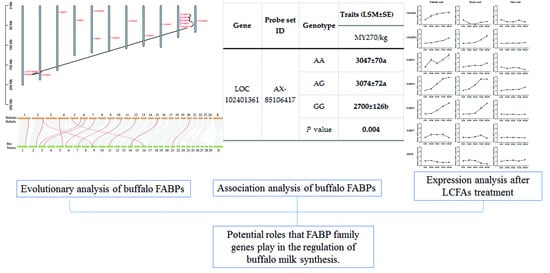
Evolutionary and Association Analysis of Buffalo FABP Family Genes Reveal Their Potential Role in Milk Performance
Abstract
:1. Introduction
2. Methods and Materials
2.1. Genome-Wide Identification of FABP Genes
2.2. Phylogenetic Analysis of FABPs in Different Organism
2.3. Structural Features Analysis
2.4. Chromosomal Distribution and Gene Duplication Analysis
2.5. Association Analysis of SNP and Buffalo Milk Traits
2.6. Cell Culture and Fatty Acid Treatment
2.7. Isolation and Culture of BuMECs
2.8. qRT-PCR Analysis
3. Results
3.1. Genome Identification of FABP Family Members
3.2. Structural Features of Buffalo FABP Family Members
3.3. Phylogenetic Relationship Analysis of FABP Protein in Five Mammals
3.4. Chromosomal Distribution and Collinearity Analysis of FABP Genes
3.5. Analyses of Association between Traits Related to Buffalo Milk Production
3.6. Effect of LCFAs on Expression of FABPs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Z.; Yue, Y.X.; Liu, Z.M.; Yang, L.Y.; Li, H.; Li, Z.J.; Li, G.X.; Wang, Y.B.; Tian, Y.D.; Kang, X.T.; et al. Genome-Wide Analysis of the FABP Gene Family in Liver of Chicken (Gallus gallus): Identification, Dynamic Expression Profile, and RegulatoryMechanism. Int. J. Mol. Sci. 2019, 20, 5948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marion, M.; Hamilton, J.; Richardson, B.; Roeder, N.; Figueiredo, A.; Nubelo, A.; Hetelekides, E.; Penman, S.; Owada, Y.; Kagawa, Y.; et al. Environmental enrichment sex-dependently rescues memory impairment in FABP5 KO mice not mediated by brain-derived neurotrophic factor. Behav. Brain Res. 2022, 425, 113814. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, K.L.; Smith, C.I.; Schwarzenberg, S.J.; Jessurun, J.; Boldt, M.D.; Parks, E.J. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J. Clin. Investig. 2005, 115, 1343–1351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.; Blair, D.; Bradley, J.E. Phyletic distribution of fatty acid-binding protein genes. PLoS ONE 2013, 8, e77636. [Google Scholar] [CrossRef] [Green Version]
- Glatz, J.F.; van der Vusse, G.J. Cellular fatty acid-binding proteins: Their function and physiological significance. Prog. Lipid Res. 1996, 35, 243–282. [Google Scholar] [CrossRef]
- Venkatachalam, A.B.; Thisse, C.; Thisse, B.; Wright, J.M. Differential tissue-specific distribution of transcripts for the duplicated fatty acid-binding protein 10 (fabp10) genes in embryos, larvae and adult zebrafish (Danio rerio). FEBS J. 2009, 276, 6787–6797. [Google Scholar] [CrossRef]
- Agulleiro, M.J.; André, M.; Morais, S.; Cerdà, J.; Babin, P.J. High transcript level of fatty acid-binding protein 11 but not of very low-density lipoprotein receptor is correlated to ovarian follicle atresia in a teleost fish (Solea senegalensis). Biol. Reprod. 2007, 77, 504–516. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.Z.; Li, X.; Godbout, R. A novel fatty acid-binding protein (FABP) gene resulting from tandem gene duplication in mammals: Transcription in rat retina and testis. Genomics 2008, 92, 436–445. [Google Scholar] [CrossRef] [Green Version]
