1. Introduction
Precision agriculture (PA) is a farming management technique that requires detailed information on crop status. One of the important crop status indicators is the crop nitrogen (N) weight because nitrogen is the main plant nutrient needed for producing the chlorophyll, which has a direct impact on the plant photosynthesis and thus on crop growth and yield [
1]. Therefore, there is a need to understand the spatial distribution of crop nitrogen for better use of fertilizers. Ultimately, this information leads to a better yield among the crops and reduces costs for the farmer by matching the fertilizer supply to its demand [
2,
3]. Traditionally, farmers used to rely on historical weather data, such as precipitation and temperature, and their past experiences, such as crop yields, to make decisions on their fertilizer operations for the upcoming season [
4]. Today, there have been many advances in technology, such as remote sensing data and machine learning algorithms, that can potentially aid farmers’ decision making on fertilizer application. Remote sensing-based methods used to measure crop nitrogen are typically better than the traditional ground-based methods. Ground-based methods require intensive field data collection, which can be time-consuming, destructive, and limited to a small spatial area, making it impractical for fast and efficient results. Remote sensing-based methods are required for most agricultural fields in Canada, given that they can reach up to hundreds of ha in size. Remote sensing methods are non-destructive, can cover large spatial areas, and have been increasingly used for crop monitoring in precision agriculture. Crop monitoring based on remote sensing data can use spaceborne or airborne images, but these types of high-resolution imagery are either costly or difficult to obtain [
5]. Also, they have limited applicability in precision agriculture because of the too coarse temporal and spatial resolutions of the imagery [
6,
7]. Alternatively, free-of-charge Sentinel-1 Synthetic Aperture Radar (SAR) imagery could be used to monitor nitrogen status but does not provide enough spatial resolution (10 m) for precise small-scaled applications [
8]. Bagheri et al. [
9] also found it difficult to differentiate levels of nitrogen status in corn fields using a 15 m Advanced Spaceborne Thermal Emission and Reflection Radiometer (ASTER) multispectral satellite imagery. An alternative is to use Unmanned Aerial Vehicle (UAV) imagery. The development and application of UAV imagery have increased in the past decade, filling in gaps between satellite imagery, aerial photography, and field samples [
10,
11]. Image acquisition with UAV can be deployed quickly and repeatedly, at a low cost, and with greater flexibility [
12,
13]. The temporal resolution of UAVs is superior to the satellite and aerial photography platforms, which is easily defined by the user [
14,
15]. The low cost of UAVs could also be convinced without the use of purchasing ground control points (GCPs) [
14].
The focus of this study is to test whether UAV multispectral imagery can be used to retrieve the crop nitrogen status over corn fields from a perspective of precision nitrogen fertilization. The spatial and temporal variations of the images we acquired were determined in order to match the crop requirements of nitrogen as closely as possible. One common remote sensing technique to estimate nitrogen content at the canopy level is the Radiative Transfer Model (RTM), which estimates the chlorophyll or nitrogen content by describing the interaction between the sun’s light and the crop canopy. An example of an RTM is the PROSAIL model, which uses various parameters at the leaf and canopy level and can be mathematically inverted to retrieve both chlorophyll and nitrogen content from spectral data [
1,
16,
17,
18,
19]. Other sets of remote sensing methods are empirical methods, such as machine learning algorithms, or simple/multiple-linear regression to retrieve crop nitrogen from canopy spectral data [
16]. This paper used the second set of methods because empirical estimation has already been proven to be easier in estimating nitrogen than the PROSAIL model, given that an RTM requires the calibration of numerous parameters [
20]. In this study, we tested three empirical methods: (1) linear regression, (2) Random Forests, and (3) support vector regression (SVR) to statistically relate spectral measurements and canopy-level crop nitrogen weight in the case of two corn fields located in southwest Ontario, Canada. Many studies on predicting nitrogen were conducted in a controlled experimental condition and predicted nitrogen values at the leaf level [
21,
22,
23,
24], while few studies have been performed on real field conditions [
25]. As already shown in Liu et al. [
3], it is better to use more than one spectral variable for estimating crop nitrogen. Therefore, multiple linear regression, Random Forests, and SVR was used in this paper to predict canopy nitrogen using a variety of spectral variables.
