3.2. Reaction of Heterocyclic Amines 3, 4, 9–11 and 2-Aminothioplenol (5) with 2-Bromo-1-(4-methyl-2-(methylamino)thiazol-5-yl)ethan-1-one (2)
A mixture of equimolar quantities of 2-bromo-1-(4-methyl-2-(methylamino)thiazol-5-yl)ethan-1-one (2, 1.25 gm, 5 mmol) and heterocyclic amines 3, 4, 9–11 or 2-aminothioplenol 5 (5 mmol) in ethanol (30 mL) was refluxed for 10 h (the progress of the reactions was monitored by TLC). After the reaction was complete, the reaction mixture was filtered, the filtrate washed with methanol, and crystallized from a suitable solvent to give derivatives 6, 7, 12–14 and 8, respectively.
3.2.1. 5-(Imidazo[2,1-b]thiazol-6-yl)-N,4-dimethylthiazol-2-amine (6)
Yellow solid, (78% yield), mp 243–244 °C; (EtOH); IR (KBr) νmax 3417 (NH), 3038 (sp2 C-H), 2997 (sp3 C-H), 1634 (C=N) cm−1; 1H-NMR (DMSO-d6) δ 2.45 (s, 3H, CH3), 3.56 (s, 3H, CH3), 7.37 (d, J = 3.6 Hz, 1H, thiazole-H), 7.94 (d, J = 3.6 Hz, 1H, thiazole-H), 8.20 (s, 1H, imidazole-H), 9.60 (s, 1H, NH); MS m/z (%) 250 (M+, 26), 239 (53), 234 (47), 224 (89), 220 (16), 206 (62), 191 (45), 188 (57), 183 (100), 150 (30), 119 (34), 91 (33), 65 (42), 48 (54). Anal. Calcd. for C10H10N4S2 (250.34): C, 47.98; H, 4.03; N, 22.38. Found: C, 48.05; H, 3.89; N, 22.24%.
3.2.2. 5-(Benzo[d]imidazo[2,1-b]thiazol-2-yl)-N,4-dimethylthiazol-2-amine (7)
Yellow solid, (83% yield), mp 265–266 °C; (EtOH); IR (KBr) νmax 3238 (NH), 3047 (sp2 C-H), 2987 (sp3 C-H), 1623 (C=N) cm−1; 1H-NMR (DMSO-d6) δ 2.59 (s, 3H, CH3), 3.56 (s, 3H, CH3), 7.40 (t, J = 7 Hz, 1H, Ar-H), 7.54 (t, J = 7 Hz, 1H, Ar-H), 8.01 (d, J = 7.7 Hz, 1H, Ar-H), 8.09 (d, J = 7.7 Hz, 1H, Ar-H), 8.76 (s, 1H, imidazole-H), 9.62 (s, 1H, NH); MS m/z (%) 300 (M+, 34), 285 (22), 270 (29), 254 (40), 175 (26), 153 (27), 147 (23), 150 (28), 138 (71), 129 (29), 77 (17). Anal. Calcd. for C14H12N4S2 (300.40): C, 55.98; H, 4.03; N, 18.65. Found: C, 55.87; H, 3.87; N, 18.58%.
3.2.3. 5-(4H-Benzo[b][1,4]thiazin-3-yl)-N,4-dimethylthiazol-2-amine (8)
Yellow solid, (88% yield), mp 110–112 °C; (EtOH/Dioxane); IR (KBr) νmax 3374, 3293 (2NH), 3059 (sp2 C-H), 2926 (sp3 C-H), 1612 (C=N) cm−1; 1H-NMR (DMSO-d6) δ 2.51 (s, 3H, CH3), 2.89 (s, 3H, CH3), 5.45 (s, 1H, benzothiazine-H), 6.42 (t, J = 7 Hz, 1H, Ar-H), 6.73 (d, J = 7 Hz, 1H, Ar-H), 7.01 (d, J = 7 Hz, 1H, Ar-H), 7.07 (t, J = 7 Hz, 1H, Ar-H), 7.4 (s, 1H, NH), 9.07 (s, 1H, NH); MS m/z (%) 275 (M+, 13), 272 (100), 256 (7), 166 (10), 154 (3), 152 (17), 147 (4), 139 (86), 127 (7), 113 (37), 102 (23), 91 (31), 83 (72), 76 (17). Anal. Calcd. for C13H13N3S2 (275.39): C, 56.70; H, 4.76; N, 15.26. Found: C, 56.85; H, 4.58; N, 15.17%.
