Terrestrial Laser Scanning for Vegetation Analyses with a Special Focus on Savannas
Abstract
:1. Introduction
1.1. The Role and Importance of Savannas
1.2. Monitoring of Savannas Using Remote Sensing
1.3. Terrestrial Laser Scanning (TLS)
1.4. Overall Goal and Objectives of the Review
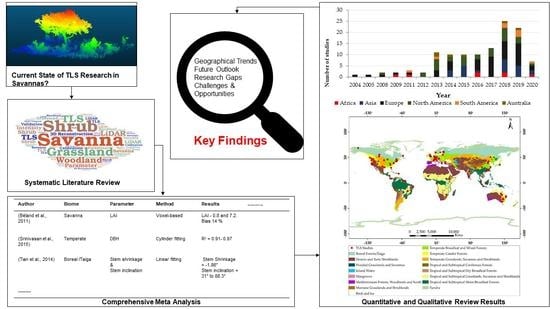
2. Materials and Methods
- ⮚
- The study under consideration focused on the extraction of one or more vegetation parameters from TLS point clouds. For example, a study was only considered if parameters like the DBH or AGB were derived on the basis of TLS point clouds [75];
- ⮚
- ⮚
- The study under consideration reviewed TLS as a remote sensing technology [39];
- ⮚
3. Results
3.1. General Overview
3.2. Temporal and Spatial Patterns
3.3. TLS Instruments and Data Used
3.4. TLS Point Cloud Pre-Processing
3.5. Methods Used with TLS Point Clouds
3.6. Vegetation Parameters Extracted from TLS Data
3.7. Primary Vegetation Attributes
3.7.1. DBH
3.7.2. Vegetation Height
3.7.3. Stem Detection
3.7.4. Crown Attributes
3.7.5. Tree Volume and Branch Parameters
3.7.6. Stem Curve Profiles
3.8. Secondary Vegetation Attributes
3.8.1. AGB
3.8.2. LAI
3.8.3. Basal Area
3.8.4. Stem Density
4. Discussion
4.1. Geographical Trends in the TLS Literature
4.2. Challenges & Opportunities
4.3. Future Outlook
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ALS | Airborne Laser Scanning |
| 3D | Three Dimensional |
| AGB | Above Ground Biomass |
| CHM | Canopy Height Model |
| DBH | Diameter at Breast Height |
| DEM | Digital Elevation Model |
| ePAI | effective Plant Area Index |
| eWAI | effective Wood Area Index |
| GPS | Global Positioning System |
| GNSS | Global Navigation Satellite System |
| IGARSS | International Geoscience and Remote Sensing Symposium |
| LAI | Leaf Area Index |
| LANDSAT | Land Remote-Sensing Satellite |
| L-Architect | LiDAR to tree Architecture |
| LiDAR | Light Detection And Ranging |
| MODIS | Moderate Resolution Imaging Spectroradiometer |
| QSM | Quantitative Structure Models |
| R2 | Coefficient of determination |
| RADAR | Radio Detection and Ranging |
| RANSAC | RANdom SAmple Consensus |
| RGB | Red, Green, Blue |
| RMSE | Root Mean Squared Error |
| TLS | Terrestrial Laser Scanner |
| UAV | Unmanned Aerial Vehicle |
References
- Scholes, R.J.; Archer, S.R. Tree-grass interactions in savannas. Annu. Rev. Ecol. Syst. 1997, 28, 517–544. [Google Scholar] [CrossRef]
- Ma, X.; Huete, A.; Moore, C.E.; Cleverly, J.; Hutley, L.B.; Beringer, J.; Leng, S.; Xie, Z.; Yu, Q.; Eamus, D. Spatiotemporal partitioning of savanna plant functional type productivity along NATT. Remote Sens. Environ. 2020, 246, 111855. [Google Scholar] [CrossRef]
- Sankaran, M.; Hanan, N.P.; Scholes, R.J.; Ratnam, J.; Augustine, D.J.; Cade, B.S.; Gignoux, J.; Higgins, S.I.; Le Roux, X.; Ludwig, F.; et al. Determinants of woody cover in African savannas. Nature 2005, 438, 846–849. [Google Scholar] [CrossRef] [PubMed]
- Grace, J.; Jose, J.S.; Meir, P.; Miranda, H.S.; Montes, R.A. Productivity and carbon fluxes of tropical savannas. J. Biogeogr. 2006, 33, 387–400. [Google Scholar] [CrossRef]
- Hutley, L.B.; Setterfield, S.A. Savanna. In Encyclopedia of Ecology, 2nd ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2018. [Google Scholar] [CrossRef]
- Galvin, K.A.; Reid, R.S. People in savanna ecosystems: Land use, change, and sustainability. In Ecosystem Function in Savannas; CRC Press: Boca Raton, FL, USA, 2010; pp. 521–536. [Google Scholar]
- Egoh, B.N.; O’Farrell, P.J.; Charef, A.; Gurney, L.J.; Koellner, T.; Abi, H.N.; Egoh, M.; Willemen, L. An African account of ecosystem service provision: Use, threats and policy options for sustainable livelihoods. Ecosyst. Serv. 2012, 2, 71–81. [Google Scholar] [CrossRef]
- Pritchard, R.; Grundy, I.M.; Van Der Horst, D.; Ryan, C.M. Environmental incomes sustained as provisioning ecosystem service availability declines along a woodland resource gradient in Zimbabwe. World Dev. 2019, 122, 325–338. [Google Scholar] [CrossRef]
- Stevens, N.; Lehmann, C.E.R.; Murphy, B.P.; Durigan, G. Savanna woody encroachment is widespread across three continents. Glob. Chang. Biol. 2017, 23, 235–244. [Google Scholar] [CrossRef] [Green Version]
- Guuroh, R.T.; Ruppert, J.C.; Ferner, J.; Čanak, K.; Schmidtlein, S.; Linstädter, A. Drivers of forage provision and erosion control in West African savannas—A macroecological perspective. Agric. Ecosyst. Environ. 2018, 251, 257–267. [Google Scholar] [CrossRef]
- Mograbi, P.J.; Asner, G.P.; Witkowski, E.T.; Erasmus, B.F.; Wessels, K.J.; Mathieu, R.; Vaughn, N.R. Humans and elephants as treefall drivers in African savannas. Ecography 2017, 40, 1274–1284. [Google Scholar] [CrossRef]
- Conradi, T. Woody encroachment in African savannas: Towards attribution to multiple drivers and a mechanistic model. J. Biogeogr. 2018, 45, 1231–1233. [Google Scholar] [CrossRef] [Green Version]
- Ciais, P.; Bombelli, A.; Williams, M.; Piao, S.L.; Chave, J.; Ryan, C.M.; Henry, M.; Brender, P.; Valentini, R. The carbon balance of Africa: Synthesis of recent research studies. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2011, 369, 2038–2057. [Google Scholar] [CrossRef] [PubMed]
- Cook, G.D.; Liedloff, A.C.; Eager, R.W.; Chen, X.; Williams, R.J.; O’Grady, A.P.; Hutley, L.B. The estimation of carbon budgets of frequently burnt tree stands in savannas of northern Australia, using allometric analysis and isotopic discrimination. Aust. J. Bot. 2005, 53, 621–630. [Google Scholar] [CrossRef]
