Mimicking Elementary Reactions of Manganese Lipoxygenase Using Mn-hydroxo and Mn-alkylperoxo Complexes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Kinetic Studies of TEMPOH and Xanthene Oxidation by [MnIII(OH)(6Medpaq)](OTf)
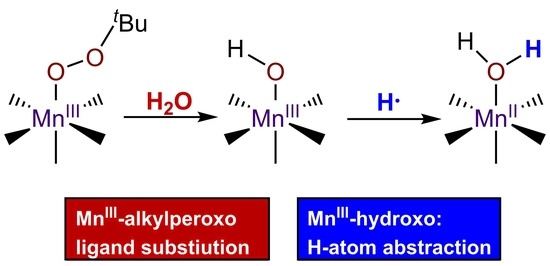
2.2. Reaction of [MnIII(OOtBu)(6Medpaq)]+ with Protic Solvents and Kinetic Investigations with Water
2.3. Cyclic Voltammetry
2.4. Electronic Structure Calculations
3. Results and Discussion
3.1. Formation of [MnIII(OH)(6Medpaq)]+ by Aerobic Oxidation
3.2. Spectroscopic Properties and Electronic Structure of [MnIII(OH)(6Medpaq)]+
3.3. Oxidative Reactivity of [MnIII(OH)(6Medpaq)](OTf)
3.4. Thermodynamic Driving Force for TEMPOH Oxidation Using Experimental and Computational Methods
3.5. Ligand Substitution Reactions of [MnIII(OOtBu)(6Medpaq)]+ with Water
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Brash, A.R. Lipoxygenases: Occurrence, Functions, Catalysis, and Acquisition of Substrate. J. Biol. Chem. 1999, 274, 23679–23682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliw, E.H.; Jernerén, F.; Hoffmann, I.; Sahlin, M.; Garscha, U. Manganese lipoxygenase oxidizes bis-allylic hydroperoxides and octadecenoic acids by different mechanisms. Biochim. Biophys. Acta (Bba)-Mol. Cell Biol. Lipids 2011, 1811, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Oliw, E.H. Manganese Lipoxygenase: Purification and Characterization. J. Biol. Chem. 1998, 273, 13072–13079. [Google Scholar] [CrossRef] [Green Version]
- Su, C.; Sahlin, M.; Oliw, E.H. Kinetics of Manganese Lipoxygenase with a Catalytic Mononuclear Redox Center. J. Biol. Chem. 2000, 275, 18830–18835. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Wennman, A.; Karkehabadi, S.; Engström, Å.; Oliw, E.H. Crystal structure of linoleate 13R-manganese lipoxygenase in complex with an adhesion protein1. J. Lipid Res. 2016, 57, 1574–1588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, W.; Richards, N.G.J. Biological functions controlled by manganese redox changes in mononuclear Mn-dependent enzymes. Essays Biochem. 2017, 61, 259–270. [Google Scholar] [PubMed] [Green Version]
- Wilson, R.A.; Talbot, N.J. Under pressure: Investigating the biology of plant infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 2009, 7, 185–195. [Google Scholar] [CrossRef]
- Wennman, A.; Jernerén, F.; Magnuson, A.; Oliw, E.H. Expression and characterization of manganese lipoxygenase of the rice blast fungus reveals prominent sequential lipoxygenation of α-linolenic acid. Arch. Biochem. Biophys. 2015, 583, 87–95. [Google Scholar] [CrossRef]
- Kostenko, A.; Ray, K.; Iavarone, A.T.; Offenbacher, A.R. Kinetic Characterization of the C–H Activation Step for the Lipoxygenase from the Pathogenic Fungus Magnaporthe oryzae: Impact of N-Linked Glycosylation. Biochemistry 2019, 58, 3193–3203. [Google Scholar] [CrossRef]
- Wennman, A.; Oliw, E.H.; Karkehabadi, S.; Chen, Y. Crystal Structure of Manganese Lipoxygenase of the Rice Blast Fungus Magnaporthe oryzae. J. Biol. Chem. 2016, 291, 8130–8139. [Google Scholar] [CrossRef] [Green Version]