3.2.4. 5-(4H-Imidazo[1,2-d]tetrazol-5-yl)-N,4-dimethylthiazol-2-amine (12)
Yellow solid, (76% yield), mp 155–157 °C; (EtOH); IR (KBr) νmax 3428, 3203 (2NH), 3106, 2924 (sp3 C-H), 1646, 1599 (C=N) cm−1; 1H-NMR (DMSO-d6) δ 2.51 (s, 3H, CH3), 2.89 (s, 3H, CH3), 6.92 (s, 1H, NH), 7.03 (s, 1H, NH), 8.85 (s, 1H, imidazole-H); MS m/z (%) 235 (M+, 25), 220 (16), 210 (55), 192 (24), 191 (33), 189 (38), 170 (23), 123 (32), 106 (24), 103 (26), 96 (34), 82 (20), 64 (100). Anal. Calcd. for C8H9N7S (235.27): C, 40.84; H, 3.86; N, 41.68. Found: C, 41.10; H, 3.65; N, 41.59%.
3.2.5. 5-(1H-Benzo[d]imidazo[1,2-a]imidazol-2-yl)-N,4-dimethylthiazol-2-amine (13)
Yellow solid, (65% yield), mp 140–142 °C; (EtOH/Dioxane); IR (KBr) νmax 3438, 3165 (2NH), 3094 (sp2 C-H), 2991 (sp3 C-H), 1649 (C=N) cm−1; 1H-NMR (DMSO-d6) δ 2.25 (s, 3H, CH3), 2.85 (s, 3H, CH3), 7.12 (d, J = 7 Hz, 1H, Ar-H), 7.24 (d, J = 7 Hz, 1H, Ar-H), 8.0–8.21 (m, 2H, Ar-H), 8.29 (s, 1H, imidazole-H), 10.44 (s, 1H, NH), 10.58 (s, 1H, NH); MS m/z (%) 283 (M+, 22), 264 (41), 245 (56), 238 (17), 230 (49), 207 (100), 181 (66), 167 (41), 137 (83), 124 (22), 115 (42), 98 (62), 67 (45). Anal. Calcd. for C14H13N5S (283.35): C, 59.34; H, 4.62; N, 24.72. Found: C, 59.17; H, 4.55; N, 24.60%.
3.2.6. 5-(4H-Imidazo[1,2-b][1,2,4]triazol-5-yl)-N,4-dimethylthiazol-2-amine (14)
Yellow crystals, (91% yield), mp 245–146 °C; (EtOH); IR (KBr) νmax 3436, 3124(2NH), 2978 (sp3 C-H), 1629 (C=N) cm−1; 1H-NMR (DMSO-d6) δ 2.41 (s, 3H, CH3), 3.03 (s, 3H, CH3), 6.75 (s, 1H, NH), 6.85 (s, 1H, NH), 9.55 (s, 2H, 2C-H); MS m/z (%) 234 (M+, 21), 224 (7), 221 (15), 187 (5), 166 (12), 153 (5), 145 (25), 127 (14), 118 (28), 107 (4), 84 (11), 73 (28), 67 (6), 65 (54), 63 (100). Anal. Calcd. for C9H10N6S (234.28): C, 46.14; H, 4.30; N, 35.87. Found: C, 46.25; H, 4.29; N, 35.75%.