- Parmar, M.B.; Venkatachalam, A.B.; Wright, J.M. The evolutionary relationship of the transcriptionally active fabp11a (intronless) and fabp11b genes of medaka with fabp11 genes of other teleost fishes. FEBS J. 2012, 279, 2310–2321. [Google Scholar] [CrossRef]
- Schaap, F.G.; van der Vusse, G.J.; Glatz, J.F. Evolution of the family of intracellular lipid binding proteins in vertebrates. Mol. Cell. Biochem. 2002, 239, 69–77. [Google Scholar] [CrossRef]
- Furuhashi, M.; Hotamisligil, G.S. Fatty acid-binding proteins: Role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 2008, 7, 489–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ono, T.; Odani, S. Initial studies of the cytoplasmic FABP superfamily. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010, 86, 220–228. [Google Scholar] [CrossRef] [Green Version]
- Chmurzyńska, A. The multigene family of fatty acid-binding proteins (FABPs): Function, structure and polymorphism. J. Appl. Genet. 2006, 47, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Bionaz, M.; Loor, J.J. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genomics 2008, 9, 366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whetstone, H.D.; Hurley, W.L.; Davis, C.L. Identification and characterization of a fatty acid binding protein in bovine mammary gland. Comp. Biochem. Physiol. B 1986, 85, 687–692. [Google Scholar] [CrossRef]
- Liang, M.Y.; Hou, X.M.; Qu, B.; Zhang, N.; Li, N.; Cui, Y.J.; Li, Q.Z.; Gao, X.J. Functional analysis of FABP3 in the milk fat synthesis signaling pathway of dairy cow mammary epithelial cells. Vitr. Cell. Dev. Biol. Anim. 2014, 50, 865–873. [Google Scholar] [CrossRef]
- Nafikov, R.A.; Schoonmaker, J.P.; Korn, K.T.; Noack, K.; Garrick, D.J.; Koehler, K.J.; Minick-Bormann, J.; Reecy, J.M.; Spurlock, D.E.; Beitz, D.C. Association of polymorphisms in solute carrier family 27, isoform A6 (SLC27A6) and fatty acid-binding protein-3 and fatty acid-binding protein-4 (FABP3 and FABP4) with fatty acid composition of bovine milk. J. Dairy Sci. 2013, 96, 6007–6021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, S.; Park, T.S.; Yoon, D.H.; Cheong, H.S.; Namgoong, S.; Park, B.L.; Lee, H.W.; Han, C.S.; Kim, E.M.; Cheong, I.C.; et al. Identification of genetic polymorphisms in FABP3 and FABN and putative association with back fat thickness in Korean native cattle. BMB Rep. 2008, 41, 29–34. [Google Scholar] [CrossRef] [Green Version]
- Zhihua, J.; Michal, J.J. Polymorphisms in Fatty Acid Binding Protein 4 (“fabp4”) Gene and their Associations with Measures of Marbling and Subcutaneous Fat Depth in Beef Cattle. US 2007/0020658 A1. 2007. Available online: https://patentscope2.wipo.int/search/en/detail.jsf;jsessionid=C98272504AA78E3E22B622E9D1098F7B.wapp1nC?docId=AU181382563_cid=P12-K6CZY1-99124-1 (accessed on 1 September 2021).
