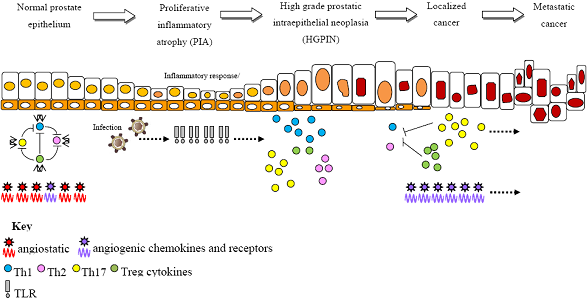
Inflammatory Genetic Markers of Prostate Cancer Risk
Abstract
:1. Introduction

2. Non-Targeted Genome-Wide Scans for Prostate Cancer Risk
3. Targeted Candidate Gene Analysis for Prostate Cancer Risk
3.1. Toll-like Receptor (TLR) Variants
| Gene family | Gene | Variant ID | # No. of publications | * Population | Ref. | Reported functional effect | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| P ≤ 0.05 | Null | EU | AA | AS | OT | |||||
| TLRs | TLR1 | rs4624663 | 3 | √ | √ | √ | √ | [32,33,34] | ||
| rs4833095 | 1 | √ | √ | √ | √ | [32] | ||||
| rs5743551 | 1 | 2 | √ | √ | √ | √ | [32,33,34] | |||
| rs5743556 | 1 | 2 | √ | √ | √ | √ | [32,33,34] | |||
| rs5743594 | 1 | √ | √ | √ | √ | [32] | ||||
| rs5743595 | 1 | √ | √ | √ | √ | [32] | √ [35] | |||
| rs5743596 | 1 | √ | √ | √ | √ | [32] | ||||
| rs5743604 | 2 | 2 | √ | √ | √ | √ | [32,33,34,36] | |||
| rs5743611 | 3 | √ | √ | √ | √ | [32,33,34] | ||||
| TLR2 | rs3804100 | 1 | √ | [36] | ||||||
| TLR3 | rs3775296 | 1 | √ | [36] | ||||||
| rs5743305 | 1 | √ | [36] | |||||||
| rs5743313 | 1 | √ | [36] | |||||||
| TLR4 | rs1927911 | 2 | 1 | √ | √ | [29,30,31] | ||||
| rs1927914 | 1 | 4 | √ | √ | [29,30,31,36,37] | |||||
| rs2149356 | 1 | 3 | √ | √ | [28,30,31,37] | |||||
| rs2737190 | 1 | √ | [30] | |||||||
| rs2770150 | 1 | √ | [31] | |||||||
| rs4986790 | 1 | 4 | √ | √ | [28,30,31,36,37] | √ [38,39] | ||||
| rs5030717 | 1 | √ | [31] | |||||||
| rs5030721 | 1 | √ | [36,37] | |||||||
| rs5030728 | 1 | √ | √ | [28] | ||||||
| rs6478317 | 1 | √ | [31] | |||||||
| rs7873784 | 1 | 3 | √ | √ | [28,30,31,36] | |||||
| rs10116253 | 2 | √ | [30,31] | |||||||
| rs10759932 | 2 | 2 | √ | √ | [28,31,36,37] | √ [40] | ||||
| rs10759933 | 1 | √ | [37] | |||||||
| rs11536871 | 2 | √ | [36,37] | |||||||
| rs11536858 | 1 | 1 | √ | √ | [29,31] | |||||
| rs11536878 | 1 | √ | [31] | |||||||
| rs11536889 | 2 | 3 | √ | √ | [28,30,31,36,37] | |||||
| rs11536891 | 1 | 2 | √ | √ | [29,30,31] | |||||
| rs11536897 | 2 | √ | √ | [29,31] | ||||||
| rs11536898 | 1 | 1 | √ | [30,31] | ||||||
| TLR5 | rs1053954 | 1 | √ | [36] | ||||||
| rs2072493 | 1 | √ | [36] | |||||||
| rs5744113 | 1 | √ | [36] | |||||||
| rs5744174 | 1 | √ | [36] | |||||||
| TLR6 | rs1039599 | 1 | √ | [34] | ||||||
| rs3821985 | 1 | √ | [34] | |||||||
| rs5743788 | 2 | √ | [33,34] | |||||||
| rs5743795 | 1 | 2 | √ | √ | √ | √ | [32,33,34] | |||
| rs5743806 | 1 | 2 | √ | √ | √ | √ | [32,33,34] | |||
| rs5743810 | 3 | √ | √ | √ | √ | [32,33,34] | ||||
| rs5743815 | 3 | √ | √ | √ | √ | [32,33,34] | ||||
| TLR7 | rs179008 | 1 | √ | [36] | √ [41] | |||||
| rs179019 | 1 | √ | [36] | |||||||