3.3. Reaction of 2-Bromo-1-(4-methyl-2-(methylamino)thiazol-5-yl)ethan-1-one (2) with Pyrimidinethione Derivatives 15a,b and thiosemicarbazones 18a–f, 20a,b 22, or 23
To a stirred hot solution of pyrimidinethione derivatives 15a or 15b or thiosemicarbazone derivatives 18a–f, 20a,b 22, or 23 (5 mmole) in dioxane (30 mL) was added 2-bromo-1-(4-methyl-2-(methylamino)thiazol-5-yl)ethan-1-one (2, 1.25 g, 5 mmol) and triethylamine (0.7 mL) then the reaction mixture was refluxed for 6 h. After cooling, the formed solid product was collected by filtration and washed with aqueous ethanol to dissolve any triethylamine hydrochloride crystals, dried and crystallized from ethanol/dioxane to give thiazolopyrimidines 17a,b or hydrazine-thiazole derivatives 19a–f, 21a,b, 24 and 25.
3.3.1. 3-(4-Methyl-2-(methylamino)thiazol-5-yl)-7-phenyl-5H-thiazolo[3,2-a]pyrimidin-5-one (17a)
Yellow solid, (81% yield), mp 200–201 °C; IR (KBr) νmax 3430 (NH), 2926 (sp3 C-H), 1653 (C=O), 1564 (C=N) cm−1; 1H-NMR (DMSO-d6) δ 2.51 (s, 3H, CH3), 3.30 (s, 3H, CH3), 4.44 (s, 2H, CH2), 6.69 (s, 1H, pyrimidine-H), 7.36–7.95 (m, 5H, Ar-H); 13C-NMR (DMSO-d6) δ 18.9 (CH3), 30.1 (CH2), 34.0 (N-CH3), 110.9, 112.0, 114.3, 119.8, 125.5, 127.32, 129.6, 131.9, 132.6, 145.7, 147.6, 166.7 (C=O) ppm; MS m/z (%) 354 (M+, 40), 338 (28), 331 (40), 327 (30), 311 (41), 269 (44), 258 (69), 201 (30), 173 (43), 155 (56), 111 (83), 86 (47), 60 (100). Anal. Calcd. for C17H14N4OS2 (354.45): C, 57.61; H, 3.98; N, 15.81. Found: C, 57.73; H, 3.76; N, 15.72%.
3.3.2. 7-Methyl-3-(4-methyl-2-(methylamino)thiazol-5-yl)-5H-thiazolo[3,2-a]pyrimidin-5-one (17b)
Dark orange solid, (92% yield), mp 259–260 °C; IR (KBr) νmax 3430 (NH), 2928 (sp3 C-H), 1636 (C=O), 1560 (C=N) cm−1; 1H-NMR (DMSO-d6) δ 2.07 (s, 3H, CH3), 2.51 (s, 3H, CH3), 3.37 (s, 3H, CH3), 5.68 (s, 2H, pyrimidine-H and thiazole-H), 12.24 (s, 1H, NH). MS m/z (%) 292 (M+, 100), 284 (59), 277 (23), 265 (21), 256 (63), 246 (32), 231 (25), 202 (44), 163 (25), 139 (16), 127 (40), 108 (53), 87 (35), 82 (25). Anal. Calcd. for C12H12N4OS2 (292.38): C, 49.30; H, 4.14; N, 19.16. Found: C, 49.45; H, 4.05; N, 19.02%.
3.3.3. N,4′-dimethyl-2-(2-(1-phenylethylidene)hydrazineyl)-[4,5′-bithiazol]-2′-amine (19a)
Green solid, (77% yield), mp 235–237 °C; IR (KBr) νmax br. 3419 (2NH), 2929 (sp3 C-H), 1554 (C=N) cm−1; 1H-NMR (DMSO-d6) δ 2.31 (s, 3H, CH3), 2.51 (s, 3H, CH3), 2.89 (s, 3H, CH3), 7.39–8.27 (m, 7H, Ar-H, thiazole-H, NH), 10.22 (s, 1H, NH); MS m/z (%) 343 (M+, 14), 334 (14), 303 (17), 282 (30), 218 (20), 163 (10), 127 (100), 93 (61), 91 (49), 85 (12). Anal. Calcd. for C16H17N5S2 (343.47): C, 55.95; H, 4.99; N, 20.39. Found: C, 56.14; H, 4.80; N, 20.27%.