- Williams, R.J.; Carter, J.; Duff, G.A.; Woinarski, J.C.Z.; Cook, G.D.; Farrer, S.L. Carbon accounting, land management, science and policy uncertainty in Australian savanna landscapes: Introduction and overview. Aust. J. Bot. 2005, 53, 583–588. [Google Scholar] [CrossRef] [Green Version]
- Tsalyuk, M.; Kelly, M.; Getz, W.M. Improving the prediction of African savanna vegetation variables using time series of MODIS products. ISPRS J. Photogramm. Remote Sens. 2017, 131, 77–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patenaude, G.; Milne, R.; Dawson, T.P. Synthesis of remote sensing approaches for forest carbon estimation: Reporting to the Kyoto Protocol. Environ. Sci. Policy 2005, 8, 161–178. [Google Scholar] [CrossRef]
- Asner, G.P.; Levick, S.R.; Smit, I.P.J. Remote sensing of fractional cover and biochemistry in Savannas. In Ecosystem Function in Savannas: Measurement and Modeling at Landscape to Global Scales; CRC Press: Boca Raton, FL, USA, 2010; pp. 195–217. [Google Scholar] [CrossRef]
- Viergever, K.M.; Woodhouse, I.H.; Stuart, N. Monitoring the world’s savanna biomass by earth observation. Scott. Geogr. J. 2008, 124, 218–225. [Google Scholar] [CrossRef]
- Hirata, Y.; Takao, G.; Sato, T.; Toriyama, J. (Eds.) REDD-Plus Cookbook; REDD Research and Development Centre, Forestry and Forest Products Research Institute: Tsukuba, Japan, 2012; 156p, ISBN 9784905304135. [Google Scholar]
- Feng, X.; He, L.; Cheng, Q.; Long, X.; Yuan, Y. Hyperspectral and Multispectral Remote Sensing Image Fusion Based on Endmember Spatial Information. Remote Sens. 2020, 12, 1009. [Google Scholar] [CrossRef] [Green Version]
- Gwenzi, D. Lidar remote sensing of savanna biophysical attributes: Opportunities, progress, and challenges. Int. J. Remote Sens. 2016, 38, 235–257. [Google Scholar] [CrossRef]
- Bioucas-Dias, J.M.; Plaza, A.; Camps-Valls, G.; Scheunders, P.; Nasrabadi, N.; Chanussot, J. Hyperspectral remote sensing data analysis and future challenges. IEEE Geosci. Remote Sens. Mag. 2013, 1, 6–36. [Google Scholar] [CrossRef] [Green Version]
- Cho, M.A.; Mathieu, R.; Asner, G.P.; Naidoo, L.; Van Aardt, J.A.N.; Ramoelo, A.; Debba, P.; Wessels, K.; Main, R.; Smit, I.P.; et al. Mapping tree species composition in South African savannas using an integrated airborne spectral and LiDAR system. Remote Sens. Environ. 2012, 125, 214–226. [Google Scholar] [CrossRef]
- Sinha, S.; Jeganathan, C.; Sharma, L.K.; Nathawat, M.S. A review of radar remote sensing for biomass estimation. Int. J. Environ. Sci. Technol. 2015, 12, 1779–1792. [Google Scholar] [CrossRef] [Green Version]
- Lim, K.; Treitz, P.; Wulder, M.; St-Onge, B.; Flood, M. LiDAR remote sensing of forest structure. Prog. Phys. Geogr. Earth Environ. 2003, 27, 88–106. [Google Scholar] [CrossRef] [Green Version]
- Beland, M.; Parker, G.; Sparrow, B.; Harding, D.; Chasmer, L.; Phinn, S.; Antonarakis, A.; Strahler, A. On promoting the use of lidar systems in forest ecosystem research. For. Ecol. Manag. 2019, 450, 450. [Google Scholar] [CrossRef]
- Gwenzi, D.; Lefsky, M.A. Plot-level aboveground woody biomass modeling using canopy height and auxiliary remote sensing data in a heterogeneous savanna. J. Appl. Remote Sens. 2016, 10, 16001. [Google Scholar] [CrossRef]
- Hill, R.; Broughton, R. Mapping the understorey of deciduous woodland from leaf-on and leaf-off airborne LiDAR data: A case study in lowland Britain. ISPRS J. Photogramm. Remote Sens. 2009, 64, 223–233. [Google Scholar] [CrossRef]
- Singh, J.; Levick, S.R.; Guderle, M.; Schmullius, C. Moving from plot-based to hillslope-scale assessments of savanna vegetation structure with long-range terrestrial laser scanning (LR-TLS). Int. J. Appl. Earth Obs. Geoinf. 2020, 90, 102070. [Google Scholar] [CrossRef]
- Bauwens, S.; Bartholomeus, H.M.; Calders, K.; Lejeune, P. Forest Inventory with Terrestrial LiDAR: A Comparison of Static and Hand-Held Mobile Laser Scanning. Forests 2016, 7, 127. [Google Scholar] [CrossRef] [Green Version]
- Brede, B.; Calders, K.; Lau, A.; Raumonen, P.; Bartholomeus, H.M.; Herold, M.; Kooistra, L. Non-destructive tree volume estimation through quantitative structure modelling: Comparing UAV laser scanning with terrestrial LIDAR. Remote Sens. Environ. 2019, 233, 111355. [Google Scholar] [CrossRef]
- Boukoberine, M.N.; Zhou, Z.; Benbouzid, M. A critical review on unmanned aerial vehicles power supply and energy management: Solutions, strategies, and prospects. Appl. Energy 2019, 255, 113823. [Google Scholar] [CrossRef]
- Eisfelder, C.; Kuenzer, C.; Dech, S. Derivation of biomass information for semi-arid areas using remote-sensing data. Int. J. Remote Sens. 2011, 33, 2937–2984. [Google Scholar] [CrossRef]
- Galidaki, G.; Zianis, D.; Gitas, I.; Radoglou, K.; Karathanassi, V.; Tsakiri–Strati, M.; Woodhouse, I.; Mallinis, G. Vegetation biomass estimation with remote sensing: Focus on forest and other wooded land over the Mediterranean ecosystem. Int. J. Remote Sens. 2016, 38, 1940–1966. [Google Scholar] [CrossRef] [Green Version]
- Dassot, M.; Constant, T.; Fournier, M. The use of terrestrial LiDAR technology in forest science: Application fields, benefits and challenges. Ann. For. Sci. 2011, 68, 959–974. [Google Scholar] [CrossRef] [Green Version]
- Burt, A.; Disney, M.; Raumonen, P.; Armston, J.; Calders, K.; Lewis, P. Rapid characterisation of forest structure from TLS and 3D modelling. In Proceedings of the 2013 IEEE International Geoscience and Remote Sensing Symposium—IGARSS, Melbourne, VIC, Australia, 21–26 July 2013; pp. 3387–3390. Available online: http://128.197.168.195/wp-content/uploads/2013/08/Burt-Disney-IGARSS.pdf (accessed on 12 October 2020).