- Barendse, W.; Bunch, R.J.; Thomas, M.B.; Harrison, B.E. A splice site single nucleotide polymorphism of the fatty acid binding protein 4 gene appears to be associated with intramuscular fat deposition in longissimus muscle in Australian cattle. Anim. Genet. 2009, 40, 770–773. [Google Scholar] [CrossRef]
- Zhang, C.; Yan, C.; Xu, A.; Chan, B.; Chiu, K. Knocking out or pharmaceutical inhibition of fatty acid binding protein 4 (fabp-4) alleviates osteoarthritis induced by a very high fat diet in mouse. OsCar 2017, 25, S14. [Google Scholar] [CrossRef]
- Majchrzak, K.; Piotrowska, M.; Krajewska, J.; Fichna, J. Adipocyte fatty acid binding protein (A-FABP) as a potential new therapeutic target for treatment of obesity—associated cancers. Curr. Drug Targets 2021, 22, 1. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari, M.H.; Alaedin, M.T.; Sadri, H.; Hofs, I.; Koch, C.; Sauerwein, H. Longitudinal changes in fatty acid metabolism and in the mitochondrial protein import system in overconditioned and normal conditioned cows: A transcriptional study using microfluidic quantitative PCR. J. Dairy Sci. 2021, 104, 10338–10354. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Rickers-Haunerland, J.; Haunerland, N.H. Induction of cardiac FABP gene expression by long chain fatty acids in cultured rat muscle cells. Mol. Cell. Biochem. 2001, 221, 127–132. [Google Scholar] [CrossRef] [PubMed]
- van Breda, E.; Keizer, H.A.; Vork, M.M.; Surtel, D.A.; de Jong, Y.F.; van der Vusse, G.J.; Glatz, J.F. Modulation of fatty-acid-binding protein content of rat heart and skeletal muscle by endurance training and testosterone treatment. Pflugers Arch. 1992, 421, 274–279. [Google Scholar] [CrossRef]
- van der Lee, K.A.; Vork, M.M.; De Vries, J.E.; Willemsen, P.H.; Glatz, J.F.; Reneman, R.S.; Van der Vusse, G.J.; Van Bilsen, M. Long-chain fatty acid-induced changes in gene expression in neonatal cardiac myocytes. J. Lipid Res. 2000, 41, 41–47. [Google Scholar] [CrossRef]
- Johnsen, G.M.; Weedon-Fekjaer, M.S.; Tobin, K.A.; Staff, A.C.; Duttaroy, A.K. Long-chain polyunsaturated fatty acids stimulate cellular fatty acid uptake in human placental choriocarcinoma (BeWo) cells. Placenta 2009, 30, 1037–1044. [Google Scholar] [CrossRef]
- Kadegowda, A.K.; Bionaz, M.; Piperova, L.S.; Erdman, R.A.; Loor, J.J. Peroxisome proliferator-activated receptor-gamma activation and long-chain fatty acids alter lipogenic gene networks in bovine mammary epithelial cells to various extents. J. Dairy Sci. 2009, 92, 4276–4289. [Google Scholar] [CrossRef] [Green Version]