3.3.4. 2-(2-(1-(4-Chlorophenyl)ethylidene)hydrazineyl)-N,4′-dimethyl-[4,5′-bithiazol]-2′-amine (19b)
Green solid, (68% yield), mp 226–228 °C; IR (KBr) νmax br. 3423 (2NH), 2925 (sp3 C-H),1552 (C=N) cm−1; 1H-NMR (DMSO-d6) δ 2.29 (s, 3H, CH3), 2.33 (s, 3H, CH3), 3.37 (s, 3H, CH3), 7.42–7.98 (m, 6H, Ar-H, thiazole-H, NH), 10.26 (s, 1H, NH); MS m/z (%) 379 (M++2, 12), 378 (M++1, 4), 377 (M+, 38), 364 (28), 346 (25), 337 (34), 328 (100), 265 (50), 251 (49), 239 (43), 225 (22), 216 (30), 188 (49), 128 (52), 83 (53). Anal. Calcd. for C16H16ClN5S2 (377.91): C, 50.85; H, 4.27; N, 18.53. Found: C, 51.02; H, 4.13; N, 18.44%.
3.3.5. 2-(2-(1-(4-Bromophenyl)ethylidene)hydrazineyl)-N,4′-dimethyl-[4,5′-bithiazol]-2′-amine (19c)
Green solid, (64% yield), mp 220–222 °C; IR (KBr) νmax br. 3416 (2NH), 2919 (sp3 C-H), 1552 cm-1; 1H-NMR (DMSO-d6) δ 2.28 (s, 3H, CH3), 2.33 (s, 3H, CH3), 3.36 (s, 3H, CH3), 7.56–7.92 (m, 5H, Ar-H, thiazole-H), 8.31 (s, 1H, NH), 10.26 (s, 1H, NH); MS m/z (%) 422 (M+, 25), 406 (52), 390 (100), 371 (56), 347 (32), 325 (37), 302 (38), 270 (67), 238 (36), 111 (24), 72 (20). Anal. Calcd. for C16H16BrN5S2 (422.36): C, 45.50; H, 3.82; N, 16.58. Found: C, 45.75; H, 3.68; N, 16.46%.
3.3.6. 2-(2-(1-(4-Fluorophenyl)ethylidene)hydrazineyl)-N,4′-dimethyl-[4,5′-bithiazol]-2′-amine (19d)
Green solid, (72% yield), mp 249–250 °C; IR (KBr) νmax 3423, 3249 (2NH), 3161, 2936 (sp3 C-H), 1602, 1560 (C=N) cm−1; 1H-NMR (DMSO-d6) δ 2.28 (s, 3H, CH3), 2.51 (s, 3H, CH3), 3.51 (s, 3H, CH3), 7.17–8.00 (m, 5H, Ar-H, thiazole-H), 8.24 (s, 1H, NH), 10.19 (s, 1H, NH); MS m/z (%) 361 (M+, 12), 338 (22), 299 (66), 273 (45), 271 (100), 252 (16), 202 (16), 189 (15), 136 (39). Anal. Calcd. for C16H16FN5S2 (361.46): C, 53.17; H, 4.46; N, 19.38. Found: C, 53.24; H, 4.31; N, 19.29%.