| rs2302267 | 1 | √ | [36] | |||||||
| TLR8 | rs1548731 | 1 | √ | [36] | ||||||
| rs4830806 | 1 | √ | [36] | |||||||
| rs5744068 | 1 | √ | [36] | |||||||
| TLR9 | rs187084 | 1 | √ | [36] | ||||||
| TLR10 | rs4274855 | 1 | 2 | √ | √ | √ | √ | [32,33,34] | ||
| rs4129009 | 1 | 2 | √ | √ | √ | √ | [32,33,34] | √ [35] | ||
| rs7653908 | 1 | √ | √ | √ | √ | [32] | ||||
| rs7658893 | 1 | √ | √ | √ | √ | [32] | ||||
| rs10856838 | 1 | √ | √ | √ | √ | [32] | ||||
| rs11096955 | 2 | 1 | √ | √ | √ | √ | [32,33,34] | |||
| rs11096957 | 2 | 1 | √ | √ | √ | √ | [32,33,34] | |||
| rs11466617 | 1 | 2 | √ | √ | √ | √ | [32,33,34] | |||
| rs11466640 | 1 | 2 | √ | √ | √ | √ | [32,33,34] | |||
| rs11466649 | 1 | √ | √ | √ | √ | [32] | ||||
| rs11466651 | 1 | √ | √ | √ | √ | [32] | ||||
| rs11466653 | 1 | √ | √ | √ | √ | [32] | ||||
| rs11466655 | 1 | √ | √ | √ | √ | [32] | ||||
| rs11096956 | 1 | √ | √ | √ | √ | [32] | ||||
| rs11466657 | 3 | √ | √ | √ | √ | [32,33,34] | ||||
| Epistasis | ||||||||||
| TLR1/ TLR6/ TLR10 | rs11096955/ rs11096957/ rs4833095/ rs5743596/ rs5743595/ rs5743551 | 1 | √ | √ | √ | √ | [32] | |||
| 11 SNPs | 1 | √ | [33] | |||||||
| TLR4 | 15 SNPs | 1 | √ | [31] | ||||||
3.2. T helper (Th) Cytokine Variants
| Gene familyGene Variant ID | # No. of publications | * Population | Reference | Reported functional effect | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P ≤ 0.05 | Null | EU | AA | AS | OT | |||||
| Th Cytokines | IL-1α | rs1800587 | 1 | √ | [61] | √ [78] | ||||
| IL-1β | rs1143627 | 3 | √ | [30] | √ [79,80] | |||||
| rs16944 | 4 | √ | √ | [46,53,54,81] | √ [82,83] | |||||
| rs1143634 | 1 | 2 | √ | √ | [46,54,81] | |||||
| IL-1RN | rs878972 | 1 | √ | [84] | ||||||
| rs315934 | 1 | √ | [84] | |||||||
| rs3087263 | 2 | √ | √ | [84,85] | ||||||
| rs380092 | 1 | √ | √ | [85] | ||||||
| rs4252019 | 1 | √ | √ | [85] | ||||||
| rs579543 | 1 | √ | √ | [85] | ||||||
| rs315951 | 2 | √ | √ | [84,85] | ||||||
| rs4252041 | 1 | √ | √ | [85] | ||||||
| rs9005 | 1 | √ | √ | [85] | ||||||
| IL-2 | rs2069762 | 1 | √ | [86] | √ [87,88] | |||||
| rs2069763 | 1 | √ | [47] | |||||||
| rs3136534 | 1 | √ | [86] | |||||||
| IL-4 | Intron 3, 70bp VNTR | 1 | √ | [48] | √ [89] | |||||
| IL-6 | rs1800797 | 3 | √ | √ | [30,46,90] | |||||
| rs1800796 | 3 | √ | √ | [30,90,91] | √ [66,92] | |||||
| rs1800795 | 3 | 6 | √ | √ | √ | [30,46,48,49,50,81,90,91] | √ [64,65,66,67,68] | |||
| rs2069830 | 1 | √ | [91] | |||||||
| rs2069832 | 2 | √ | √ | [46,49] | ||||||
| rs1474348 | 1 | √ | [90] | |||||||
| rs2069837 | 2 | √ | √ | [90,91] | ||||||
| rs2069860 | 1 | √ | [90] | |||||||
| rs1474347 | 1 | √ | [91] | |||||||
| rs1524107 | 1 | √ | [91] | |||||||