- Khedkar, C.; Kalyankar, S.; Deosarkar, S. Buffalo milk. In Encyclopedia of Food and Health; Academic Press: Waltham, MA, USA, 2016; pp. 522–528. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Z.; Li, J.; Li, H.; Yang, L. Genome-wide identification of Diacylglycerol Acyltransferases (DGAT) family genes influencing Milk production in Buffalo. BMC Genet. 2020, 21, 26. [Google Scholar] [CrossRef] [Green Version]
- Mintoo, A.A.; Zhang, H.; Chen, C.; Moniruzzaman, M.; Deng, T.; Anam, M.; Emdadul Huque, Q.M.; Guang, X.; Wang, P.; Zhong, Z. Draft genome of the river water buffalo. Ecol. Evol. 2019, 9, 3378–3388. [Google Scholar] [CrossRef]
- Li, Y.; He, L.; Li, J.; Chen, J.; Liu, C. Genome-Wide Identification, Characterization, and Expression Profiling of the Legume BZR Transcription Factor Gene Family. Front. Plant Sci. 2018, 9, 1332. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strimmer, K.; von Haeseler, A. Quartet Puzzling: A Quartet Maximum-Likelihood Method for Reconstructing Tree Topologies. Mol. Biol. Evol. 1996, 13, 964. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Liang, S.; Liang, A.; Rushdi, H.E.; Deng, T. Evolutionary Analysis of OAT Gene Family in River and Swamp Buffalo: Potential Role of SLCO3A1 Gene in Milk Performance. Genes 2021, 12, 1394. [Google Scholar] [CrossRef]
- Liu, J.; Liang, A.; Campanile, G.; Plastow, G.; Zhang, C.; Wang, Z.; Salzano, A.; Gasparrini, B.; Cassandro, M.; Yang, L. Genome-wide association studies to identify quantitative trait loci affecting milk production traits in water buffalo. J. Dairy Sci. 2018, 101, 433–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.C.; Zhao, J.; Li, W.; Liu, Y.H.; Wei, C.Q.; Wang, Y.K.; Ye, Z.Y.; Zhu, X.P. Development and characterization of 27 SNP markers in the Mauremys mutica transcriptome. Conserv. Genet. Resour. 2018, 10, 667–670. [Google Scholar] [CrossRef]
- Ye, T.; Deng, T.; Hosseini, S.M.; Raza, S.H.A.; Du, C.; Chen, C.; Zhang, X.; Hu, X.; Yang, L. Association analysis between FASN genotype and milk traits in Mediterranean buffalo and its expression among different buffalo tissues. Trop. Anim. Health Prod. 2021, 53, 366. [Google Scholar] [CrossRef]
- Anand, V.; Dogra, N.; Singh, S.; Kumar, S.N.; Jena, M.K.; Malakar, D.; Dang, A.K.; Mishra, B.P.; Mukhopadhyay, T.K.; Kaushik, J.K. Establishment and characterization of a buffalo (Bubalus bubalis) mammary epithelial cell line. PLoS ONE 2012, 7, e40469. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Warriach, H.; McGill, D.; Bush, R.; Wynn, P.; Chohan, K. A review of recent developments in buffalo reproduction—A review. Asian-Australas. J. Anim. Sci. 2015, 28, 451. [Google Scholar] [CrossRef]
- Du, C.; Deng, T.; Zhou, Y.; Ye, T.; Zhou, Z.; Zhang, S.; Shao, B.; Wei, P.; Sun, H.; Khan, F. Systematic analyses for candidate genes of milk production traits in water buffalo (Bubalus Bubalis). Anim. Genet. 2019, 50, 207–216. [Google Scholar] [CrossRef]