The first regression method is a traditional regression approach that has two major assumptions (multicollinearity and linear relationship). Most studies using UAV imagery have used linear regressions to estimate nitrogen status from vegetation indices (VIs) [
13]. The latter two approaches are machine learning techniques that have been advanced in recent years and are unaffected by the multicollinearity and linear relationship assumptions. In addition, they can handle overfitting [
13,
26,
27]. Overfitting is a common problem in machine learning where the models produced perform poorly on unseen data. Random Forests has become popular recently within the remote sensing research community for classification and regression purposes. The variable importance plot provided by the Random Forests algorithm is very successful in identifying the most relevant input data in the model [
27,
28]. Therefore, we used the Random Forests variable importance plots to identify the most important variables for canopy nitrogen estimation and adjusted the model parameters accordingly. Random Forests modeling has been found to perform very well out of all the non-parametric methods in various studies to monitor nitrogen content in wheat [
3], rice [
13], and citrus trees [
28]. Random Forests was also the most accurate machine learning approach for monitoring corn yield, which is related to crop nitrogen status [
26]. Support vector modeling has also been popular in estimating nitrogen status due to its strong ability to decorrelate the input variables and can work with non-linear relationships [
29,
30,
31]. SVR modeling has been found to predict nitrogen concentration with high accuracy in bokchoy [
29], wheat [
30], and corn [
31].
Ideally, the relationship between the spectral data and the canopy nitrogen weight should be linear because there should be only one canopy nitrogen weight estimation for each level of input data, given that the relationship will be used to calibrate an N fertilizer sprayer that needs to have an exact determination of the crop N fertilization level. However, when the canopy becomes dense, the relationship can saturate and become non-linear. This is the case when using some VIs, such as the standard normalized difference vegetation index (NDVI), green NDVI, red edge NDVI, and modified transformed vegetation index 2 (MTVI2), which has been shown to saturate at high canopy densities [
32]. Therefore, this study tested the ability of Random Forests and SVR to work with non-linear saturated data, particularly when combining datasets throughout the growing cycle.
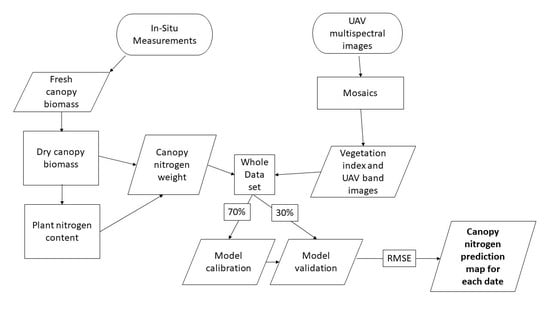
The best performing model was applied to the UAV imagery for mapping crop nitrogen content at the field level. Deng et al. [
33] found that mounting narrowband multispectral cameras on UAVs acquire images with far better quality than broadband multispectral cameras. Therefore, the objectives of this study are to (i) generate machine learning models to predict crop nitrogen weight in corn fields using UAV multispectral imagery, (ii) determine which individual MicaSense spectral bands and VIs have the most influence on the Random Forests decision tree when predicting nitrogen, (iii) generate nitrogen prediction maps by applying the best model to the entire UAV image and analyze whether the UAV images are able to detect any spatial variation of nitrogen within the fields. The study evaluates three different modeling approaches for predicting nitrogen in corn over different dates and growth stages using UAV multispectral imagery. The best algorithm not only fills the research gap between monitoring nitrogen and UAVs, but also has practical meaning for future modeling study designs. Ultimately, these images should be given to farmers in highly accurate, quick, and timely manner field information for their precision nitrogen fertilization management.
4. Discussion
Our study used five different MicaSense multispectral bands to derive various VIs to predict canopy nitrogen weight. Values of the in-situ canopy nitrogen weight saw an increase in variation on 10 July and 18 July. This is due to the rapid increase in biomass between the dates. This could be explained by the crop biomass variation increasing due to the factors that contribute to the crop’s growth, such as the absorption, utilization, and transformation of solar energy; climate; and nutrient/water management [
69,
70].
Individual bands were tested to predict canopy nitrogen in all the models along with the VIs. Both the MicaSense green band and red-edge band performed poorly in the model compared to the other bands. The green wavelength is closely related to the leaf chlorophyll
a and
b contents (Zhao et al. [
71]) in which nitrogen is used for plant photosynthesis. The poor performance in the model may be due to the chlorophyll saturating in the middle to the late growth stages, causing the crops to reflect the same amount of green wavelength. However, our results are not in agreement with Schlemmer et al. [
72] and Li et al. [
73], who observed a good relationship between the green reflectance and corn nitrogen weight. The red-edge spectral region is an interesting region, in which the position of the sharp change in reflectance (known as the red-edge position) is particularly known to be a sensitive indicator of leaf chlorophyll content [
74,
75,
76]. The red-edge position changes in the wavelength of 680–800 nm depending on the strength of the absorption of chlorophyll [
77]. Therefore, the narrowband of 10nm in the MicaSense red-edge band may have not fully captured the red-edge position throughout the growing season, whereas another sensor with different band ranges could have captured it. A possible consideration in the future would be to fly two cameras simultaneously and compare the results. Furthermore, the red-edge reflectance has been also found to be significantly related to corn nitrogen weight in Schlemmer et al. [
72] and Li et al. [
73]. Such difference in both green and red-edge reflectance can be explained by the fact that their study focused more on predicting nitrogen at individual growth stages, while our study considered all the growth stages in the model. All the top six VIs in the variable importance table use the near-infrared and red bands, indicating that they are both critical to the prediction of nitrogen. This is probably due to the chlorophyll absorption present in the red region and high reflectance of near-infrared energy for leaf development of healthy vegetation. However, the near-infrared band alone does not have a good relationship with canopy nitrogen weight, and therefore it must be included with other bands under the form of VIs. Interestingly, NDVI (the most commonly used VI in literature) did not perform as well as the top VIs in the variable importance plot. This is because NDVI is known to saturate with canopy nitrogen weight once the canopy of the crop becomes dense [
32].