- Wennman, A.; Karkehabadi, S.; Oliw, E.H. Kinetic investigation of the rate-limiting step of manganese- and iron-lipoxygenases. Arch. Biochem. Biophys. 2014, 555–556, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Klinman, J.P. Importance of Protein Dynamics during Enzymatic C–H Bond Cleavage Catalysis. Biochemistry 2013, 52, 2068–2077. [Google Scholar] [CrossRef] [PubMed]
- Glickman, M.H.; Klinman, J.P. Nature of Rate-Limiting Steps in the Soybean Lipoxygenase-1 Reaction. Biochemistry 1995, 34, 14077–14092. [Google Scholar] [CrossRef] [PubMed]
- Klinman, J.P.; Offenbacher, A.R. Understanding Biological Hydrogen Transfer Through the Lens of Temperature Dependent Kinetic Isotope Effects. Acc. Chem. Res. 2018, 51, 1966–1974. [Google Scholar] [CrossRef]
- Skrzypczak-Jankun, E.; Bross, R.A.; Carroll, R.T.; Dunham, W.R.; Funk, M.O. Three-Dimensional Structure of a Purple Lipoxygenase. J. Am. Chem. Soc. 2001, 123, 10814–10820. [Google Scholar] [CrossRef]
- Goldsmith, C.R.; Cole, A.P.; Stack, T.D.P. C−H Activation by a Mononuclear Manganese(III) Hydroxide Complex: Synthesis and Characterization of a Manganese-Lipoxygenase Mimic? J. Am. Chem. Soc. 2005, 127, 9904–9912. [Google Scholar] [CrossRef]
- Rice, D.B.; Wijeratne, G.B.; Burr, A.D.; Parham, J.D.; Day, V.W.; Jackson, T.A. Steric and Electronic Influence on Proton-Coupled Electron-Transfer Reactivity of a Mononuclear Mn(III)-Hydroxo Complex. Inorg. Chem. 2016, 55, 8110–8120. [Google Scholar] [CrossRef]
- Wijeratne, G.B.; Corzine, B.; Day, V.W.; Jackson, T.A. Saturation Kinetics in Phenolic O–H Bond Oxidation by a Mononuclear Mn(III)–OH Complex Derived from Dioxygen. Inorg. Chem. 2014, 53, 7622–7634. [Google Scholar] [CrossRef]
- Rice, D.B.; Massie, A.A.; Jackson, T.A. Manganese–Oxygen Intermediates in O–O Bond Activation and Hydrogen-Atom Transfer Reactions. Acc. Chem. Res. 2017, 50, 2706–2717. [Google Scholar] [CrossRef]
- Rice, D.B.; Munasinghe, A.; Grotemeyer, E.N.; Burr, A.D.; Day, V.W.; Jackson, T.A. Structure and Reactivity of (μ-Oxo)dimanganese(III,III) and Mononuclear Hydroxomanganese(III) Adducts Supported by Derivatives of an Amide-Containing Pentadentate Ligand. Inorg. Chem. 2019, 58, 622–636. [Google Scholar] [CrossRef]
- Mayfield, J.R.; Grotemeyer, E.N.; Jackson, T.A. Concerted proton–electron transfer reactions of manganese–hydroxo and manganese–oxo complexes. Chem. Commun. 2020, 56, 9238–9255. [Google Scholar] [CrossRef] [PubMed]
- Warren, J.J.; Mayer, J.M. Moving protons and electrons in biomimetic systems. Biochemistry 2015, 54, 1863–1878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warren, J.J.; Tronic, T.A.; Mayer, J.M. Thermochemistry of Proton-Coupled Electron Transfer Reagents and its Implications. Chem. Rev. 2010, 110, 6961–7001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coggins, M.K.; Brines, L.M.; Kovacs, J.A. Synthesis and Structural Characterization of a Series of MnIIIOR Complexes, Including a Water-Soluble MnIIIOH That Promotes Aerobic Hydrogen-Atom Transfer. Inorg. Chem. 2013, 52, 12383–12393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Opalade, A.A.; Hessefort, L.; Day, V.W.; Jackson, T.A. Controlling the Reactivity of a Metal-Hydroxo Adduct with a Hydrogen Bond. J. Am. Chem. Soc. 2021, 143, 15159–15175. [Google Scholar] [CrossRef]
- Coggins, M.K.; Kovacs, J.A. Structural and Spectroscopic Characterization of Metastable Thiolate-Ligated Manganese(III)–Alkylperoxo Species. J. Am. Chem. Soc. 2011, 133, 12470–12473. [Google Scholar] [CrossRef] [Green Version]