3.3.7. N,4′-Dimethyl-2-(2-(1-(p-tolyl)ethylidene)hydrazineyl)-[4,5′-bithiazol]-2′-amine (19e)
Green solid, (68% yield), mp 229–230 °C; IR (KBr) νmax br. 3411 (2NH), 2919 (sp3 C-H), 1552 cm−1; 1H-NMR (DMSO-d6) δ 2.25 (s, 3H, CH3), 2.35 (s, 3H, CH3), 2.50 (s, 3H, CH3), 3.26 (s, 3H, CH3), 6.94 (d, J = 7 Hz, 2H, Ar-H), 7.07 (s, 1H, thiazole-H), 7.76 (d, J = 7 Hz, 2H, Ar-H), 8.50 (s, 1H, NH), 10.21 (s, 1H, NH); MS m/z (%) 357 (M+, 35), 346 (53), 324 (100), 298 (60), 270 (45), 229 (34), 174 (68), 145 (44), 135 (81), 115 (50), 65 (56). Anal. Calcd. for C17H19N5S2 (357.49): C, 57.12; H, 5.36; N, 19.59. Found: C, 57.30; H, 5.33; N, 19.46%.
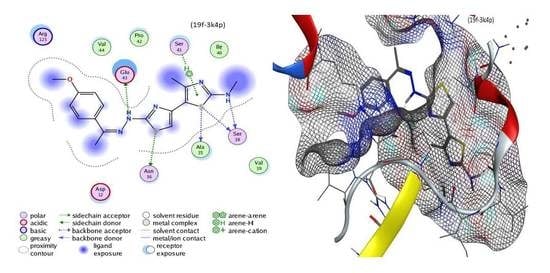
3.3.8. 2-(2-(1-(4-Methoxyphenyl)ethylidene)hydrazineyl)-N,4′-dimethyl-[4,5′-bithiazol]-2′-amine (19f)
Green solid, (75% yield), mp 195–197 °C; IR (KBr) νmax 3386, 3265 (2NH), 2926 (sp3 C-H), 1597 (C=N) cm−1; 1H-NMR (DMSO-d6) δ 2.00 (s, 3H, CH3), 2.51 (s, 3H, CH3), 2.80 (s, 3H, CH3), 3.86 (s, 3H, OCH3), 7.30 (s, 1H, thiazole-H), 7.44 (d, J = 7 Hz, 2H, Ar-H), 7.57 (d, J = 7 Hz, 2H, Ar-H), 8.29 (s, 1H, NH), 10.39 (s, 1H, NH); MS m/z (%) 373 (M+, 24), 369 (52), 346 (100), 332 (55), 284 (24), 270 (63), 250 (34), 177 (36), 123 (37), 111 (63), 76 (38). Anal. Calcd. for C17H19N5OS2 (373.49): C, 54.67; H, 5.13; N, 18.75. Found: C, 54.76; H, 5.24; N, 18.65%.
3.3.9. 2-(2-(1-(5-Imino-4-(p-tolyl)-4,5-dihydro-1,3,4-thiadiazol-2-yl)ethylidene)hydrazineyl)-N,4′-dimethyl-[4,5′-bithiazol]-2′-amine (21a)
Green solid, (68% yield), mp 270–271 °C; IR (KBr) νmax br. 3423 (3NH), 2927 (sp3 C-H), 1590 (C=N) cm−1; 1H-NMR (DMSO-d6) δ 2.34 (s, 3H, CH3), 2.39 (s, 3H, CH3), 2.51 (s, 3H, CH3), 3.35 (s, 3H, CH3), 6.69–8.08 (m, 6H, Ar-H, thiazole-H, NH), 9.39 (s, 1H, NH), 10.91 (s, 1H, NH); MS m/z (%) 456 (M+, 34), 420 (24), 367 (24), 267 (26), 249 (52), 200 (44), 139 (33), 104 (21), 85 (60), 76 (29). Anal. Calcd. for C19H20N8S3 (456.61): C, 49.98; H, 4.42; N, 24.54. Found: C, 50.11; H, 4.27; N, 24.35%.