- Newnham, G.J.; Armston, J.D.; Calders, K.; Disney, M.I.; Lovell, J.L.; Schaaf, C.B.; Strahler, A.H.; Danson, F.M. Terrestrial laser scanning for plot-scale forest measurement. Curr. For. Rep. 2015, 1, 239–251. [Google Scholar] [CrossRef] [Green Version]
- Béland, M.; Widlowski, J.-L.; Fournier, R.A.; Côté, J.-F.; Verstraete, M.M. Estimating leaf area distribution in savanna trees from terrestrial LiDAR measurements. Agric. For. Meteorol. 2011, 151, 1252–1266. [Google Scholar] [CrossRef]
- Calders, K.; Newnham, G.; Burt, A.; Murphy, S.; Raumonen, P.; Herold, M.; Culvenor, D.S.; Avitabile, V.; Disney, M.; Armston, J.; et al. Nondestructive estimates of above-ground biomass using terrestrial laser scanning. Methods Ecol. Evol. 2014, 6, 198–208. [Google Scholar] [CrossRef]
- Wang, Y.; Spiecker, H.; Calders, K.; Disney, M.I.; Raumonen, P. SimpleTree—An efficient open source tool to build tree models from TLS clouds. Forests 2015, 6, 4245–4294. [Google Scholar] [CrossRef]
- Lemmens, M. Geo-Information: Technologies, Applications and the Environment; Springer Science & Business Media: Dordrecht, The Netherlands, 2011; ISBN 9400716672. [Google Scholar] [CrossRef]
- Wilkes, P.; Lau, A.; Disney, M.; Calders, K.; Burt, A.; De Tanago, J.G.; Bartholomeus, H.; Brede, B.; Herold, M. Data acquisition considerations for Terrestrial Laser Scanning of forest plots. Remote Sens. Environ. 2017, 196, 140–153. [Google Scholar] [CrossRef]
- Liang, X.; Kankare, V.; Hyyppä, J.; Wang, Y.; Kukko, A.; Haggrén, H.; Yu, X.; Kaartinen, H.; Jaakkola, A.; Guan, F.; et al. Terrestrial laser scanning in forest inventories. ISPRS J. Photogramm. Remote Sens. 2016, 115, 63–77. [Google Scholar] [CrossRef]
- Fan, G.; Nan, L.; Chen, F.; Dong, Y.; Wang, Z.; Li, H.; Chen, D. A new quantitative approach to tree attributes estimation based on LIDAR point clouds. Remote Sens. 2020, 12, 1779. [Google Scholar] [CrossRef]
- Fang, R.; Strimbu, B.M. Comparison of mature douglas-firs’ crown structures developed with two quantitative structural models using TLS point clouds for neighboring trees in a natural regime stand. Remote Sens. 2019, 11, 1661. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Wang, T.; Darvishzadeh, R.; Skidmore, A.K.; Niemann, K.O. 3D leaf water content mapping using terrestrial laser scanner backscatter intensity with radiometric correction. ISPRS J. Photogramm. Remote Sens. 2015, 110, 14–23. [Google Scholar] [CrossRef]
- Yrttimaa, T.; Saarinen, N.; Kankare, V.; Liang, X.; Hyyppä, J.; Holopainen, M.; Vastaranta, M. Investigating the feasibility of multi-scan terrestrial laser scanning to characterize tree communities in southern boreal forests. Remote Sens. 2019, 11, 1423. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, S.; Popescu, S.C.; Eriksson, M.; Sheridan, R.D.; Ku, N.-W. Multi-temporal terrestrial laser scanning for modeling tree biomass change. For. Ecol. Manag. 2014, 318, 304–317. [Google Scholar] [CrossRef]
- Olivier, M.-D.; Robert, S.; Fournier, R.A. A method to quantify canopy changes using multi-temporal terrestrial lidar data: Tree response to surrounding gaps. Agric. For. Meteorol. 2017, 237, 184–195. [Google Scholar] [CrossRef]
- Liang, X.; Hyyppä, J.; Kaartinen, H.; Lehtomäki, M.; Pyörälä, J.; Pfeifer, N.; Holopainen, M.; Brolly, G.; Francesco, P.; Trochta, J.; et al. International benchmarking of terrestrial laser scanning approaches for forest inventories. ISPRS J. Photogramm. Remote Sens. 2018, 144, 137–179. [Google Scholar] [CrossRef]
- Tian, W.; Lin, Y.; Liu, Y.; Niu, Z. Derivation of tree stem structural parameters from static terrestrial laser scanning data. Lidar Remote Sens. Environ. Monit. Xiv 2014, 9262, 92620Z. [Google Scholar] [CrossRef]
- Dassot, M.; Colin, A.; Santenoise, P.; Fournier, M.; Constant, T. Terrestrial laser scanning for measuring the solid wood volume, including branches, of adult standing trees in the forest environment. Comput. Electron. Agric. 2012, 89, 86–93. [Google Scholar] [CrossRef]
- Olsoy, P.J.; Glenn, N.F.; Clark, P.E.; Derryberry, D.R. Aboveground total and green biomass of dryland shrub derived from terrestrial laser scanning. ISPRS J. Photogramm. Remote Sens. 2014, 88, 166–173. [Google Scholar] [CrossRef] [Green Version]
- Danson, F.M.; Sasse, F.; Schofield, L.A. Spectral and spatial information from a novel dual-wavelength full-waveform terrestrial laser scanner for forest ecology. Interface Focus 2018, 8, 20170049. [Google Scholar] [CrossRef]
- Calders, K.; Origo, N.; Burt, A.; Disney, M.I.; Nightingale, J.; Raumonen, P.; Åkerblom, M.; Malhi, Y.; Lewis, P. Realistic forest stand reconstruction from terrestrial LIDAR for radiative transfer modelling. Remote Sens. 2018, 10, 933. [Google Scholar] [CrossRef] [Green Version]
- De Tanago, J.G.; Lau, A.; Bartholomeus, H.; Herold, M.; Avitabile, V.; Raumonen, P.; Martius, C.; Goodman, R.C.; Disney, M.I.; Manuri, S.; et al. Estimation of above-ground biomass of large tropical trees with terrestrial LiDAR. Methods Ecol. Evol. 2018, 9, 223–234. [Google Scholar] [CrossRef] [Green Version]
- Maas, H.-G.; Bienert, A.; Scheller, S.; Keane, E. Automatic forest inventory parameter determination from terrestrial laser scanner data. Int. J. Remote Sens. 2008, 29, 1579–1593. [Google Scholar] [CrossRef]
- Greaves, H.E.; Vierling, L.A.; Eitel, J.U.H.; Boelman, N.T.; Magney, T.S.; Prager, C.M.; Griffin, K.L. Applying terrestrial lidar for evaluation and calibration of airborne lidar-derived shrub biomass estimates in Arctic tundra. Remote Sens. Lett. 2017, 8, 175–184. [Google Scholar] [CrossRef]
- Levick, S.R.; Whiteside, T.; Loewensteiner, D.A.; Rudge, M.; Bartolo, R. Leveraging TLS as a calibration and validation tool for MLS and ULS mapping of savanna structure and biomass at landscape-scales. Remote Sens. 2021, 13, 257. [Google Scholar] [CrossRef]
- Indirabai, I.; Nair, M.H.; Jaishanker, R.N.; Nidamanuri, R.R. Terrestrial laser scanner based 3D reconstruction of trees and retrieval of leaf area index in a forest environment. Ecol. Inform. 2019, 53, 100986. [Google Scholar] [CrossRef]
- Odipo, V.O.; Nickless, A.; Berger, C.; Baade, J.; Urbazaev, M.; Walther, C.; Schmullius, C. Assessment of aboveground woody biomass dynamics using terrestrial laser scanner and L-band ALOS PALSAR Data in South African Savanna. Forests 2016, 7, 294. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Gao, J.; Hu, Q.; Li, Y.; Tian, J.; Liao, C.; Ma, W.; Xu, Y. Assessing revegetation effectiveness on an extremely degraded grassland, southern Qinghai-Tibetan Plateau, using terrestrial LiDAR and field data. Agric. Ecosyst. Environ. 2019, 282, 13–22. [Google Scholar] [CrossRef]
- Zimbres, B.; Shimbo, J.Z.; Bustamante, M.M.D.C.; Levick, S.R.; De Miranda, S.D.C.; Roitman, I.; Silvério, D.V.; Gomes, L.; Fagg, C.W.; Alencar, A. Savanna vegetation structure in the Brazilian Cerrado allows for the accurate estimation of aboveground biomass using terrestrial laser scanning. For. Ecol. Manag. 2020, 458, 117798. [Google Scholar] [CrossRef]
- Luck, L.; Hutley, L.B.; Calders, K.; Levick, S.R. Exploring the variability of tropical savanna tree structural allometry with terrestrial laser scanning. Remote Sens. 2020, 12, 3893. [Google Scholar] [CrossRef]
- Lindenmayer, D.B.; Laurance, W.F.; Franklin, J.F. Global Decline in Large Old Trees. Science 2012, 338, 1305–1306. [Google Scholar] [CrossRef]
- Estornell, J.; Velázquez-Martí, A.; Fernández-Sarría, A.; Cortés, I.L.; Martí-Gavilá, J.; Salazar, D. Estimation of structural attributes of walnut trees based on terrestrial laser scanning. Rev. Teledetec. 2017, 2017, 67–76. [Google Scholar] [CrossRef]
- Richardson, J.J.; Moskal, L.M.; Bakker, J.D. Terrestrial laser scanning for vegetation sampling. Sensors 2014, 14, 20304–20319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marselis, S.M.; Tang, H.; Armston, J.D.; Calders, K.; Labrière, N.; Dubayah, R. Distinguishing vegetation types with airborne waveform lidar data in a tropical forest-savanna mosaic: A case study in Lopé National Park, Gabon. Remote Sens. Environ. 2018, 216, 626–634. [Google Scholar] [CrossRef]
- Cuni-Sanchez, A.; White, L.J.T.; Calders, K.; Jeffery, K.J.; Abernethy, K.; Burt, A.; Disney, M.; Gilpin, M.; Gomez-Dans, J.L.; Lewis, S.L. African savanna-forest boundary dynamics: A 20-year study. PLoS ONE 2016, 11, e0156934. [Google Scholar] [CrossRef] [PubMed]
- Calders, K.; Adams, J.; Armston, J.; Bartholomeus, H.; Bauwens, S.; Bentley, L.P.; Chave, J.; Danson, F.M.; Demol, M.; Disney, M.; et al. Terrestrial laser scanning in forest ecology: Expanding the horizon. Remote Sens. Environ. 2020, 251, 112102. [Google Scholar] [CrossRef]
- Disney, M.I.; Burt, A.; Calders, K.; Schaaf, C.; Stovall, A. Innovations in Ground and Airborne Technologies as Reference and for Training and Validation: Terrestrial Laser Scanning (TLS). Surv. Geophys. 2019, 40, 937–958. [Google Scholar] [CrossRef] [Green Version]
- Calders, K.; Disney, M.I.; Armston, J.; Burt, A.; Brede, B.; Origo, N.; Muir, J.; Nightingale, J. Evaluation of the range accuracy and the radiometric calibration of multiple terrestrial laser scanning instruments for data interoperability. IEEE Trans. Geosci. Remote Sens. 2017, 55, 2716–2724. [Google Scholar] [CrossRef]
- Clarivate Analytics. Web of Science Core Collection. Web Sci. 2020. Available online: https://apps.webofknowledge.com/WOS_GeneralSearch_input.do?product=WOS&search_mode=GeneralSearch&SID=F2RTSaN12sn1d9ppMkH&preferencesSaved= (accessed on 23 September 2020).