- Coggins, M.K.; Martin-Diaconescu, V.; DeBeer, S.; Kovacs, J.A. Correlation Between Structural, Spectroscopic, and Reactivity Properties Within a Series of Structurally Analogous Metastable Manganese(III)–Alkylperoxo Complexes. J. Am. Chem. Soc. 2013, 135, 4260–4272. [Google Scholar] [CrossRef] [Green Version]
- Downing, A.N.; Coggins, M.K.; Poon, P.C.Y.; Kovacs, J.A. Influence of Thiolate versus Alkoxide Ligands on the Stability of Crystallographically Characterized Mn(III)-Alkylperoxo Complexes. J. Am. Chem. Soc. 2021, 143, 6104–6113. [Google Scholar] [CrossRef]
- Parham, J.D.; Wijeratne, G.B.; Rice, D.B.; Jackson, T.A. Spectroscopic and Structural Characterization of Mn(III)-Alkylperoxo Complexes Supported by Pentadentate Amide-Containing Ligands. Inorg. Chem. 2018, 57, 2489–2502. [Google Scholar] [CrossRef]
- Opalade, A.A.; Parham, J.D.; Day, V.W.; Jackson, T.A. Characterization and chemical reactivity of room-temperature-stable MnIII–alkylperoxo complexes. Chem. Sci. 2021, 12, 12564–12575. [Google Scholar] [CrossRef]
- Neese, F.; Wennmohs, F.; Becker, U.; Riplinger, C. The ORCA quantum chemistry program package. J. Chem. Phys. 2020, 152, 224108. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef] [Green Version]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef] [Green Version]
- Weigend, F.; Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297–3305. [Google Scholar] [CrossRef]
- Weigend, F. Accurate Coulomb-fitting basis sets for H to Rn. Phys. Chem. Chem. Phys. 2006, 8, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Grimme, S.; Ehrlich, S.; Goerigk, L. Effect of the damping function in dispersion corrected density functional theory. J. Comput. Chem. 2011, 32, 1456–1465. [Google Scholar] [CrossRef]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132, 154104. [Google Scholar] [CrossRef] [Green Version]
- Grimme, S. Semiempirical GGA-type density functional constructed with a long-range dispersion correction. J. Comput. Chem. 2006, 27, 1787–1799. [Google Scholar] [CrossRef] [PubMed]
- Grimme, S. Accurate description of van der Waals complexes by density functional theory including empirical corrections. J. Comput. Chem. 2004, 25, 1463–1473. [Google Scholar] [CrossRef] [PubMed]
- Marenich, A.V.; Cramer, C.J.; Truhlar, D.G. Universal Solvation Model Based on Solute Electron Density and on a Continuum Model of the Solvent Defined by the Bulk Dielectric Constant and Atomic Surface Tensions. J. Phys. Chem. B 2009, 113, 6378–6396. [Google Scholar] [CrossRef]
- Izsák, R.; Neese, F. An overlap fitted chain of spheres exchange method. J. Chem. Phys. 2011, 135, 144105. [Google Scholar] [CrossRef] [PubMed]
- Petrenko, T.; Kossmann, S.; Neese, F. Efficient time-dependent density functional theory approximations for hybrid density functionals: Analytical gradients and parallelization. J. Chem. Phys. 2011, 134, 054116. [Google Scholar] [CrossRef]
- Hubin, T.J.; McCormick, J.M.; Alcock, N.W.; Busch, D.H. Topologically Constrained Manganese(III) and Iron(III) Complexes of Two Cross-Bridged Tetraazamacrocycles. Inorg. Chem. 2001, 40, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Shirin, Z.; Borovik, A.S.; Young, V.G., Jr. Synthesis and structure of a MnIII(OH) complex generated from dioxygen. Chem. Commun. 1997, 1967–1968. [Google Scholar] [CrossRef]
