road | 0.1166 | 0.8953 | 0.8953 | 0.1069 | 0.1336 | 0.0443 |
| Mean | 0.1525 | 0.3129 | 0.3134 | 0.1900 | 0.1654 | 0.1444 |
| RMSE | 0.1560 | 0.2190 | 0.2324 | 0.1350 | 0.1612 | 0.1257 |
| RE | 0.0207 | 0.3104 | 0.3148 | 0.0074 | 0.0069 | 0.0048 |
| Algorithms | VCA | ATGP | SGA | ECSO | SENMAV | SPEW |
|---|---|---|---|---|---|---|
| Asphalt | 0.1416 | 0.1227 | 0.0798 | 0.1227 | 0.1740 | 0.3069 |
| Grass | 0.1517 | 0.1462 | 0.1227 | 0.1740 | 0.1160 | 0.1587 |
| Tree | 1.1779 | 0.1480 | 0.2112 | 0.3893 | 1.1990 | 1.0581 |
| Roof | 0.1388 | 0.5432 | 0.5432 | 0.8092 | 0.1956 | 0.0741 |
| Dirt | 0.1127 | 1.3748 | 1.3748 | 1.1410 | 0.1227 | 0.1062 |
| Mean | 0.3445 | 0.4669 | 0.4663 | 0.5272 | 0.3615 | 0.3408 |
| RMSE | 0.3288 | 0.3272 | 0.3244 | 0.3331 | 0.2789 | 0.3158 |
| RE | 0.1362 | 0.2792 | 0.2792 | 0.1358 | 0.1658 | 0.0162 |
| Algorithms | VCA | ATGP | SGA | ECSO | SENMAV | SPEW |
|---|---|---|---|---|---|---|
| Alunite | 0.0962 | 0.0824 | 0.9675 | 0.9543 | 0.0889 | 0.1413 |
| Andradite | 0.0691 | 1.1329 | 0.0824 | 0.1046 | 0.0797 | 1.0480 |
| Buddingtonite | 0.0896 | 0.0848 | 0.0848 | 0.1150 | 0.1114 | 0.0622 |
| Dumortierite | 0.8286 | 0.0859 | 0.0859 | 0.0892 | 0.0948 | 0.1895 |
| Kaolinite_1 | 0.0838 | 0.0618 | 0.0618 | 0.0701 | 0.0603 | 0.0743 |
| Kaolinite_2 | 0.1133 | 0.1159 | 0.1159 | 0.1702 | 0.1982 | 0.0824 |
| Muscovite | 0.0741 | 0.0736 | 0.0736 | 0.0859 | 0.1195 | 0.0704 |
| Montmorillonite | 0.1500 | 0.2400 | 0.1695 | 0.1204 | 1.0240 | 0.0812 |
| Nontronite | 0.0615 | 0.0802 | 0.0802 | 0.2337 | 0.1057 | 0.1604 |
| Pyrope | 0.1248 | 0.0852 | 0.0852 | 1.1614 | 0.0680 | 0.0998 |
| Sphene | 0.0797 | 0.1207 | 0.1027 | 1.1696 | 0.0736 | 0.0734 |
| Chalcedony | 0.0583 | 0.0994 | 0.8724 | 1.1330 | 0.9396 | 0.0704 |
| Mean | 0.1524 | 0.1886 | 0.2318 | 0.4506 | 0.2467 | 0.1794 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, M.; Li, Y.; Yang, T.; Xu, D. Spatial Potential Energy Weighted Maximum Simplex Algorithm for Hyperspectral Endmember Extraction. Remote Sens. 2022, 14, 1192. https://doi.org/10.3390/rs14051192
Song M, Li Y, Yang T, Xu D. Spatial Potential Energy Weighted Maximum Simplex Algorithm for Hyperspectral Endmember Extraction. Remote Sensing. 2022; 14(5):1192. https://doi.org/10.3390/rs14051192
Chicago/Turabian StyleSong, Meiping, Ying Li, Tingting Yang, and Dayong Xu. 2022. "Spatial Potential Energy Weighted Maximum Simplex Algorithm for Hyperspectral Endmember Extraction" Remote Sensing 14, no. 5: 1192. https://doi.org/10.3390/rs14051192