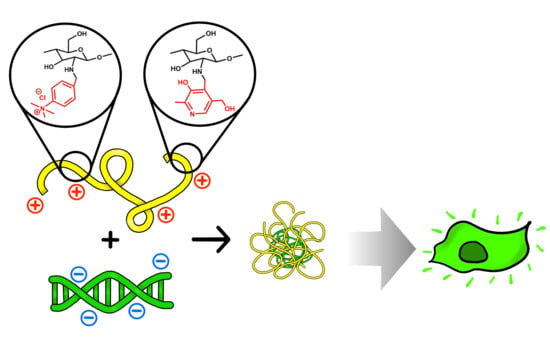
Effect of Double Substitution in Cationic Chitosan Derivatives on DNA Transfection Efficiency
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of n-[(3-hydroxy-5-(hydroxymethyl)-2-methyl-4-pyridine)methyl]chitosan Chloride (Pyr-CS)
2.3. Synthesis of n-[4-(n’,n’,n’-trimethylammonium)benzyl]chitosan Chloride (TMAB-CS)
2.4. Synthesis of n-[4-(n’,n’,n’-trimethylammonium)benzyl]-N-[(3-hydroxy-5-(hydroxymethyl)-2-methyl-4-pyridine)methyl]chitosan Chloride (PyrTMAB-CS)
2.5. Characterization of Polymers
2.6. Spectrophotometric Measurements and pKa Calculations
2.7. Preparation of Polymer: DNA Polyplexes
2.8. Dynamic Light Scattering (DLS)
2.9. Polymer:DNA Binding Assay
2.10. Atomic Force Microscopy (AFM)
2.11. Transfection of the HEK293 Cell Line
2.12. Statistical Analysis
3. Results and Discussion
3.1. Synthesis and NMR Characterization of Cationic Chitosan Derivatives
3.2. Spectrophotometric pKa Determination
3.3. Preparation and Hydrodynamic Parameters of Polymer:DNA Polyplexes
3.4. DNA Binding Assay
3.5. AFM Morphology of Polymer:DNA Polyplexes
3.6. Transfection of the HEK293 Cell Line
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Naldini, L. Gene therapy returns to centre stage. Nature 2015, 526, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Gardlík, R.; Pálffy, R.; Hodosy, J.; Lukács, J.; Turna, J.; Celec, P. Vectors and delivery systems in gene therapy. Med Sci. Monit. Int. Med. J. Exp. Clin. Res. 2005, 11, RA110–RA121. [Google Scholar]
- Nayak, S.; Herzog, R.W. Progress and prospects: Immune responses to viral vectors. Gene Ther. 2010, 17, 295–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marshall, E. Gene therapy death prompts review of adenovirus vector. Science 1999, 286, 2244–2245. [Google Scholar] [CrossRef] [PubMed]
- Hacein-Bey-Abina, S.; Von Kalle, C.; Schmidt, M.; Le Deist, F.; Wulffraat, N.; McIntyre, E.; Radford, I.; Villeval, J.-L.; Fraser, C.C.; Cavazzana-Calvo, M. A serious adverse event after successful gene therapy for x-linked severe combined immunodeficiency. N. Engl. J. Med. 2003, 348, 255–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, Y.; Yue, Y.; Duan, D. Evidence for the failure of adeno-associated virus serotype 5 to package a viral genome ≥8.2 kb. Mol. Ther. 2010, 18, 75–79. [Google Scholar] [CrossRef]
- Ghosh, A.; Duan, D. Expanding adeno-associated viral vector capacity: A tale of two vectors. Biotechnol. Genet. Eng. Rev. 2007, 24, 165–178. [Google Scholar] [CrossRef] [Green Version]
- Chira, S.; Jackson, C.S.; Oprea, I.; Ozturk, F.; Pepper, M.S.; Diaconu, I.; Braicu, C.; Raduly, L.-Z.; Calin, G.A.; Berindan-Neagoe, I. Progresses towards safe and efficient gene therapy vectors. Oncotarget 2015, 6, 30675–30703. [Google Scholar] [CrossRef] [Green Version]
- Zylberberg, C.; Gaskill, K.; Pasley, S.; Matosevic, S. Engineering liposomal nanoparticles for targeted gene therapy. Gene Ther. 2017, 24, 441–452. [Google Scholar] [CrossRef]