- Moskal, L.M.; Zheng, G. Retrieving forest inventory variables with terrestrial laser scanning (TLS) in urban heterogeneous forest. Remote Sens. 2011, 4, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Hancock, S.; Anderson, K.; Disney, M.; Gaston, K.J. Measurement of fine-spatial-resolution 3D vegetation structure with airborne waveform lidar: Calibration and validation with voxelised terrestrial lidar. Remote Sens. Environ. 2017, 188, 37–50. [Google Scholar] [CrossRef] [Green Version]
- Tan, K.; Zhang, W.; Shen, F.; Cheng, X. Investigation of TLS intensity data and distance measurement errors from target specular reflections. Remote Sens. 2018, 10, 1077. [Google Scholar] [CrossRef] [Green Version]
- Aijazi, A.K.; Checchin, P.; Malaterre, L.; Trassoudaine, L. Automatic detection and parameter estimation of trees for forest inventory applications using 3D terrestrial LiDAR. Remote Sens. 2017, 9, 946. [Google Scholar] [CrossRef] [Green Version]
- Calders, K.; Origo, N.; Disney, M.I.; Nightingale, J.; Woodgate, W.; Armston, J.; Lewis, P. Variability and bias in active and passive ground-based measurements of effective plant, wood and leaf area index. Agric. For. Meteorol. 2018, 252, 231–240. [Google Scholar] [CrossRef]
- Zheng, G.; Moskal, L.M.; Kim, S.-H. Retrieval of effective leaf area index in heterogeneous forests with terrestrial laser scanning. IEEE Trans. Geosci. Remote Sens. 2013, 51, 777–786. [Google Scholar] [CrossRef]
- Bordin, F.; Teixeira, E.C.; Rolim, S.B.A.; Tognoli, F.M.W.; Souza, C.N.; Veronez, M.R. Analysis of the Influence of Distance on Data Acquisition Intensity Forestry Targets by a LIDAR Technique with Terrestrial Laser Scanner. ISPRS—Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2013, XL-2/W1, 99–103. [Google Scholar] [CrossRef] [Green Version]
- Heinzel, J.; Huber, M.O. TLS field data based intensity correction for forest environments. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, 41, 643–649. [Google Scholar] [CrossRef]
- Tan, K.; Cheng, X. Intensity data correction based on incidence angle and distance for terrestrial laser scanner. J. Appl. Remote Sens. 2015, 9, 94094. [Google Scholar] [CrossRef]
- Vaccari, S.; Van Leeuwen, M.; Calders, K.; Coops, N.C.; Herold, M. Bias in lidar-based canopy gap fraction estimates. Remote Sens. Lett. 2013, 4, 391–399. [Google Scholar] [CrossRef]
- Tansey, K.; Selmes, N.; Anstee, A.; Tate, N.J.; Denniss, A. Estimating tree and stand variables in a Corsican Pine woodland from terrestrial laser scanner data. Int. J. Remote Sens. 2009, 30, 5195–5209. [Google Scholar] [CrossRef]
- Olofsson, K.; Holmgren, J.; Olsson, H. Tree stem and height measurements using terrestrial laser scanning and the RANSAC algorithm. Remote Sens. 2014, 6, 4323–4344. [Google Scholar] [CrossRef] [Green Version]
- Magney, T.S.; Eitel, J.U.; Griffin, K.L.; Boelman, N.T.; Greaves, H.E.; Prager, C.M.; Logan, B.A.; Zheng, G.; Ma, L.; Fortin, E.A.; et al. LiDAR canopy radiation model reveals patterns of photosynthetic partitioning in an Arctic shrub. Agric. For. Meteorol. 2016, 221, 78–93. [Google Scholar] [CrossRef]
- Sun, H.; Wang, G.; Lin, H.; Li, J.; Zhang, H.; Ju, H. Retrieval and Accuracy Assessment of Tree and Stand Parameters for Chinese Fir Plantation Using Terrestrial Laser Scanning. IEEE Geosci. Remote Sens. Lett. 2015, 12, 1993–1997. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, L.; Fang, T.; Mathiopoulos, P.T.; Qu, H.; Chen, D.; Wang, Y. A structure-aware global optimization method for reconstructing 3-D tree models from terrestrial laser scanning data. IEEE Trans. Geosci. Remote Sens. 2014, 52, 5653–5669. [Google Scholar] [CrossRef]
- Xu, K.; Su, Y.; Liu, J.; Hu, T.; Jin, S.; Ma, Q.; Zhai, Q.; Wang, R.; Zhang, J.; Li, Y.; et al. Estimation of degraded grassland aboveground biomass using machine learning methods from terrestrial laser scanning data. Ecol. Indic. 2020, 108, 105747. [Google Scholar] [CrossRef]
- Singh, J.; Levick, S.R.; Guderle, M.; Schmullius, C.; Trumbore, S.E. Variability in fire-induced change to vegetation physiognomy and biomass in semi-arid savanna. Ecosphere 2018, 9, e02514. [Google Scholar] [CrossRef] [Green Version]
- Muir, J.; Phinn, S.; Eyre, T.; Scarth, P. Measuring plot scale woodland structure using terrestrial laser scanning. Remote Sens. Ecol. Conserv. 2018, 4, 320–338. [Google Scholar] [CrossRef] [Green Version]
- Olson, D.M.; Dinerstein, E.; Wikramanayake, E.D.; Burgess, N.D.; Powell, G.V.N.; Underwood, E.C.; D’Amico, J.A.; Itoua, I.; Strand, H.E.; Morrison, J.C.; et al. Terrestrial ecoregions of the worlds: A new map of life on Earth. Bioscience 2001, 51, 933–938. [Google Scholar] [CrossRef]
- Srinivasan, S.; Popescu, S.; Eriksson, M.; Sheridan, R.D.; Ku, N.-W. Terrestrial laser scanning as an effective tool to retrieve tree level height, crown width, and stem diameter. Remote Sens. 2015, 7, 1877–1896. [Google Scholar] [CrossRef] [Green Version]
- Watt, P.J.; Donoghue, D.N.M. Measuring forest structure with terrestrial laser scanning. Int. J. Remote Sens. 2005, 26, 1437–1446. [Google Scholar] [CrossRef]
- Krooks, A.; Kaasalainen, S.; Kankare, V.; Joensuu, M.; Raumonen, P.; Kaasalainen, M. Tree structure vs. height from terrestrial laser scanning and quantitative structure models. Silva Fenn. 2014, 48, 48. [Google Scholar] [CrossRef] [Green Version]
- Pitkänen, T.P.; Raumonen, P.; Kangas, A. Measuring stem diameters with TLS in boreal forests by complementary fitting procedure. ISPRS J. Photogramm. Remote Sens. 2019, 147, 294–306. [Google Scholar] [CrossRef]
- Yu, X.; Liang, X.; Hyyppä, J.; Kankare, V.; Vastaranta, M.; Holopainen, M. Stem biomass estimation based on stem reconstruction from terrestrial laser scanning point clouds. Remote Sens. Lett. 2013, 4, 344–353. [Google Scholar] [CrossRef]
- Lau, A.; Martius, C.; Bartholomeus, H.; Shenkin, A.; Jackson, T.D.; Malhi, Y.; Herold, M.; Bentley, L.P. Estimating architecture-based metabolic scaling exponents of tropical trees using terrestrial LiDAR and 3D modelling. For. Ecol. Manag. 2019, 439, 132–145. [Google Scholar] [CrossRef]
- Moorthy, S.M.K.; Bao, Y.; Calders, K.; Schnitzer, S.A.; Verbeeck, H. Semi-automatic extraction of liana stems from terrestrial LiDAR point clouds of tropical rainforests. ISPRS J. Photogramm. Remote Sens. 2019, 154, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Paynter, I.; Genest, D.; Peri, F.; Schaaf, C. Bounding uncertainty in volumetric geometric models for terrestrial lidar observations of ecosystems. Interface Focus 2018, 8, 20170043. [Google Scholar] [CrossRef] [Green Version]
- RIEGL Laser Measurement Systems GmbH. RIEGL—About RIEGL. 2020. Available online: http://www.riegl.com/company/about-riegl/ (accessed on 30 September 2020).
