Detection and Quantitation of Adulterated Paprika Samples Using Second-Order HPLC-FLD Fingerprints and Chemometrics
Abstract
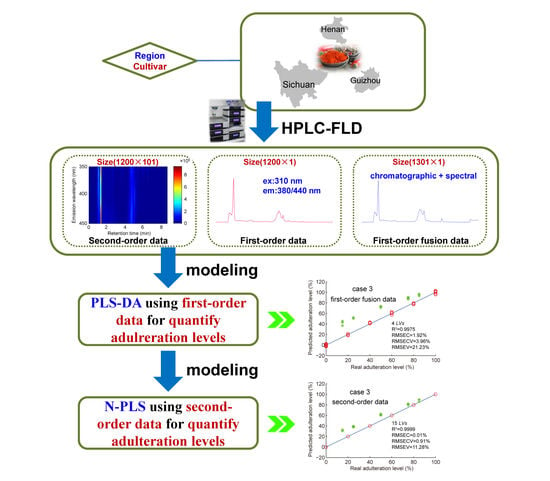
:1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. Samples
2.3. Instrumentation
2.4. Chemometrics
3. Results and Discussion
3.1. General Concerns
3.2. Variable Reduction
3.3. Detection and Quantitation of Adulteration by PLS
3.4. Detection and Quantitation of Adulteration by N-PLS
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Topuz, A.; Dincer, C.; Özdemir, K.S.; Feng, H.; Kushad, M. Influence of Different Drying Methods on Carotenoids and Capsaicinoids of Paprika (Cv., Jalapeno). Food Chem. 2011, 129, 860–865. [Google Scholar] [CrossRef] [PubMed]
- Van Asselt, E.D.; Banach, J.L.; van der Fels-Klerx, H.J. Prioritization of Chemical Hazards in Spices and Herbs for European Monitoring Programs. Food Control 2018, 83, 7–17. [Google Scholar] [CrossRef]
- Oliveira, M.M.; Cruz-Tirado, J.P.; Roque, J.V.; Teófilo, R.F.; Barbin, D.F. Portable Near-Infrared Spectroscopy for Rapid Authentication of Adulterated Paprika Powder. J. Food Compos. Anal. 2020, 87, 103403. [Google Scholar] [CrossRef]
- Galvin-King, P.; Haughey, S.A.; Elliott, C.T. The Detection of Substitution Adulteration of Paprika with Spent Paprika by the Application of Molecular Spectroscopy Tools. Foods 2020, 9, 944. [Google Scholar] [CrossRef]
- Monago-Maraña, O.; Durán-Merás, I.; Muñoz de la Peña, A.; Galeano-Díaz, T. Analytical Techniques and Chemometrics Approaches in Authenticating and Identifying Adulteration of Paprika Powder Using Fingerprints: A Review. Microchem. J. 2022, 178, 107382. [Google Scholar] [CrossRef]
- Farzaneh, V.; Gominho, J.; Pereira, H.; Carvalho, I.S. Screening of the Antioxidant and Enzyme Inhibition Potentials of Portuguese Pimpinella anisum L. Seeds by GC-MS. Food Anal. Methods 2018, 11, 2645–2656. [Google Scholar] [CrossRef]
- Cetó, X.; Sánchez, C.; Serrano, N.; Díaz-Cruz, J.M.; Núñez, O. Authentication of Paprika Using HPLC-UV Fingerprints. LWT 2020, 124, 109153. [Google Scholar] [CrossRef]
- De Sá Mendes, N.; Branco de Andrade Gonçalves, É.C. The Role of Bioactive Components Found in Peppers. Trends Food Sci. Technol. 2020, 99, 229–243. [Google Scholar] [CrossRef]
- Oliveira, M.M.; Cruz-Tirado, J.P.; Barbin, D.F. Nontargeted Analytical Methods as a Powerful Tool for the Authentication of Spices and Herbs: A Review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 670–689. [Google Scholar] [CrossRef] [Green Version]
- Kucharska-Ambrożej, K.; Karpinska, J. The Application of Spectroscopic Techniques in Combination with Chemometrics for Detection Adulteration of Some Herbs and Spices. Microchem. J. 2020, 153, 104278. [Google Scholar] [CrossRef]
- Marcone, M.F. Characterization of the Edible Bird’s Nest the “Caviar of the East”. Food Res. Int. 2005, 38, 1125–1134. [Google Scholar] [CrossRef]
- Núñez, N.; Saurina, J.; Núñez, O. Non-Targeted HPLC-FLD Fingerprinting for the Detection and Quantitation of Adulterated Coffee Samples by Chemometrics. Food Control 2021, 124, 107912. [Google Scholar] [CrossRef]
- Di Anibal, C.; Rodriguez, M.S.; Albertengo, L. UV-Visible Spectroscopy and Multivariate Classification as a Screening Tool to Identify Adulteration of Culinary Spices with Sudan I and Blends of Sudan I + IV Dyes. Food Anal. Methods 2014, 7, 1090–1096. [Google Scholar] [CrossRef]