| rs1554606 | 1 | √ | √ | [91] | ||||||
| rs2069849 | 2 | √ | √ | [49,91] | ||||||
| rs1818879 | 1 | √ | √ | [91] | ||||||
| IL-10 | rs1800896 | 5 | 3 | √ | √ | √ | [30,46,51,52,53,54,81] | √ [62] | ||
| rs1800871 | 2 | 3 | √ | √ | √ | [46,51,52,54,81] | ||||
| rs1800872 | 2 | 3 | √ | √ | [30,46,51,54,93] | |||||
| rs3024496 | 2 | √ | √ | [51,81] | ||||||
| IL-18 | rs1946518 | 1 | √ | [55] | √ [63] | |||||
| rs187238 | 1 | √ | [55] | √ [63] | ||||||
| IL-21 | rs6822844 | 1 | √ | [86] | ||||||
| rs6840978 | 1 | √ | [86] | |||||||
| TGF-β1 | rs1800468 | 1 | √ | √ | [94] | √ [69] | ||||
| rs1800469 | 2 | 1 | √ | √ | [56,57,94] | √ [69,70,71] | ||||
| rs1800470 | 2 | 3 | √ | √ | √ | [57,58,59,95,94,95,94,95] | √ [72,73,74] | |||
| rs1800471 | 1 | √ | √ | [94] | √ [96] | |||||
| rs1800472 | 1 | √ | √ | [94] | ||||||
| TNF | rs1799964 | 1 | 1 | √ | √ | [60,97] | ||||
| rs1800630 | 2 | √ | √ | [60,97] | ||||||
| rs1799724 | 1 | 3 | √ | √ | √ | [46,54,60,97] | √ [75] | |||
| rs1800629 | 1 | 6 | √ | √ | √ | [30,46,49,54,60,61,97] | √ [76,77,98] | |||
| rs361525 | 3 | √ | √ | [46,54,97] | √ [99] | |||||
| rs3093661 | 1 | √ | [49] | |||||||
| rs1800610 | 1 | √ | [97] | |||||||
| rs3093668 | 1 | √ | [49] | |||||||
| Epistasis | ||||||||||
| IL-1β/IL-10 | rs1143627/ rs1800896 | 1 | √ | [46] | ||||||
| rs1143627/ rs1800896 | 1 | √ | [54] | |||||||
| rs16944/ rs1800872 | 1 | √ | [54] | |||||||
| IL-1RN | rs878972/ rs315934/ rs3087263/ rs315951 | 1 | √ | [84] | ||||||
| IL-10/TNF | rs1800872/ rs361525 | 1 | √ | [54] | ||||||
| IL-10 | rs1800896/ rs1800871/ rs1800872/ rs3024496 | 1 | √ | [51] | √ [62,100,101] | |||||
| rs1800896/ rs1800871 | 1 | √ | ||||||||
| IL-18 | rs1946518/ rs187238 | 1 | √ | [55] | √ [63] | |||||
| TNF | rs1799964/ rs1800630/ rs1799724/ rs1800629 | 2 | √ | √ | [60,97] | |||||
3.3. Chemokine variants
| Gene family | Gene | Variant ID | # No. of publications | * Population | Ref. | Reported functional effect | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| P ≤ 0.05 | Null | EU | AA | AS | OT | |||||
| Chemokines | CCL2 (MCP1) | rs1024611 | 1 | √ | [61] | √ [113] | ||||
| CCL5 (RANTES) | rs2107538 | 1 | 1 | √ | [61,111] | √ [114] | ||||
| CCR5 | rs333 | 1 | 1 | √ | [111,112] | √ [115] | ||||
| CCR2 | rs1799864 | 1 | √ | [111] | √ [116] | |||||
| CXCL8 (IL-8) | rs4073 | 1 | 1 | √ | [53,109] | √ [117] | ||||
| CXCL12 (SDF1) | rs1801157 | 1 | 1 | √ | √ | [110,111] | √ [118] | |||
| CXCR1 | rs2230054 | 1 | √ | [109] | ||||||
| CXCR2 | rs11226580 | 1 | √ | [109] | ||||||
| CX3CR1 | rs3732378 | 1 | √ | [111] | √ [119] | |||||
| rs3732379 | 1 | √ | [111] | √ [119] | ||||||
4. Conclusions
Acknowledgements
References