- Sánchez-Gurmaches, J.; Cruz-Garcia, L.; Gutiérrez, J.; Navarro, I. mRNA expression of fatty acid transporters in rainbow trout: In vivo and in vitro regulation by insulin, fasting and inflammation and infection mediators. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2012, 163, 177–188. [Google Scholar] [CrossRef]
- Zenker, J.; Stettner, M.; Ruskamo, S.; Domènech-Estévez, E.; Baloui, H.; Médard, J.J.; Verheijen, M.H.; Brouwers, J.F.; Kursula, P.; Kieseier, B.C.; et al. A role of peripheral myelin protein 2 in lipid homeostasis of myelinating Schwann cells. Glia 2014, 62, 1502–1512. [Google Scholar] [CrossRef]
- Kitamura, K.; Suzuki, M.; Suzuki, A.; Uyemura, K. The complete amino acid sequence of the P2 protein in bovine peripheral nerve myelin. FEBS Lett. 1980, 115, 27–30. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.E.; Ventura, M.; Cellamare, A.; Chen, L.; Cheng, Z.; Zhu, B.; Li, C.; Song, J.; Eichler, E.E. Analysis of recent segmental duplications in the bovine genome. BMC Genom. 2009, 10, 571. [Google Scholar] [CrossRef] [Green Version]
- Liao, X.; Bao, H.; Meng, Y.; Plastow, G.; Moore, S.; Stothard, P. Sequence, structural and expression divergence of duplicate genes in the bovine genome. PLoS ONE 2014, 9, e102868. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Zhao, X.Q.; Wang, J.; Wong, G.K.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Wang, Y.; Zhang, R.; Shi, C.; Lu, W.; Li, J.; Bai, M. Three Complete Mitochondrial Genomes of Erotylidae (Coleoptera: Cucujoidea) with Higher Phylogenetic Analysis. Insects 2021, 12, 524. [Google Scholar] [CrossRef] [PubMed]
- Mori, S.; Matsunami, M. Signature of positive selection in mitochondrial DNA in Cetartiodactyla. Genes Genet. Syst. 2018, 93, 65–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bionaz, M.; Loor, J.J. ACSL1, AGPAT6, FABP3, LPIN1, and SLC27A6 are the most abundant isoforms in bovine mammary tissue and their expression is affected by stage of lactation. J. Nutr. 2008, 138, 1019–1024. [Google Scholar] [CrossRef]
- Cohen-Zinder, M.; Lipkin, E.; Shor-Shimoni, E.; Ben-Meir, Y.; Agmon, R.; Asher, A.; Miron, J.; Shabtay, A. FABP4 gene has a very large effect on feed efficiency in lactating Israeli Holstein cows. Physiol. Genom. 2019, 51, 481–487. [Google Scholar] [CrossRef]
- Calvo, J.H.; Marcos, S.; Jurado, J.J.; Serrano, M. Association of the heart fatty acid-binding protein (FABP3) gene with milk traits in Manchega breed sheep. Anim. Genet. 2004, 35, 347–349. [Google Scholar] [CrossRef]
- Sun, Y.; Luo, J.; Zhu, J.; Shi, H.; Li, J.; Qiu, S.; Wang, P.; Loor, J.J. Effect of short-chain fatty acids on triacylglycerol accumulation, lipid droplet formation and lipogenic gene expression in goat mammary epithelial cells. Anim. Sci. J. 2016, 87, 242–249. [Google Scholar] [CrossRef]