- FARO Technologies Inc. High-Precision 3D Acquisition, Measurement and Analysis. 2020. Available online: https://www.faro.com/de-de/faro-im-uberblick/ (accessed on 30 September 2020).
- Leica Geosystems. When it has to be Right|Leica Geosystems. 2020. Available online: https://leica-geosystems.com/ (accessed on 30 September 2020).
- Kim, A.M.; Olsen, R.C.; Béland, M. Simulated full-waveform lidar compared to Riegl VZ-400 terrestrial laser scans. Laser Radar Technol. Appl. XXI 2016, 9832, 98320. [Google Scholar] [CrossRef]
- Wu, D.; Phinn, S.; Johansen, K.; Robson, A.; Muir, J.; Searle, C. Estimating Changes in Leaf Area, Leaf Area Density, and Vertical Leaf Area Profile for Mango, Avocado, and Macadamia Tree Crowns Using Terrestrial Laser Scanning. Remote Sens. 2018, 10, 1750. [Google Scholar] [CrossRef] [Green Version]
- Ferrara, R.; Virdis, S.G.; Ventura, A.; Ghisu, T.; Duce, P.; Pellizzaro, G. An automated approach for wood-leaf separation from terrestrial LIDAR point clouds using the density based clustering algorithm DBSCAN. Agric. For. Meteorol. 2018, 262, 434–444. [Google Scholar] [CrossRef]
- Åkerblom, M.; Raumonen, P.; Casella, E.; Disney, M.I.; Danson, F.M.; Gaulton, R.; Schofield, L.A.; Kaasalainen, M. Non-intersecting leaf insertion algorithm for tree structure models. Interface Focus 2018, 8, 20170045. [Google Scholar] [CrossRef]
- Wan, P.; Wang, T.; Zhang, W.; Liang, X.; Skidmore, A.K.; Yan, G. Quantification of occlusions influencing the tree stem curve retrieving from single-scan terrestrial laser scanning data. For. Ecosyst. 2019, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Anderson, K.E.; Glenn, N.F.; Spaete, L.P.; Shinneman, D.J.; Pilliod, D.S.; Arkle, R.S.; McIlroy, S.K.; Derryberry, D.R. Estimating vegetation biomass and cover across large plots in shrub and grass dominated drylands using terrestrial lidar and machine learning. Ecol. Indic. 2018, 84, 793–802. [Google Scholar] [CrossRef]
- Bailey, B.N.; Ochoa, M.H. Semi-direct tree reconstruction using terrestrial LiDAR point cloud data. Remote Sens. Environ. 2018, 208, 133–144. [Google Scholar] [CrossRef] [Green Version]
- Reddy, R.S.; Rakesh, A.; Jha, C.S.; Rajan, K.S. Automatic estimation of tree stem attributes using terrestrial laser scanning in central Indian dry deciduous forests. Curr. Sci. 2018, 114, 201–206. [Google Scholar] [CrossRef]
- Vaaja, M.T.; Virtanen, J.-P.; Kurkela, M.; Lehtola, V.; Hyyppä, J.; Hyyppä, H. The Effect of Wind on Tree Stem Parameter Estimation Using Terrestrial Laser Scanning. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, III-8, 117–122. [Google Scholar] [CrossRef]
- Ghimire, S.; Xystrakis, F.; Koutsias, N. Using terrestrial laser scanning to measure forest inventory parameters in a mediterranean coniferous stand of western Greece. PFG-J. Photogramm. Remote Sens. Geoinf. Sci. 2017, 85, 213–225. [Google Scholar] [CrossRef]
- Guimarães-Steinicke, C.; Weigelt, A.; Ebeling, A.; Eisenhauer, N.; Duque-Lazo, J.; Reu, B.; Roscher, C.; Schumacher, J.; Wagg, C.; Wirth, C. Terrestrial laser scanning reveals temporal changes in biodiversity mechanisms driving grassland productivity. Adv. Ecol. Res. 2019, 61, 133–161. [Google Scholar] [CrossRef]
- RIEGL Laser Measurement Systems GmbH. RIEGL VZ-400; RIEGL Laser Measurement Systems GmbH: Vienna, Austria, 2017. [Google Scholar]
- Leica Geosystems. Leica HDS6100 Latest Generation of Ultra-High Speed Laser Scanner. 2009. Available online: https://w3.leica-geosystems.com/downloads123/hds/hds/HDS6100/brochures/Leica_HDS6100_brochure_us.pdf (accessed on 6 October 2020).
- RIEGL Laser Measurement Systems GmbH. RIEGL VZ-1000; RIEGL Laser Measurement Systems GmbH: Vienna, Austria, 2017. [Google Scholar]
- FARO Technologies Inc. FARO Focus 3D Features, Benefits & Technical Specifications; FARO Technologies, Inc.: Lake Mary, FL, USA, 2013; Available online: http://www.faro.com/en-us/products/3d-surveying/faro-focus3d/overview (accessed on 6 October 2020).