- Márquez, C.; Ruisánchez, I.; Callao, M.P. Qualitative and Quantitative Multivariate Strategies for Determining Paprika Adulteration with SUDAN I and II Dyes. Microchem. J. 2019, 145, 686–692. [Google Scholar] [CrossRef]
- Maraña, O.M.; García, T.d.J.B.; Díaz, T.G. Characterization of Spanish Paprika by Multivariate Analysis of Absorption and Fluorescence Spectra. Anal. Lett. 2016, 49, 1184–1197. [Google Scholar] [CrossRef]
- Kolašinac, S.; Pećinar, I.; Danojević, D.; Stevanović, Z.D. Raman Spectroscopy Coupled with Chemometric Modeling Approaches for Authentication of Different Paprika Varieties at Physiological Maturity. LWT 2022, 162, 113402. [Google Scholar] [CrossRef]
- Monago-Maraña, O.; Eskildsen, C.E.; Galeano-Díaz, T.; Muñoz de la Peña, A.; Wold, J.P. Untargeted Classification for Paprika Powder Authentication Using Visible—Near Infrared Spectroscopy (VIS-NIRS). Food Control 2021, 121, 107564. [Google Scholar] [CrossRef]
- Campmajó, G.; Rodríguez-Javier, L.R.; Saurina, J.; Núñez, O. Assessment of Paprika Geographical Origin Fraud by High-Performance Liquid Chromatography with Fluorescence Detection (HPLC-FLD) Fingerprinting. Food Chem. 2021, 352, 129397. [Google Scholar] [CrossRef]
- Serrano, N.; Cetó, X.; Núñez, O.; Aragó, M.; Gámez, A.; Ariño, C.; Díaz-Cruz, J.M. Characterization and Classification of Spanish Paprika (Capsicum annuum L.) by Liquid Chromatography Coupled to Electrochemical Detection with Screen-Printed Carbon-Based Nanomaterials Electrodes. Talanta 2018, 189, 296–301. [Google Scholar] [CrossRef]
- Barbosa, S.; Saurina, J.; Puignou, L.; Núñez, O. Classification and Authentication of Paprika by UHPLC-HRMS Fingerprinting and Multivariate Calibration Methods (PCA and PLS-DA). Foods 2020, 9, 486. [Google Scholar] [CrossRef] [Green Version]
- Biancolillo, A.; Maggi, M.A.; De Martino, A.; Marini, F.; Ruggieri, F.; D’Archivio, A.A. Authentication of PDO Saffron of L’Aquila (Crocus sativus L.) by HPLC-DAD Coupled with a Discriminant Multi-Way Approach. Food Control 2020, 110, 107022. [Google Scholar] [CrossRef]
- Cetó, X.; Serrano, N.; Aragó, M.; Gámez, A.; Esteban, M.; Díaz-Cruz, J.M.; Núñez, O. Determination of HPLC-UV Fingerprints of Spanish Paprika (Capsicum annuum L.) for Its Classification by Linear Discriminant Analysis. Sensors 2018, 18, 4479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Consonni, V.; Baccolo, G.; Gosetti, F.; Todeschini, R.; Ballabio, D. A MATLAB Toolbox for Multivariate Regression Coupled with Variable Selection. Chemom. Intell. Lab. Syst. 2021, 213, 104313. [Google Scholar] [CrossRef]
- Ballabio, D.; Consonni, V. Classification Tools in Chemistry. Part 1: Linear Models. PLS-DA. Anal. Methods 2013, 5, 3790–3798. [Google Scholar] [CrossRef]
- Andersson, C.A.; Bro, R. The N-Way Toolbox for MATLAB. Chemom. Intell. Lab. Syst. 2000, 52, 1–4. [Google Scholar] [CrossRef]
- Bro, R. Multiway Calibration. Multilinear PLS. J. Chemom. 1996, 10, 47–61. [Google Scholar] [CrossRef]
- Seo, S.-W.; Kim, J.-M.; Han, D.-G.; Geum, D.; Yun, H.; Yoon, I.-S. A Sensitive HPLC-FLD Method for the Quantification of Alpelisib, a Novel Phosphatidylinositol 3-Kinase Inhibitor, in Rat Plasma: Drug Metabolism and Pharmacokinetic Evaluation in Vitro and in Vivo. J. Chromatogr. B 2021, 1163, 122508. [Google Scholar] [CrossRef]