The use of the variable importance plot in Random Forests to eliminate spectral variable inputs was also performed in Osco et al. [
28] on predicting canopy nitrogen in citrus trees. The authors used the top five and ten variables of 33 spectral variables and found there was a slight decrease in the model performance. Similar to our results, Osco et al. [
28] found a decrease in performance relatively small, and the trade-off between the number of spectral indices used and obtained accuracy is something that should be considered. However, Osco et al. [
28] used a different list of VIs, but the application of the variable importance plot was the same. Random Forests performed poorly on corn in Fan et al. [
24] (R
2 = 0.60) compared to partial least squares regression (R
2 = 0.80) on the validation dataset. However, the authors used nitrogen content percentage at the leaf level and found weak correlation with most of the spectral variables, while our study incorporated our nitrogen values at the canopy level (g/m
2). Random Forests with spectral variables on the validation set performed much better at the canopy level (R
2 = 0.85), probably due to the spectral variables having a good correlation with nitrogen at the canopy level.
Our study on SVR modeling lines up with the results of Karimi et al. [
31]. The authors found that SVR performed better and more consistent than its multiple linear regression counterpart in their study. The difference in results of Random Forests and SVR in our study are not too far apart in model performance. However, the concepts and outputs of Random Forests are a lot easier to interpret than the concepts and outputs of support vector machines.
Zha et al. [
13] found Random Forests performed better than SVR, multiple linear regression and artificial neural networks on predicting nitrogen content in rice using spectral indices. A model comparison study using spectral indices in Liu et al. [
3] also found Random Forests to perform better than the other non-parametric machine learning models in wheat. This could mean that the performance of Random Forests on nitrogen using spectral indices could be consistent on other types of crops and possibly give consistent results in different regions of Canada. Since different regions provide different climates, a comparison of the soil and nitrogen status in the crops could be studied. A consideration of future study could involve comparing Random Forests to other machine learning models or deep learning using spectral indices on other types of crops. However, delving into deep learning requires a huge training dataset in order to be effective. Another drawback is the computational cost such as memory and processing power in order to tackle the datasets effectively with deep learning.
5. Conclusions
In this study, different regression methods were used to predict canopy nitrogen weight of corn using UAV MicaSense multispectral images. These models were established using the individual MicaSense bands and their associated VIs derived from the UAV reflectance values. Using the top 12 variables (in order: MSR, WDRVI, RVI, NDVI, ISR, BNDVI, Red band, OSAVI, RGBVI, CI Red edge, Blue band, and Red edge NDVI) derived with the Random Forests, the importance plot performed the best on estimating canopy nitrogen weight throughout the three dates in corn crops. Using the Random Forests model applied to the top 12 variables (RMSE = 4.52 g/m2) on the validation performed marginally worse than the Random Forests model using all the variables (RMSE = 4.51 g/m2), indicating that adding more variables into the model does not always improve its accuracy. However, because the difference of the accuracy is marginally different, this removes the unnecessary processing time of generating the 22 other VI images.
The UAV nitrogen prediction map can also detect spatial nitrogen variations within the field, especially in the 18 July image where the canopy nitrogen weight showed a large variation with the field data. In practice, these results could be useful for farmers in retrieving fast information about a field’s nitrogen status, as they will know exactly which parts of their fields are in excess or deficient in the amount of nitrogen present. Practically, these results could be obtained on the day of the UAV flight, depending on the size of the field and the number of images acquired. Ultimately, this will lead to a much more efficient fertilizer application program for the farmers as they will know precisely how much nitrogen is needed in a particular spot with their GPS-enabled fertilizer spreader.
This study used data acquired over southwest Ontario fields in 2019 and there is the need to test the method over other datasets, such as different zones in Canada or a different crop. This will give an idea of how the developed method can be generalized and applied to different parts of Canada and whether it can be used on different crops. The study used MicaSense images with five spectral bands and there is a need to test different cameras that capture different wavelengths to understand which multispectral bands perform the best on predicting nitrogen using empirical regression techniques. Finally, another future consideration of this study can involve comparing the canopy nitrogen prediction map with other field spatial information, such as drainage and soil. This information can give a better idea on the contributions of nitrogen content that occur below the canopy.