- FARO Technologies Inc. FARO Laser Scanner Focus 3D X 330 Features, Benefits & Technical Specifications; FARO Technologies, Inc.: Lake Mary, FL, USA, 2013. [Google Scholar]
- Heinzel, J.; Huber, M.O. Detecting tree stems from volumetric tls data in forest environments with rich understory. Remote Sens. 2016, 9, 9. [Google Scholar] [CrossRef] [Green Version]
- Soma, M.; Pimont, F.; Allard, D.; Fournier, R.; Dupuy, J.-L. Mitigating occlusion effects in Leaf Area Density estimates from Terrestrial LiDAR through a specific kriging method. Remote Sens. Environ. 2020, 245, 111836. [Google Scholar] [CrossRef]
- Stovall, A.E.L.; Shugart, H.H. Improved biomass calibration and validation with terrestrial lidar: Implications for future LiDAR and SAR missions. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 3527–3537. [Google Scholar] [CrossRef]
- Delagrange, S.; Rochon, P. Reconstruction and analysis of a deciduous sapling using digital photographs or terrestrial-LiDAR technology. Ann. Bot. 2011, 108, 991–1000. [Google Scholar] [CrossRef] [Green Version]
- Schulze-Brüninghoff, D.; Hensgen, F.; Wachendorf, M.; Astor, T. Methods for LiDAR-based estimation of extensive grassland biomass. Comput. Electron. Agric. 2019, 156, 693–699. [Google Scholar] [CrossRef]
- Cooper, S.D.; Roy, D.; Schaaf, C.; Paynter, I. Examination of the potential of terrestrial laser scanning and structure-from-motion photogrammetry for rapid nondestructive field measurement of grass biomass. Remote Sens. 2017, 9, 531. [Google Scholar] [CrossRef] [Green Version]
- Bremer, M.; Wichmann, V.; Rutzinger, M. Multi-temporal fine-scale modelling of Larix decidua forest plots using terrestrial LiDAR and hemispherical photographs. Remote Sens. Environ. 2018, 206, 189–204. [Google Scholar] [CrossRef]
- Pfeifer, N.; Winterhalder, D.; Gorte, B.G.H. Three-dimensional reconstruction of stems for assessment of taper, sweep and lean based on laser scanning of standing trees. Scand. J. For. Res. 2004, 19, 571–581. [Google Scholar] [CrossRef]
- Tan, K.; Cheng, X. Correction of incidence angle and distance effects on TLS intensity data based on reference targets. Remote Sens. 2016, 8, 251. [Google Scholar] [CrossRef] [Green Version]
- Zheng, G.; Moskal, L.M. Retrieving Leaf Area Index (LAI) Using Remote Sensing: Theories, Methods and Sensors. Sensors 2009, 9, 2719–2745. [Google Scholar] [CrossRef] [Green Version]
- Beyer, R.; Bayer, D.; Letort, V.; Pretzsch, H.; Cournède, P.-H. Validation of a functional-structural tree model using terrestrial Lidar data. Ecol. Model. 2017, 357, 55–57. [Google Scholar] [CrossRef]
- LaRue, E.A.; Wagner, F.W.; Fei, S.; Atkins, J.W.; Fahey, R.T.; Gough, C.; Hardiman, B.S. Compatibility of Aerial and Terrestrial LiDAR for Quantifying Forest Structural Diversity. Remote Sens. 2020, 12, 1407. [Google Scholar] [CrossRef]
- Oveland, I.; Hauglin, M.; Gobakken, T.; Naesset, E.; Maalen-Johansen, I. Automatic estimation of tree position and stem diameter using a moving terrestrial laser scanner. Remote Sens. 2017, 9, 350. [Google Scholar] [CrossRef] [Green Version]
- Grotti, M.; Calders, K.; Origo, N.; Puletti, N.; Alivernini, A.; Ferrara, C.; Chianucci, F. An intensity, image-based method to estimate gap fraction, canopy openness and effective leaf area index from phase-shift terrestrial laser scanning. Agric. For. Meteorol. 2020, 280, 107766. [Google Scholar] [CrossRef]
- Kaasalainen, S.; Jaakkola, A.; Kaasalainen, M.; Krooks, A.; Kukko, A. Analysis of incidence angle and distance effects on terrestrial laser scanner intensity: Search for correction methods. Remote Sens. 2011, 3, 2207–2221. [Google Scholar] [CrossRef] [Green Version]
- Xiangyu, W.; Donghui, X.; Guangjian, Y.; Wuming, Z.; Yan, W.; Yiming, C. 3D reconstruction of a single tree from terrestrial LiDAR data. In Proceedings of the International Geoscience and Remote Sensing Symposium (IGARSS), Quebec City, QC, Canada, 13–18 July 2014; IEEE: New York, NY, USA, 2014; pp. 796–799. [Google Scholar] [CrossRef]
- Kong, J.; Ding, X.; Liu, J.; Yan, L.; Wang, J. New Hybrid Algorithms for estimating tree stem diameters at breast height using a two dimensional terrestrial laser scanner. Sensors 2015, 15, 15661–15683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelbe, D.; Van Aardt, J.; Romanczyk, P.; Van Leeuwen, M.; Cawse-Nicholson, K. Single-scan stem reconstruction using low-resolution terrestrial laser scanner data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 3414–3427. [Google Scholar] [CrossRef]
- Yang, X.; Strahler, A.H.; Schaaf, C.B.; Jupp, D.L.; Yao, T.; Zhao, F.; Wang, Z.; Culvenor, D.S.; Newnham, G.J.; Lovell, J.L.; et al. Three-dimensional forest reconstruction and structural parameter retrievals using a terrestrial full-waveform lidar instrument (Echidna®). Remote Sens. Environ. 2013, 135, 36–51. [Google Scholar] [CrossRef]
- Bazezew, M.N.; Hussin, Y.A.; Kloosterman, E. Integrating Airborne LiDAR and Terrestrial Laser Scanner forest parameters for accurate above-ground biomass/carbon estimation in Ayer Hitam tropical forest, Malaysia. Int. J. Appl. Earth Obs. Geoinf. 2018, 73, 638–652. [Google Scholar] [CrossRef]
- Riegl. Training Material for RIEGL VZ-400 8. Project Planning. 2014. Available online: www.riegl.com: (accessed on 18 December 2020).
- Greaves, H.E.; Vierling, L.A.; Eitel, J.U.H.; Boelman, N.T.; Magney, T.S.; Prager, C.M.; Griffin, K.L. Estimating aboveground biomass and leaf area of low-stature Arctic shrubs with terrestrial LiDAR. Remote Sens. Environ. 2015, 164, 26–35. [Google Scholar] [CrossRef]
- Hu, C.; Pan, Z.; Li, P. A 3D point cloud filtering method for leaves based on manifold distance and normal estimation. Remote Sens. 2019, 11, 198. [Google Scholar] [CrossRef] [Green Version]
- Han, X.-F.; Jin, J.S.; Wang, M.-J.; Jiang, W.; Gao, L.; Xiao, L. A review of algorithms for filtering the 3D point cloud. Signal Process. Image Commun. 2017, 57, 103–112. [Google Scholar] [CrossRef]
- Kirton, A.; Scholes, B.; Verstraete, M.M.; Archibald, S.; Mennell, K.; Asner, G.; Nickless, A.; Scholes, R.; Asner, G.P. Detailed structural characterisation of the savanna flux site at Skukuza, South Africa. In Proceedings of the 2009 IEEE International Geoscience and Remote Sensing Symposium, Cape Town, South Africa, 12–17 July 2009; Volume 2, p. II-186. [Google Scholar] [CrossRef]
- Calders, K.; Armston, J.; Newnham, G.; Herold, M.; Goodwin, N.R. Implications of sensor configuration and topography on vertical plant profiles derived from terrestrial LiDAR. Agric. For. Meteorol. 2014, 194, 104–117. [Google Scholar] [CrossRef]
- Burt, A.; Disney, M.I.; Calders, K. Extracting individual trees from lidar point clouds using treeseg. Methods Ecol. Evol. 2018, 10, 438–445. [Google Scholar] [CrossRef] [Green Version]
- Yurtseven, H.; Çoban, S.; Akgul, M.; Akay, A.O. Individual tree measurements in a planted woodland with terrestrial laser scanner. Turk. J. Agric. For. 2019, 43, 192–208. [Google Scholar] [CrossRef]
- Côté, J.-F.; Fournier, R.A.; Luther, J.E.; Van Lier, O.R. Fine-scale three-dimensional modeling of boreal forest plots to improve forest characterization with remote sensing. Remote Sens. Environ. 2018, 219, 99–114. [Google Scholar] [CrossRef]
- Yang, X.; Schaaf, C.; Strahler, A.; Li, Z.; Wang, Z.; Yao, T.; Zhao, F.; Saenz, E.; Paynter, I.; Douglas, E.S.; et al. Studying canopy structure through 3-D reconstruction of point clouds from full-waveform terrestrial lidar. In Proceedings of the 2013 IEEE International Geoscience and Remote Sensing Symposium-IGARSS, Melbourne, VIC, Australia, 21–26 July 2013; IEEE: New York, NY, USA, 2013; pp. 3375–3378. [Google Scholar]
- Lau, A.; Bentley, L.P.; Martius, C.; Shenkin, A.; Bartholomeus, H.; Raumonen, P.; Malhi, Y.; Jackson, T.D.; Herold, M. Quantifying branch architecture of tropical trees using terrestrial LiDAR and 3D modelling. Trees Struct. Funct. 2018, 32, 1219–1231. [Google Scholar] [CrossRef] [Green Version]
- Lau, A.; Calders, K.; Bartholomeus, H.M.; Martius, C.; Raumonen, P.; Herold, M.; Vicari, M.B.; Sukhdeo, H.; Singh, J.; Goodman, R.C. Tree Biomass equations from terrestrial LiDAR: A Case study in Guyana. Forests 2019, 10, 527. [Google Scholar] [CrossRef] [Green Version]
- Isenburg, M. LAStools-Efficient LiDAR Processing Software. 2014. Available online: https://rapidlasso.com/lastools/ (accessed on 28 October 2020).