- Lächelt, U.; Wagner, E. Nucleic acid therapeutics using polyplexes: A journey of 50 years (and beyond). Chem. Rev. 2015, 115, 11043–11078. [Google Scholar] [CrossRef]
- Xue, H.Y.; Liu, S.; Wong, H.L. Nanotoxicity: A key obstacle to clinical translation of sirna-based nanomedicine. Nanomedicine 2014, 9, 295–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, R.C.F.; Ng, T.B.; Wong, J.H.; Chan, W.Y. Chitosan: An update on potential biomedical and pharmaceutical applications. Mar. Drugs 2015, 13, 5156–5186. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.Q.; Roy, K.; Troung-Le, V.L.; Janes, K.A.; Lin, K.Y.; Wang, Y.; August, J.T.; Leong, K.W. Chitosan-DNA nanoparticles as gene carriers: Synthesis, characterization and transfection efficiency. J. Control. Release Off. J. Control. Release Soc. 2001, 70, 399–421. [Google Scholar] [CrossRef]
- Ishii, T.; Okahata, Y.; Sato, T. Mechanism of cell transfection with plasmid/chitosan complexes. Biochim. Biophys. Acta 2001, 1514, 51–64. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Tan, Y.F.; Wong, Y.S.; Liew, M.W.J.; Venkatraman, S. Recent advances in chitosan-based carriers for gene delivery. Mar. Drugs 2019, 17, 381. [Google Scholar] [CrossRef] [Green Version]
- Kritchenkov, A.S.; Andranovitš, S.; Skorik, Y.A. Chitosan and its derivatives: Vectors in gene therapy. Russ. Chem. Rev. 2017, 86, 231. [Google Scholar] [CrossRef]
- Jiang, H.-L.; Xing, L.; Luo, C.-Q.; Zhou, T.-J.; Li, H.-S.; Cho, C.-S. Chemical modification of chitosan as a gene transporter. Curr. Org. Chem. 2018, 22, 668–689. [Google Scholar] [CrossRef]
- Rabea, E.I.; Badawy, M.E.; Steurbaut, W.; Stevens, C.V. In vitro assessment of N-(benzyl)chitosan derivatives against some plant pathogenic bacteria and fungi. Eur. Polym. J. 2009, 45, 237–245. [Google Scholar] [CrossRef]
- Raik, S.V.; Poshina, D.N.; Lyalina, T.A.; Polyakov, D.S.; Vasilyev, V.B.; Kritchenkov, A.S.; Skorik, Y.A. N-[4-(n,n,n-trimethylammonium)benzyl]chitosan chloride: Synthesis, interaction with DNA and evaluation of transfection efficiency. Carbohydr. Polym. 2018, 181, 693–700. [Google Scholar] [CrossRef]
- Raik, S.V.; Andranovitš, S.; Petrova, V.A.; Xu, Y.; Lam, J.K.-W.; Morris, G.A.; Brodskaia, A.V.; Casettari, L.; Kritchenkov, A.S.; Skorik, Y.A. Comparative study of diethylaminoethyl-chitosan and methylglycol-chitosan as potential non-viral vectors for gene therapy. Polymers 2018, 10, 442. [Google Scholar] [CrossRef] [Green Version]
- Sajomsang, W.; Ruktanonchai, U.; Gonil, P.; Mayen, V.; Opanasopit, P. Methylated N-aryl chitosan derivative/DNA complex nanoparticles for gene delivery: Synthesis and structure–activity relationships. Carbohydr. Polym. 2009, 78, 743–752. [Google Scholar] [CrossRef]
- Sajomsang, W.; Gonil, P.; Ruktanonchai, U.R.; Petchsangsai, M.; Opanasopit, P.; Puttipipatkhachorn, S. Effect of N-pyridinium positions of quaternized chitosan on transfection efficiency in gene delivery system. Carbohydr. Polym. 2014, 104, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Du, Y.-Z.; Chen, F.-Y.; You, J.; Yuan, H.; Hu, F.-Q. Effect of proteins with different isoelectric points on the gene transfection efficiency mediated by stearic acid grafted chitosan oligosaccharide micelles. Mol. Pharm. 2013, 10, 2568–2577. [Google Scholar] [CrossRef] [PubMed]