| List | Protein Isoform | Gene ID | Protein ID | Amino Acids | Isoelectric Point | Mw/kDa | Product |
|---|---|---|---|---|---|---|---|
| 1 | CRABP1 | 102392457 | XP_006044444.1 | 137 | 5.26 | 15.59 | Cellular retinoic acid-binding protein 1 |
| 2 | CRABP2 | 102406669 | XP_006052022.1 | 138 | 5.37 | 15.73 | Cellular retinoic acid-binding protein 2 |
| 3 | FABP1 | 102407733 | XP_006074804.1 | 127 | 7.78 | 14.19 | Fatty acid-binding protein%2C liver |
| 4 | FABP12 | 102410779 | XP_006068112.2 | 121 | 5.51 | 13.8 | Fatty acid-binding protein 12 |
| 5 | FABP2 | 102414836 | XP_006067721.1 | 132 | 5.94 | 15.09 | Fatty acid-binding protein%2C intestinal |
| 6 | FABP3.1 | 102394447 | NP_001277811.1 | 133 | 6.73 | 14.77 | Fatty acid-binding protein%2C heart |
| 7 | FABP3.2 | 102394447 | XP_025150979.1 | 171 | 8.63 | 18.91 | Fatty acid-binding protein%2C heart isoform X1 |
| 8 | FABP4 | 102410448 | NP_001277890.1 | 132 | 5.04 | 14.76 | Fatty acid-binding protein%2C adipocyte |
| 9 | FABP5 | 102409117 | XP_006068107.1 | 135 | 7.58 | 15.07 | Fatty acid-binding protein%2C epidermal |
| 10 | FABP6 | 102412445 | XP_006073509.1 | 128 | 6.91 | 14.37 | gastrotropin |
| 11 | FABP7-X1 | 102409019 | XP_006047648.1 | 132 | 5.38 | 14.95 | Fatty acid-binding protein%2C brain isoform X1 |
| 12 | FABP7-X2 | 102409019 | XP_025150573.1 | 118 | 5.17 | 13.42 | Fatty acid-binding protein%2C brain isoform X2 |
| 13 | FABP7-X3 | 102409019 | XP_006047649.1 | 116 | 5.17 | 13.16 | Fatty acid-binding protein%2C brain isoform X3 |
| 14 | FABP9 | 102410109 | XP_006068110.1 | 132 | 9.07 | 14.87 | Fatty acid-binding protein 9 |
| 15 | LOC102401361 | 102401361 | XP_025146712.1 | 346 | 6.9 | 39.13 | LOW QUALITY PROTEIN: uncharacterized protein LOC102401361 |
| 16 | MP2P | 102409775 | XP_006068109.1 | 132 | 9.67 | 14.95 | Myelin P2 protein |
| 17 | RBP1 | 102389538 | XP_006055984.1 | 135 | 4.88 | 15.69 | Retinol-binding protein 1 |
| 18 | RBP2.1 | 102401674 | XP_006070028.1 | 134 | 5.76 | 15.7 | Retinol-binding protein 2 |
| 19 | RBP2.2 | 102401674 | XP_006070029.1 | 134 | 5.76 | 15.7 | Retinol-binding protein 2 |
| 20 | RBP5-X1.1 | 102393311 | XP_025138922.1 | 186 | 5.79 | 21.5 | Retinol-binding protein 5 isoform X1 |
| 21 | RBP5-X1.2 | 102393311 | XP_025138923.1 | 186 | 5.79 | 21.5 | Retinol-binding protein 5 isoform X1 |
| 22 | RBP5-X2 | 102393311 | XP_025138924.1 | 158 | 5.68 | 18.72 | Retinol-binding protein 5 isoform X2 |
| 23 | RBP5-X3 | 102393311 | XP_025138925.1 | 147 | 5.29 | 17.44 | Retinol-binding protein 5 isoform X3 |
| 24 | RBP5-X4 | 102393311 | XP_025138926.1 | 135 | 5.93 | 15.96 | Retinol-binding protein 5 isoform X4 |
| 25 | RBP5-X5 | 102393311 | XP_025138927.1 | 133 | 5.46 | 15.82 | Retinol-binding protein 5 isoform X5 |