- Pons, J.; Bedmar, À.; Núñez, N.; Saurina, J.; Núñez, O. Tea and Chicory Extract Characterization, Classification and Authentication by Non-Targeted HPLC-UV-FLD Fingerprinting and Chemometrics. Foods 2021, 10, 2935. [Google Scholar] [CrossRef]
- Núñez, N.; Martínez, C.; Saurina, J.; Núñez, O. High-Performance Liquid Chromatography with Fluorescence Detection Fingerprints as Chemical Descriptors to Authenticate the Origin, Variety and Roasting Degree of Coffee by Multivariate Chemometric Methods. J. Sci. Food Agric. 2021, 101, 65–73. [Google Scholar] [CrossRef]
- Campmajó, G.; Saez-Vigo, R.; Saurina, J.; Núñez, O. High-Performance Liquid Chromatography with Fluorescence Detection Fingerprinting Combined with Chemometrics for Nut Classification and the Detection and Quantitation of Almond-Based Product Adulterations. Food Control 2020, 114, 107265. [Google Scholar] [CrossRef]
- Bajoub, A.; Medina-Rodríguez, S.; Gómez-Romero, M.; Ajal, E.A.; Bagur-González, M.G.; Fernández-Gutiérrez, A.; Carrasco-Pancorbo, A. Assessing the Varietal Origin of Extra-Virgin Olive Oil Using Liquid Chromatography Fingerprints of Phenolic Compound, Data Fusion and Chemometrics. Food Chem. 2017, 215, 245–255. [Google Scholar] [CrossRef]
- Núñez, N.; Pons, J.; Saurina, J.; Núñez, O. Non-Targeted High-Performance Liquid Chromatography with Ultraviolet and Fluorescence Detection Fingerprinting for the Classification, Authentication, and Fraud Quantitation of Instant Coffee and Chicory by Multivariate Chemometric Methods. LWT 2021, 147, 111646. [Google Scholar] [CrossRef]
- Jiménez-Carvelo, A.M.; Cruz, C.M.; Olivieri, A.C.; González-Casado, A.; Cuadros-Rodríguez, L. Classification of Olive Oils According to Their Cultivars Based on Second-Order Data Using LC-DAD. Talanta 2019, 195, 69–76. [Google Scholar] [CrossRef]




| Calibration Set | Validation Set | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C01 | C02 | C03 | C04 | C05 | C06 | V01 | V02 | V03 | V04 | V05 | |
| X (%) | 100 | 80 | 60 | 40 | 20 | 0 | 15 | 25 | 50 | 75 | 85 |
| Y (%) | 0 | 20 | 40 | 60 | 80 | 100 | 85 | 75 | 50 | 25 | 15 |
| replicates | 10 | 4 | 4 | 4 | 4 | 6 | 4 | 4 | 4 | 4 | 4 |
| Original Paprika | Paprika Used as Adulterant | LV | Linearity (R2) | RMSEC (%) | RMSECV (%) | RMSEV (%) |
|---|---|---|---|---|---|---|
| Sichuan EJT | Guizhou EJT | 5 | 0.9991 | 1.13 | 2.19 | 5.12 |
| Guizhou LT | Henan LT | 5 | 0.9995 | 0.89 | 2.27 | 20.28 |
| Guizhou LT | Guizhou EJT | 15 | 0.9999 | 0.01 | 0.91 | 11.28 |
| Guizhou XM | Henan NH | 9 | 0.9999 | 0.30 | 0.69 | 10.90 |
| Guizhou BT | Henan BT | 5 | 0.9997 | 0.68 | 0.87 | 5.07 |
| Guizhou WZ | Guizhou PD | 5 | 0.9997 | 0.70 | 1.03 | 1.52 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, X.; Zhang, M.; Wang, P.; Chen, J.; Yang, S.; Luo, P.; Gao, X. Detection and Quantitation of Adulterated Paprika Samples Using Second-Order HPLC-FLD Fingerprints and Chemometrics. Foods 2022, 11, 2376. https://doi.org/10.3390/foods11152376
Sun X, Zhang M, Wang P, Chen J, Yang S, Luo P, Gao X. Detection and Quantitation of Adulterated Paprika Samples Using Second-Order HPLC-FLD Fingerprints and Chemometrics. Foods. 2022; 11(15):2376. https://doi.org/10.3390/foods11152376
Chicago/Turabian StyleSun, Xiaodong, Min Zhang, Pengjiao Wang, Junhua Chen, Shengjun Yang, Peng Luo, and Xiuli Gao. 2022. "Detection and Quantitation of Adulterated Paprika Samples Using Second-Order HPLC-FLD Fingerprints and Chemometrics" Foods 11, no. 15: 2376. https://doi.org/10.3390/foods11152376