- MathWorks. MATLAB-MathWorks-MATLAB & Simulink. 2020. Available online: https://www.mathworks.com/products/matlab.html (accessed on 28 October 2020).
- The R Foundation. R: The R Project for Statistical Computing. 2018. Available online: https://www.r-project.org/ (accessed on 28 October 2020).
- Python Software Foundation. Welcome to Python.org. 2020. Available online: https://www.python.org/ (accessed on 28 October 2020).
- Leica Geosystems. Leica Cyclone 3D Point Cloud Processing Software. Available online: http://leica-geosystems.com/products/laser-scanners/software/leica-cyclone (accessed on 28 January 2021).
- FARO Technologies Inc. SCENE—The Most Intuitive Data Scan Software|FARO Technologies. 2020. Available online: https://www.faro.com/products/construction-bim/faro-scene/ (accessed on 28 October 2020).
- RIEGL Laser Measurement Systems GmbH. RIEGL—RiSCAN PRO. 2020. Available online: http://www.riegl.com/products/software-packages/riscan-pro/ (accessed on 28 October 2020).
- Computree Group. The Computree Platform|Computree—Official Site. 2018. Available online: http://computree.onf.fr/?page_id=42 (accessed on 28 October 2020).
- Girardeau-Montaut, D. CloudCompare; Électricité de France S.A. (EDF) R&D: Paris, France, 2003. [Google Scholar]
- Popovas, D.; Mikalauskas, V.; Šlikas, D.; Valotka, S.; Šorys, T. Individual tree parameters estimation from terrestrial laser scanner data. In Proceedings of the 10th International Conference Environmental Engineering, ICEE, Vilnius, Lithuania, 27–28 April 2017; pp. 27–28. [Google Scholar]
- Moorthy, I.; Miller, J.R.; Hu, B.; Chen, J.; Li, Q. Retrieving crown leaf area index from an individual tree using ground-based lidar data. Can. J. Remote Sens. 2008, 34, 320–332. [Google Scholar] [CrossRef]
- Putman, E.B.; Popescu, S.; Eriksson, M.; Zhou, T.; Klockow, P.A.; Vogel, J.G.; Moore, G.W. Detecting and quantifying standing dead tree structural loss with reconstructed tree models using voxelized terrestrial lidar data. Remote Sens. Environ. 2018, 209, 52–65. [Google Scholar] [CrossRef]
- Kato, A.; Kajiwara, K.; Honda, Y.; Watanabe, M.; Enoki, T.; Yamaguchi, Y.; Kobayashi, T. Efficient field data collection of tropical forest using terrestrial laser scanner. In Proceedings of the 2014 IEEE Geoscience and Remote Sensing Symposium, Quebec City, QC, Canada, 13–18 July 2014; pp. 816–819. [Google Scholar] [CrossRef]
- Xi, Z.; Hopkinson, C.; Chasmer, L. Automating plot-level stem analysis from terrestrial laser scanning. Forests 2016, 7, 252. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Feng, Z.; Chen, P.; Khan, T.U.; Lian, Y. Nondestructive estimation of the above-ground biomass of multiple tree species in boreal forests of china using terrestrial laser scanning. Forests 2019, 10, 936. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Zhou, G.; Wei, H.; Zhang, X. Estimation of the Plot-Level Forest Parameters from Terrestrial Laser Scanning Data. In Proceedings of the IGARSS 2018-2018 IEEE International Geoscience and Remote Sensing Symposium, Valencia, Spain, 22–27 July 2018; IEEE: New York, NY, USA, 2018; pp. 9014–9017. [Google Scholar]
- Tao, S.; Wu, F.; Guo, Q.; Wang, Y.; Li, W.; Xue, B.; Hu, X.; Li, P.; Tian, D.; Li, C.; et al. Segmenting tree crowns from terrestrial and mobile LiDAR data by exploring ecological theories. ISPRS J. Photogramm. Remote Sens. 2015, 110, 66–76. [Google Scholar] [CrossRef] [Green Version]
- Vierling, L.A.; Xu, Y.; Eitel, J.U.H.; Oldow, J.S. Shrub characterization using terrestrial laser scanning and implications for airborne LiDAR assessment. Can. J. Remote Sens. 2013, 38, 709–722. [Google Scholar] [CrossRef]
- Zhang, W.; Wan, P.; Wang, T.; Cai, S.; Chen, Y.; Jin, X.; Yan, G. A Novel Approach for the Detection of Standing Tree Stems from Plot-Level Terrestrial Laser Scanning Data. Remote Sens. 2019, 11, 211. [Google Scholar] [CrossRef] [Green Version]
- Gollob, C.; Ritter, T.; Wassermann, C.; Nothdurft, A. Influence of scanner position and plot size on the accuracy of tree detection and diameter estimation using terrestrial laser scanning on forest inventory plots. Remote Sens. 2019, 11, 1602. [Google Scholar] [CrossRef] [Green Version]
- Reddy, R.S.; Jha, C.S.; Rajan, K.S. Automatic Tree Identification and Diameter Estimation Using Single Scan Terrestrial Laser Scanner Data in Central Indian Forests. J. Indian Soc. Remote Sens. 2018, 46, 937–943. [Google Scholar] [CrossRef]
- FAO. Knowledge Reference for National Forest Assessments—Modeling for Estimation and Monitoring. 2020. Available online: http://www.fao.org/forestry/17109/en/ (accessed on 3 December 2020).
- Kaasalainen, S.; Krooks, A.; Liski, J.; Raumonen, P.; Kaartinen, H.; Kaasalainen, M.; Puttonen, E.; Anttila, K.; Mäkipää, R. Change detection of tree biomass with terrestrial laser scanning and quantitative structure modelling. Remote Sens. 2014, 6, 3906–3922. [Google Scholar] [CrossRef] [Green Version]
- Disney, M.I.; Vicari, M.B.; Burt, A.; Calders, K.; Lewis, S.L.; Raumonen, P.; Wilkes, P. Weighing trees with lasers: Advances, challenges and opportunities. Interface Focus 2018, 8, 20170048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raumonen, P.; Kaasalainen, M.; Åkerblom, M.; Kaasalainen, S.; Kaartinen, H.; Vastaranta, M.; Holopainen, M.; Disney, M.I.; Lewis, P. Fast Automatic Precision Tree Models from Terrestrial Laser Scanner Data. Remote Sens. 2013, 5, 491–520. [Google Scholar] [CrossRef] [Green Version]
- Madhibha, T.P. ASSESSMENT OF ABOVE GROUND BIOMASS WITH TERRESTRIAL LiDAR USING 3D QUANTITATIVE STRUCTURE MODELLING IN TROPICAL RAIN FOREST OF AYER HITAM FOREST RESERVE, MALAYSIA. University of Twente Faculty of Geo-Information and Earth Observation (ITC). Available online: http://www.itc.nl/library/papers_2016/msc/nrm/madhibha.pdf (accessed on 28 February 2016).