| 26 | RBP7 | 102408740 | XP_006076768.1 | 13c4 | 6.82 | 15.54 | LOW QUALITY PROTEIN: retinoid-binding protein 7 |
| Seq_1 | Seq_2 | Ka | Ks | Ka_Ks | Divergence Time (Mya) |
|---|---|---|---|---|---|
| FABP12 | FABP4 | 0.271232885 | 1.293454386 | 0.209696521 | 51.328 |
| FABP4 | FABP9 | 0.261352777 | 2.49279534 | 0.104843255 | 98.92 |
| LOC102401361 | MP2P | 2.925282375 | 1.6126178 | 1.813996085 | 63.993 |
| LOC102401361 | RBP2 | 1.99354897 | 1.474584642 | 1.351939328 | 58.515 |
| MP2P | FABP9 | 0.234370241 | 2.248770297 | 0.104221512 | 89.237 |
| Buffalo | Cattle | Ka | Ks | Ka_Ks |
|---|---|---|---|---|
| LOC102401361 | RBP1 | NaN | NaN | NaN |
| FABP3 | FABP3 | 0.107671646 | 0.227999676 | 0.472244734 |
| FABP3 | FABP7 | 0.31065625 | 1.679232279 | 0.184998975 |
| RBP5 | RBP5 | 0.08500455 | 0.179602149 | 0.473293616 |
| RBP7 | RBP7 | 0.006256553 | 0.0379828 | 0.164720704 |
| CRABP2 | CRABP2 | 0.003127448 | 0.044008957 | 0.071063894 |
| RBP1 | RBP1 | 0.041334963 | 0.090922001 | 0.454620033 |
| FABP2 | FABP2 | NaN | NaN | NaN |
| FABP6 | FABP6 | 0.023564351 | 0.03739092 | 0.630215853 |
| FABP7 | FABP7 | 0 | 0.049299057 | 0 |
| FABP1 | FABP1 | 0.009955898 | 0.053355825 | 0.186594405 |
| FABP5 | FABP5 | 0 | 0.057832106 | 0 |
| FABP12 | PMP2 | 0.257783907 | 1.849116423 | 0.139409236 |
| CRABP1 | CRABP1 | NaN | NaN | NaN |
| Gene | Probe Set ID | Location | Genotype | Number | Frequency | Alleles | Rate | Observed He | Predicted He | HWE (p-Value) | PIC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| LOC102401361 | AX-85097756 | Intro | AA | 72 | 15.7% | A | 0.389 | 0.464 | 0.475 | 0.677 | 0.362 |
| AG | 214 | 46.5% | G | 0.611 | |||||||
| GG | 174 | 37.8% | |||||||||
| AX-85049047 | Intro | TT | 46 | 10.0% | T | 0.307 | 0.413 | 0.425 | 0.616 | 0.335 | |
| TC | 191 | 41.4% | C | 0.693 | |||||||
| CC | 224 | 48.6% | |||||||||
| AX-85116471 | Intro | AA | 127 | 27.5% | A | 0.508 | 0.465 | 0.500 | 0.157 | 0.375 | |
| AG | 215 | 46.5% | G | 0.492 | |||||||
| GG | 120 | 26.0% | |||||||||
| AX-85106417 | Intro | AA | 275 | 59.5% | A | 0.779 | 0.368 | 0.344 | 0.177 | 0.285 | |
| AG | 170 | 36.8% | G | 0.221 | |||||||
| GG | 17 | 3.7% | |||||||||
| AX-85072673 | Intro | AA | 19 | 4.1% | A | 0.183 | 0.282 | 0.298 | 0.306 | 0.254 | |
| AG | 130 | 28.3% | G | 0.817 | |||||||
| GG | 311 | 67.6% | |||||||||
| RBP2 | AX-85109932 | Intro | TT | 258 | 55.8% | T | 0.753 | 0.390 | 0.372 | 0.377 | 0.303 |
| TC | 180 | 39.0% | C | 0.247 | |||||||
| CC | 24 | 5.2% | |||||||||
| RBP5 | AX-85111933 | Intro | TT | 24 | 5.2% | T | 0.231 | 0.357 | 0.355 | 1.000 | 0.292 |
| TC | 165 | 35.7% | C | 0.769 | |||||||
| CC | 273 | 59.1% |
| Gene | Probe Set ID | Traits (LSM ± SE) | ||||||
|---|---|---|---|---|---|---|---|---|
| Genotype | PM270/kg | MY270/kg | PY270/kg | PP270/% | FY270/kg | FP270/% | ||