- Inamdar, N.; Mourya, V.K. Chitosan and anionic polymers—Complex formation and applications. In Polysaccharides: Development, Properties and Applications; Nova Science Publishers: Hauppauge, NY, USA, 2011; pp. 333–377. [Google Scholar]
- Pogodina, N.; Pavlov, G.; Bushin, S.; Mel’Nikov, A.; Lysenko, Y.B.; Nud’Ga, L.; Marsheva, V.; Marchenko, G.; Tsvetkov, V. Conformational characteristics of chitosan molecules as demonstrated by diffusion-sedimentation analysis and viscometry. Polym. Sci. USSR 1986, 28, 251–259. [Google Scholar] [CrossRef]
- Köping-Höggård, M.; Mel’nikova, Y.S.; Vårum, K.M.; Lindman, B.; Artursson, P. Relationship between the physical shape and the efficiency of oligomeric chitosan as a gene delivery system in vitro and in vivo. J. Gene Med. Cross Discip. J. Res. Sci. Gene Transf. Its Clin. Appl. 2003, 5, 130–141. [Google Scholar]
- Zhang, H.; Neau, S.H. In vitro degradation of chitosan by bacterial enzymes from rat cecal and colonic contents. Biomaterials 2002, 23, 2761–2766. [Google Scholar] [CrossRef]
- Weecharangsan, W.; Opanasopit, P.; Ngawhirunpat, T.; Apirakaramwong, A.; Rojanarata, T.; Ruktanonchai, U.; Lee, R.J. Evaluation of chitosan salts as non-viral gene vectors in cho-k1 cells. Int. J. Pharm. 2008, 348, 161–168. [Google Scholar] [CrossRef]
- Strand, S.P.; Danielsen, S.; Christensen, B.E.; Vårum, K.M. Influence of chitosan structure on the formation and stability of DNA−chitosan polyelectrolyte complexes. Biomacromolecules 2005, 6, 3357–3366. [Google Scholar] [CrossRef]
- Rejman, J.; Bragonzi, A.; Conese, M. Role of clathrin-and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Mol. Ther. 2005, 12, 468–474. [Google Scholar] [CrossRef]
- Rehman, Z.U.; Hoekstra, D.; Zuhorn, I.S. Mechanism of polyplex-and lipoplex-mediated delivery of nucleic acids: Real-time visualization of transient membrane destabilization without endosomal lysis. ACS Nano 2013, 7, 3767–3777. [Google Scholar] [CrossRef]
- Behr, J.-P. The proton sponge: A trick to enter cells the viruses did not exploit. CHIMIA Int. J. Chem. 1997, 51, 34–36. [Google Scholar]
- Richard, I.; Thibault, M.; De Crescenzo, G.; Buschmann, M.D.; Lavertu, M. Ionization behavior of chitosan and chitosan–DNA polyplexes indicate that chitosan has a similar capability to induce a proton-sponge effect as pei. Biomacromolecules 2013, 14, 1732–1740. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, L.M.; De Smedt, S.C.; Remaut, K.; Braeckmans, K. The proton sponge hypothesis: Fable or fact? Eur. J. Pharm. Biopharm. 2018, 129, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Sonawane, N.D.; Szoka, F.C.; Verkman, A. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J. Biol. Chem. 2003, 278, 44826–44831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sillén, L.G.; Martell, A.E. Stability Constants of Metal-Ion Complexes; The Chemical Society: London, UK, 1964; p. 754. [Google Scholar]