3.3.10. 2-(2-(1-(4-(4-Chlorophenyl)-5-imino-4,5-dihydro-1,3,4-thiadiazol-2-yl)ethylidene)-hydrazineyl)-N,4′-dimethyl-[4,5′-bithiazol]-2′-amine (21b)
Green solid, (65% yield), mp 278–280 °C; IR (KBr) νmax br. 3425 (3NH), 2991 (sp3 C-H), 1621 (C=N) cm−1; 1H-NMR (DMSO-d6) δ 1.92 (s, 3H, CH3), 2.41 (s, 3H, CH3), 2.68 (s, 3H, CH3), 7.50–8.11 (m, 6H, Ar-H, thiazole-H, NH), 8.66 (s, 1H, NH), 10.92 (s, 1H, NH); MS m/z (%) 479 (M++2, 34), 477 (M+, 73), 461 (41), 382 (33), 313 (15), 222 (26), 202 (36), 111 (15), 108 (32), 96 (100). Anal. Calcd. for C18H17ClN8S3 (477.02): C, 45.32; H, 3.59; N, 23.49. Found: C, 45.43; H, 3.41; N, 23.28%.
3.3.11. 3-(2-(4′-Methyl-2′-(methylamino)-[4,5′-bithiazol]-2-yl)hydrazineylidene)indolin-2-one (24)
Brown solid, (79% yield), mp 210–212 °C; IR (KBr) νmax 3419, 3262 (3NH), 1650 (C=O), 1600 (C=N) cm−1; 1H-NMR (DMSO-d6) δ 2.51 (s, 3H, CH3), 3.39 (s, 3H, CH3), 6.92 (d, J = 7.8 Hz, 1H, Ar-H), 7.07 (t, J = 7.45 Hz, 1H, Ar-H), 7.33 (t, J = 7.5 Hz, 1H, Ar-H), 7.65 (d, J = 7.44 Hz, 1H, Ar-H), 8.68 (s, 1H, thiazole-H), 9.03 (s, 1H, NH), 11.21 (s, 1H, NH), 12.48 (s, 1H. NH); MS m/z (%) 370 (M+, 29), 356 (50), 340 (32), 244 (45), 337 (50), 305 (40), 297 (63), 282 (25), 264 (58), 224 (32), 211 (44), 190 (100), 87 (44), 82 (19), 77 (32). Anal. Calcd. for C16H14N6OS2 (370.45): C, 51.88; H, 3.81; N, 22.69. Found: C, 52.02; H, 3.65; N, 22.53%.
3.3.12. 2,2′′-((1H-Indene-1,3(2H)-diylidene)bis(hydrazin-1-yl-2-ylidene))bis(N,4′-dimethyl-[4,5′-bithiazol]-2′-amine) (25)
Brown solid, (72% yield), mp 216–217 °C; IR (KBr) νmax 3415, 3240 (2NH), 2937 (sp3 C-H), 1575 (C=N) cm−1; 1H-NMR (DMSO-d6) δ 2.51 (s, 6H, 2CH3), 3.38 (s, 6H, 2CH3), 3.83 (s, 2H, CH2), 7.51–8.03 (m, 4H, Ar-H), 8.13(s, 2H, thiazole-H), 8.40 (s, 2H, 2NH), 10.02 (s, 2H, 2NH); MS m/z (%) 593 (M++1, 11), 567 (14), 548 (23), 531 (20), 510 (14), 439 (20), 404 (20), 360 (100), 352 (15), 276 (31), 226 (11), 211 (13), 200 (58). Anal. Calcd. for C25H24N10S4 (592.78): C, 50.66; H, 4.08; N, 23.63. Found: C, 50.76; H, 4.14; N, 23.58%.













 The tested compound has activity similar to reference drug.
The tested compound has activity similar to reference drug. The tested compound has activity exceed the reference drug.
The tested compound has activity exceed the reference drug. The tested compound has activity similar to reference drug.
The tested compound has activity similar to reference drug. The tested compound has activity exceed the reference drug.
The tested compound has activity exceed the reference drug. The tested compound has activity similar to reference drug.
The tested compound has activity similar to reference drug. The tested compound has activity exceed the reference drug.
The tested compound has activity exceed the reference drug. The tested compound has activity similar to reference drug.
The tested compound has activity similar to reference drug.