- Olofsson, K.; Holmgren, J. Single tree stem profile detection using terrestrial laser scanner data, flatness saliency features and curvature properties. Forests 2016, 7, 207. [Google Scholar] [CrossRef]
- Drake, J.B.; Knox, R.G.; Dubayah, R.O.; Clark, D.B.; Condit, R.; Blair, J.B.; Hofton, M. Above-ground biomass estimation in closed canopy Neotropical forests using lidar remote sensing: Factors. Glob. Ecol. Biogeogr. 2003, 12, 147–159. [Google Scholar] [CrossRef]
- Li, A.; Glenn, N.F.; Olsoy, P.J.; Mitchell, J.J.; Shrestha, R. Aboveground biomass estimates of sagebrush using terrestrial and airborne LiDAR data in a dryland ecosystem. Agric. For. Meteorol. 2015, 213, 138–147. [Google Scholar] [CrossRef]
- Olsoy, P.J.; Mitchell, J.J.; Levia, D.F.; Clark, P.E.; Glenn, N.F. Estimation of big sagebrush leaf area index with terrestrial laser scanning. Ecol. Indic. 2016, 61, 815–821. [Google Scholar] [CrossRef]
- McElhinny, C.; Gibbons, P.; Brack, C.; Bauhus, J. Forest and woodland stand structural complexity: Its definition and measurement. For. Ecol. Manag. 2005, 218, 1–24. [Google Scholar] [CrossRef]
- Côté, J.-F.; Fournier, R.A.; Frazer, G.W.; Niemann, K.O. A fine-scale architectural model of trees to enhance LiDAR-derived measurements of forest canopy structure. Agric. For. Meteorol. 2012, 166–167, 72–85. [Google Scholar] [CrossRef]
- Maltamo, M. Estimation of timber volume and stem density based on scanning laser altimetry and expected tree size distribution functions. Remote Sens. Environ. 2004, 90, 319–330. [Google Scholar] [CrossRef]
- Decuyper, M.; Mulatu, K.A.; Brede, B.; Calders, K.; Armston, J.; Rozendaal, D.M.; Mora, B.; Clevers, J.G.; Kooistra, L.; Herold, M.; et al. Assessing the structural differences between tropical forest types using Terrestrial Laser Scanning. For. Ecol. Manag. 2018, 429, 327–335. [Google Scholar] [CrossRef]
- Kelbe, D.; Romanczyk, P.; Van Aardt, J.; Cawse-Nicholson, K. Reconstruction of 3D tree stem models from low-cost terrestrial laser scanner data. Laser Radar Technol. Appl. XVIII 2013, 8731, 873106. [Google Scholar] [CrossRef]
- Paynter, I.; Saenz, E.; Genest, D.; Peri, F.; Erb, A.; Li, Z.; Wiggin, K.; Muir, J.; Raumonen, P.; Schaaf, E.S.; et al. Observing ecosystems with lightweight, rapid-scanning terrestrial lidar scanners. Remote Sens. Ecol. Conserv. 2016, 2, 174–189. [Google Scholar] [CrossRef]
- Kelbe, D.; Romanczyk, P. Automatic extraction of tree stem models from single terrestrial lidar scans in structurally heterogeneous forest environments. Proc. Silvilaser 2012, 2012, 1–8. [Google Scholar]
- Mukuralinda, A.; Kuyah, S.; Ruzibiza, M.; Ndoli, A.; Nabahungu, N.L.; Muthuri, C. Allometric equations, wood density and partitioning of aboveground biomass in the arboretum of Ruhande, Rwanda. Trees For. People 2021, 3, 100050. [Google Scholar] [CrossRef]
- Wang, D.; Hollaus, M.; Puttonen, E.; Pfeifer, N. Fast and robust stem reconstruction in complex environments using terrestrial laser scanning. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, 41, 411–417. [Google Scholar] [CrossRef]
- Tian, J.; Dai, T.; Li, H.; Liao, C.; Teng, W.; Hu, Q.; Ma, W.; Xu, Y. A Novel Tree Height Extraction Approach for Individual Trees by Combining TLS and UAV Image-Based Point Cloud Integration. Forests 2019, 10, 537. [Google Scholar] [CrossRef] [Green Version]
- Martens, S.N.; Ustin, S.L.; Rousseau, R.A. Estimation of tree canopy leaf area index by gap fraction analysis. For. Ecol. Manag. 1993, 61, 91–108. [Google Scholar] [CrossRef]
- Lau, A.; Bartholomeus, H.; Herold, M.; Martius, C.; Malhi, Y.; Patrick Bentley, L.; Shenkin, A.; Raumonen, P. Application of terrestrial LiDAR and modelling of tree branching structure for plantscaling models in tropical forest trees. Proc. SilviLaser 2015, 2015, 96–98. [Google Scholar]
- Raumonen, P.; Kaasalainen, S.; Kaartinen, H. Approximation of Volume and Branch Size Distribution of Trees from Laser Scanner Data. ISPRS—Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2012, 5, 79–84. [Google Scholar] [CrossRef] [Green Version]
- Levick, S.R.; Asner, G.P. The rate and spatial pattern of treefall in a savanna landscape. Biol. Conserv. 2013, 157, 121–127. [Google Scholar] [CrossRef]







| Search Aspect | Search Terms |
|---|---|
| Vegetation type | forest *; grassland *; savanna *; shrub *; woodland * |
| TLS | ground-based lidar; lidar; terrestrial laser scann *; terrestrial lidar; TLS |
| Extraction of vegetation parameters | 3D reconstruction; reconstruction; stem; tree parameter; tree structur *; |
| TLS experimental setup | full waveform; intensity; multi-temporal; reflection |
| TLS as auxiliary data | calibration; validation |
| Riegl VZ 400 | Leica HDS6100 | Riegl VZ 1000 | Faro Focus 3D | Faro Focus 3D X 330 | |
|---|---|---|---|---|---|
| Range finder | Time of flight | Phase shift | Time of flight | Phase shift | Phase shift |
| Range of Measurement [m] | 1.5–600 | 79 | 2.5–1400 | 0.6–120 | 0.6–330 |
| Scan angle range [°] | 100/360 | 360/310 | 100/360 | 305/360 | 300/360 |
| Accuracy [mm] | 5 | ±2 at 25 m | 8 | ±2 | ±2 |
| Beam divergence[mrad] | 0.35 | 0.22 | 0.3 | 0.19 | 0.19 |
| Laser wavelength[nm] | 1550 | 690 | 1550 | 905 | 1550 |
| Effective measurement rate [points/sec] | 42,000–122,000 | 508,000 | 42,000–122,000 | 122,000–976,000 | 122,000–976,000 |
| Angel measurement resolution[°] | 0.0005° (1.8arc sec) | 0.009° Hor × 0.009° Ver | 0.0005° (1.8arc sec) | 0.011° | 0.011° |
| Weight[kg] | 9.6 | 14 | 9.8 | 5.0 | 5.2 |
| Reference | [116] | [117] | [118] | [119] | [120] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muumbe, T.P.; Baade, J.; Singh, J.; Schmullius, C.; Thau, C. Terrestrial Laser Scanning for Vegetation Analyses with a Special Focus on Savannas. Remote Sens. 2021, 13, 507. https://doi.org/10.3390/rs13030507
Muumbe TP, Baade J, Singh J, Schmullius C, Thau C. Terrestrial Laser Scanning for Vegetation Analyses with a Special Focus on Savannas. Remote Sensing. 2021; 13(3):507. https://doi.org/10.3390/rs13030507
Chicago/Turabian StyleMuumbe, Tasiyiwa Priscilla, Jussi Baade, Jenia Singh, Christiane Schmullius, and Christian Thau. 2021. "Terrestrial Laser Scanning for Vegetation Analyses with a Special Focus on Savannas" Remote Sensing 13, no. 3: 507. https://doi.org/10.3390/rs13030507