- Shook, R.L.; Peterson, S.M.; Greaves, J.; Moore, C.; Rheingold, A.L.; Borovik, A.S. Catalytic Reduction of Dioxygen to Water with a Monomeric Manganese Complex at Room Temperature. J. Am. Chem. Soc. 2011, 133, 5810–5817. [Google Scholar] [CrossRef] [Green Version]
- Rice, D.B.; Jones, S.D.; Douglas, J.T.; Jackson, T.A. NMR Studies of a MnIII-hydroxo Adduct Reveal an Equilibrium between MnIII-hydroxo and μ-Oxodimanganese(III,III) Species. Inorg. Chem. 2018, 57, 7825–7837. [Google Scholar] [CrossRef]
- Geiger, R.A.; Chattopadhyay, S.; Day, V.W.; Jackson, T.A. A Series of Peroxomanganese(III) Complexes Supported by Tetradentate Aminopyridyl Ligands: Detailed Spectroscopic and Computational Studies. J. Am. Chem. Soc. 2010, 132, 2821–2831. [Google Scholar] [CrossRef]
- Colmer, H.E.; Howcroft, A.W.; Jackson, T.A. Formation, Characterization, and O–O Bond Activation of a Peroxomanganese(III) Complex Supported by a Cross-Clamped Cyclam Ligand. Inorg. Chem. 2016, 55, 2055–2069. [Google Scholar] [CrossRef]
- Bordwell, F.G.; Cheng, J.; Ji, G.Z.; Satish, A.V.; Zhang, X. Bond dissociation energies in DMSO related to the gas phase values. J. Am. Chem. Soc. 1991, 113, 9790–9795. [Google Scholar] [CrossRef]
- Company, A.; Lloret-Fillol, J.; Costas, M. Small molecule models for nonporphyrinic iron and manganese oxygenases. In Comprehensive Inorganic Chemistry II; Elsevier B.V.: Amsterdam, The Netherlands, 2013. [Google Scholar]













| [MnIII(OH)(dpaq)]+ | [MnIII(OH)(6Medpaq)]+ | |||
|---|---|---|---|---|
| XRD a | DFT | XRD b | DFT | |
| Mn−O1 (Å) | 1.806(13) | 1.829 | 1.806(6) | 1.830 |
| Mn−N1 (Å) | 2.072(14) | 2.089 | 2.041(7) | 2.048 |
| Mn−N2 (Å) | 1.975(14) | 1.980 | 1.962(6) | 1.968 |
| Mn−N3 (Å) | 2.173(14) | 2.222 | 2.130(6) | 2.134 |
| Mn−N4 (Å) | 2.260(14) | 2.213 | 2.322(6) | 2.339 |
| Mn−N5 (Å) | 2.216(15) | 2.209 | 2.381(7) | 2.422 |
| Experimental | DFT-Calculated | ||||
|---|---|---|---|---|---|
| Complex | k2 (M−1s−1) | MnIII/II Ep,c a | MnIII/II E1/2 a | MnII-OH2 pKa | BDFE b |
| [MnIII(OH)(6Medpaq)]+ | 3.4(2) | −0.63 | −0.60 | 28.6 | 80.2 |
| [MnIII(OH)(dpaq)]+ c | 1.1(1) | −0.70 | −0.70 | 29.3 | 79.1 |
| [MnIII(OH)(dpaq2Me)]+ c | 3.9(3) d | −0.62 d | −0.58 c | 28.7 c | 80.9 c |
| [MnIII(OH)(dpaq5NO2)]+ c | 7(1) | −0.57 | −0.51 | 27.8 | 81.2 |
| [MnIII(OH)(dpaq5Cl)]+ c | 2.8(2) | −0.66 | −0.62 | 28.7 | 79.8 |
| [MnIII(OH)(dpaq5OMe)]+ c | 0.8(1) | −0.72 | −0.73 | 29.5 | 78.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Opalade, A.A.; Grotemeyer, E.N.; Jackson, T.A. Mimicking Elementary Reactions of Manganese Lipoxygenase Using Mn-hydroxo and Mn-alkylperoxo Complexes. Molecules 2021, 26, 7151. https://doi.org/10.3390/molecules26237151
Opalade AA, Grotemeyer EN, Jackson TA. Mimicking Elementary Reactions of Manganese Lipoxygenase Using Mn-hydroxo and Mn-alkylperoxo Complexes. Molecules. 2021; 26(23):7151. https://doi.org/10.3390/molecules26237151
Chicago/Turabian StyleOpalade, Adedamola A., Elizabeth N. Grotemeyer, and Timothy A. Jackson. 2021. "Mimicking Elementary Reactions of Manganese Lipoxygenase Using Mn-hydroxo and Mn-alkylperoxo Complexes" Molecules 26, no. 23: 7151. https://doi.org/10.3390/molecules26237151