| LOC102401361 | AX-85097756 | AA | 15.8 ± 0.4 | 3041 ± 82 | 249 ± 7 | 8.21 ± 0.13 | 139 ± 4 | 4.59 ± 0.04 |
| AG | 15.7 ± 0.3 | 3040 ± 71 | 247 ± 6 | 8.17 ± 0.12 | 138 ± 3 | 4.57 ± 0.04 | ||
| GG | 15.6 ± 0.4 | 3062 ± 74 | 248 ± 6 | 8.16 ± 0.12 | 139 ± 3 | 4.58 ± 0.04 | ||
| P-value | 0.666 | 0.862 | 0.896 | 0.867 | 0.901 | 0.766 | ||
| AX-85049047 | CC | 15.6 ± 0.3 | 3048 ± 71 | 247 ± 6 | 8.16 ± 0.12 | 139 ± 3 | 4.58 ± 0.04 | |
| TC | 15.8 ± 0.3 | 3047 ± 73 | 249 ± 6 | 8.2 ± 0.12 | 138 ± 3 | 4.56 ± 0.04 | ||
| TT | 15.8 ± 0.4 | 3032 ± 88 | 249 ± 7 | 8.15 ± 0.14 | 140 ± 4 | 4.60 ± 0.04 | ||
| P-value | 0.646 | 0.97 | 0.771 | 0.852 | 0.852 | 0.343 | ||
| AX-85116471 | AA | 15.4 ± 0.4 | 3035 ± 75 | 245 ± 6 | 8.16 ± 0.12 | 138 ± 3 | 4.59 ± 0.04 | |
| AG | 15.8 ± 0.3 | 3053 ± 72 | 249 ± 6 | 8.17 ± 0.12 | 139 ± 3 | 4.56 ± 0.04 | ||
| GG | 15.8 ± 0.4 | 3045 ± 77 | 249 ± 6 | 8.19 ± 0.13 | 139 ± 3 | 4.58 ± 0.04 | ||
| P-value | 0.173 | 0.919 | 0.57 | 0.94 | 0.955 | 0.387 | ||
| AX-85106417 | AA | 15.6 ± 0.3 | 3047 ± 70 a | 247 ± 6 | 8.16 ± 0.12 | 138 ± 3 | 4.57 ± 0.04 | |
| AG | 15.9 ± 0.3 | 3074 ± 72 a | 250 ± 6 | 8.18 ± 0.12 | 140 ± 3 | 4.57 ± 0.04 | ||
| GG | 15.2 ± 0.6 | 2700 ± 126 b | 230 ± 10 | 8.23 ± 0.20 | 130 ± 5 | 4.61 ± 0.06 | ||
| P-value | 0.197 | 0.004 | 0.063 | 0.913 | 0.125 | 0.804 | ||
| RBP2 | AX-85072673 | AA | 15.6 ± 0.5 | 2997 ± 116 | 245 ± 9 | 8.20 ± 0.19 | 135 ± 5 | 4.54 ± 0.06 |
| AG | 15.9 ± 0.4 | 3067 ± 76 | 249 ± 6 | 8.17 ± 0.13 | 139 ± 3 | 4.57 ± 0.04 | ||
| GG | 15.6 ± 0.3 | 3042 ± 70 | 247 ± 6 | 8.17 ± 0.12 | 139 ± 3 | 4.58 ± 0.03 | ||
| P-value | 0.562 | 0.743 | 0.804 | 0.988 | 0.725 | 0.606 | ||
| RBP5 | AX-85111933 | CC | 15.7 ± 0.3 | 3034 ± 70 | 247 ± 6 | 8.17 ± 0.12 | 138 ± 3 | 4.57 ± 0.03 |
| TC | 15.7 ± 0.3 | 3070 ± 74 | 250 ± 6 | 8.18 ± 0.12 | 140 ± 3 | 4.58 ± 0.04 | ||
| TT | 15.1 ± 0.5 | 3015 ± 111 | 244 ± 9 | 8.15 ± 0.18 | 137 ± 5 | 4.55 ± 0.06 | ||
| P-value | 0.297 | 0.627 | 0.44 | 0.982 | 0.508 | 0.79 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, T.; Shaukat, A.; Yang, L.; Chen, C.; Zhou, Y.; Yang, L. Evolutionary and Association Analysis of Buffalo FABP Family Genes Reveal Their Potential Role in Milk Performance. Genes 2022, 13, 600. https://doi.org/10.3390/genes13040600
Ye T, Shaukat A, Yang L, Chen C, Zhou Y, Yang L. Evolutionary and Association Analysis of Buffalo FABP Family Genes Reveal Their Potential Role in Milk Performance. Genes. 2022; 13(4):600. https://doi.org/10.3390/genes13040600
Chicago/Turabian StyleYe, Tingzhu, Aftab Shaukat, Lv Yang, Chao Chen, Yang Zhou, and Liguo Yang. 2022. "Evolutionary and Association Analysis of Buffalo FABP Family Genes Reveal Their Potential Role in Milk Performance" Genes 13, no. 4: 600. https://doi.org/10.3390/genes13040600