- Edwards, B.K.; Ward, E.; Kohler, B.A.; Eheman, C.; Zauber, A.G.; Anderson, R.N.; Jemal, A.; Schymura, M.J.; Lansdorp-Vogelaar, I.; Seeff, L.C.; van Ballegooijen, M.; Goede, S.L.; Ries, L.A. Annual report to the nation on the status of cancer, 1975–2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates. Cancer 2009, 116, 544–573. [Google Scholar]
- Baade, P.D.; Youlden, D.R.; Krnjacki, L.J. International epidemiology of prostate cancer: geographical distribution and secular trends. Mol. Nutr. Food Res. 2009, 53, 171–184. [Google Scholar] [CrossRef]
- Dennis, L.K.; Dawson, D.V. Meta-analysis of measures of sexual activity and prostate cancer. Epidemiology 2002, 13, 72–79. [Google Scholar] [CrossRef]
- Taylor, M.L.; Mainous, A.G.; Wells, B.J. Prostate cancer and sexually transmitted diseases: a meta-analysis. Fam. Med. 2005, 37, 506–512. [Google Scholar]
- Dennis, L.K.; Lynch, C.F.; Torner, J.C. Epidemiologic association between prostatitis and prostate cancer. Urology 2002, 60, 78–83. [Google Scholar] [CrossRef]
- MacLennan, G.T.; Eisenberg, R.; Fleshman, R.L.; Taylor, J.M.; Fu, P.; Resnick, M.I.; Gupta, S. The influence of chronic inflammation in prostatic carcinogenesis: a 5-year followup study. J. Urol. 2006, 176, 1012–1016. [Google Scholar] [CrossRef]
- Jafari, S.; Etminan, M.; Afshar, K. Nonsteroidal anti-inflammatory drugs and prostate cancer: a systematic review of the literature and meta-analysis. Can. Urol. Assoc. J. 2009, 3, 323–330. [Google Scholar]
- Mahmud, S.; Franco, E.; Aprikian, A. Prostate cancer and use of nonsteroidal anti-inflammatory drugs: systematic review and meta-analysis. Br. J. Cancer 2004, 90, 93–99. [Google Scholar] [CrossRef]
- De Marzo, A.M.; Marchi, V.L.; Epstein, J.I.; Nelson, W.G. Proliferative inflammatory atrophy of the prostate: implications for prostatic carcinogenesis. Am. J. Pathol. 1999, 155, 1985–1992. [Google Scholar] [CrossRef]
- Costantini, S.; Capone, F.; Guerriero, E.; Castello, G. An approach for understanding the inflammation and cancer relationship. Immunol. Lett. 2009, 126, 91–92. [Google Scholar] [CrossRef]
- Smith, J.R.; Freije, D.; Carpten, J.D.; Gronberg, H.; Xu, J.; Isaacs, S.D.; Brownstein, M.J.; Bova, G.S.; Guo, H.; Bujnovszky, P.; Nusskern, D.R.; Damber, J.E.; Bergh, A.; Emanuelsson, M.; Kallioniemi, O.P.; Walker-Daniels, J.; Bailey-Wilson, J.E.; Beaty, T.H.; Meyers, D.A.; Walsh, P.C.; Collins, F.S.; Trent, J.M.; Isaacs, W.B. Major susceptibility locus for prostate cancer on chromosome 1 suggested by a genome-wide search. Science 1996, 274, 1371–1374. [Google Scholar] [CrossRef]