- Vrolijk, M.F.; Opperhuizen, A.; Jansen, E.; Hageman, G.J.; Bast, A.; Haenen, G. The vitamin B6 paradox: Supplementation with high concentrations of pyridoxine leads to decreased vitamin B6 function. Toxicology In Vitro 2017, 44, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Skorik, Y.A.; Gomes, C.A.; Vasconcelos, M.T.; Yatluk, Y.G. N-(2-carboxyethyl)chitosans: Regioselective synthesis, characterisation and protolytic equilibria. Carbohydr. Res. 2003, 338, 271–276. [Google Scholar] [CrossRef]
- Kudaibergenov, S.E. Behavior of polyampholytes in solutions. In Polyampholytes: Synthesis, Characterization and Application; Kudaibergenov, S.E., Ed.; Springer: Boston, MA, USA, 2002; pp. 43–89. [Google Scholar]
- Oliveira, F.d.P.P.; Dalla Picola, I.P.; Shi, Q.; Barbosa, H.F.G.; de Oliveira Tiera, V.A.; Fernandes, J.C.; Tiera, M.J. Synthesis and evaluation of diethylethylamine–chitosan for gene delivery: Composition effects on the in vitro transfection efficiency. Nanotechnology 2013, 24, 055101. [Google Scholar] [CrossRef]
- Danielsen, S.; Vårum, K.M.; Stokke, B.T. Structural analysis of chitosan mediated DNA condensation by afm: Influence of chitosan molecular parameters. Biomacromolecules 2004, 5, 928–936. [Google Scholar] [CrossRef]
- Danielsen, S.; Strand, S.; de Lange Davies, C.; Stokke, B.T. Glycosaminoglycan destabilization of DNA–chitosan polyplexes for gene delivery depends on chitosan chain length and gag properties. Biochim. Biophys. Acta Gen. Subj. 2005, 1721, 44–54. [Google Scholar] [CrossRef]
- Wan, L.; You, Y.; Zou, Y.; Oupický, D.; Mao, G. DNA release dynamics from bioreducible poly(amido amine) polyplexes. J. Phys. Chem. B 2009, 113, 13735–13741. [Google Scholar] [CrossRef] [Green Version]
- Wan, L.; Manickam, D.S.; Oupický, D.; Mao, G. DNA release dynamics from reducible polyplexes by atomic force microscopy. Langmuir 2008, 24, 12474–12482. [Google Scholar] [CrossRef] [PubMed]









| Compound | pKa1 | pKa3 |
|---|---|---|
| Pyr-CS | 5.5 | 10 |
| PyrTMAB-CS | 4.9 | - |
| Pyridoxal | 4.2 | 8.8 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Badazhkova, V.D.; Raik, S.V.; Polyakov, D.S.; Poshina, D.N.; Skorik, Y.A. Effect of Double Substitution in Cationic Chitosan Derivatives on DNA Transfection Efficiency. Polymers 2020, 12, 1057. https://doi.org/10.3390/polym12051057
Badazhkova VD, Raik SV, Polyakov DS, Poshina DN, Skorik YA. Effect of Double Substitution in Cationic Chitosan Derivatives on DNA Transfection Efficiency. Polymers. 2020; 12(5):1057. https://doi.org/10.3390/polym12051057
Chicago/Turabian StyleBadazhkova, Veronika D., Sergei V. Raik, Dmitry S. Polyakov, Daria N. Poshina, and Yury A. Skorik. 2020. "Effect of Double Substitution in Cationic Chitosan Derivatives on DNA Transfection Efficiency" Polymers 12, no. 5: 1057. https://doi.org/10.3390/polym12051057