- Urisman, A.; Molinaro, R.J.; Fischer, N.; Plummer, S.J.; Casey, G.; Klein, E.A.; Malathi, K.; Magi-Galluzzi, C.; Tubbs, R.R.; Ganem, D.; Silverman, R.H.; DeRisi, J.L. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006, 2, e25. [Google Scholar] [CrossRef]
- Carpten, J.; Nupponen, N.; Isaacs, S.; Sood, R.; Robbins, C.; Xu, J.; Faruque, M.; Moses, T.; Ewing, C.; Gillanders, E.; Hu, P.; Bujnovszky, P.; Makalowska, I.; Baffoe-Bonnie, A.; Faith, D.; Smith, J.; Stephan, D.; Wiley, K.; Brownstein, M.; Gildea, D.; Kelly, B.; Jenkins, R.; Hostetter, G.; Matikainen, M.; Schleutker, J.; Klinger, K.; Connors, T.; Xiang, Y.; Wang, Z.; De Marzo, A.; Papadopoulos, N.; Kallioniemi, O.P.; Burk, R.; Meyers, D.; Gronberg, H.; Meltzer, P.; Silverman, R.; Bailey-Wilson, J.; Walsh, P.; Isaacs, W.; Trent, J. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat. Genet. 2002, 30, 181–184. [Google Scholar] [CrossRef]
- Xu, J.; Zheng, S.L.; Komiya, A.; Mychaleckyj, J.C.; Isaacs, S.D.; Hu, J.J.; Sterling, D.; Lange, E.M.; Hawkins, G.A.; Turner, A.; Ewing, C.M.; Faith, D.A.; Johnson, J.R.; Suzuki, H.; Bujnovszky, P.; Wiley, K.E.; DeMarzo, A.M.; Bova, G.S.; Chang, B.; Hall, M.C.; McCullough, D.L.; Partin, A.W.; Kassabian, V.S.; Carpten, J.D.; Bailey-Wilson, J.E.; Trent, J.M.; Ohar, J.; Bleecker, E.R.; Walsh, P.C.; Isaacs, W.B.; Meyers, D.A. Germline mutations and sequence variants of the macrophage scavenger receptor 1 gene are associated with prostate cancer risk. Nat. Genet. 2002, 32, 321–325. [Google Scholar] [CrossRef]
- Thomas, G.; Jacobs, K.B.; Yeager, M.; Kraft, P.; Wacholder, S.; Orr, N.; Yu, K.; Chatterjee, N.; Welch, R.; Hutchinson, A.; Crenshaw, A.; Cancel-Tassin, G.; Staats, B.J.; Wang, Z.; Gonzalez-Bosquet, J.; Fang, J.; Deng, X.; Berndt, S.I.; Calle, E.E.; Feigelson, H.S.; Thun, M.J.; Rodriguez, C.; Albanes, D.; Virtamo, J.; Weinstein, S.; Schumacher, F.R.; Giovannucci, E.; Willett, W.C.; Cussenot, O.; Valeri, A.; Andriole, G.L.; Crawford, E.D.; Tucker, M.; Gerhard, D.S.; Fraumeni, J.F., Jr.; Hoover, R.; Hayes, R.B.; Hunter, D.J.; Chanock, S.J. Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 2008, 40, 310–315. [Google Scholar] [CrossRef]
- De Marzo, A.M.; Platz, E.A.; Sutcliffe, S.; Xu, J.; Grönberg, H.; Drake, C.G.; Nakai, Y.; Isaacs, W.B.; Nelson, W.G. Inflammation in prostate carcinogenesis. Nat. Rev. Cancer 2007, 7, 256–269. [Google Scholar] [CrossRef]
- Zheng, S.L.; Liu, W.; Wiklund, F.; Dimitrov, L.; Bälter, K.; Sun, J.; Adami, H.O.; Johansson, J.E.; Sun, J.; Chang, B.; Loza, M.; Turner, A.R.; Bleecker, E.R.; Meyers, D.A.; Carpten, J.D.; Duggan, D.; Isaacs, W.B.; Xu, J.; Grönberg, H. A comprehensive association study for genes in inflammation pathway provides support for their roles in prostate cancer risk in the CAPS study. Prostate 2006, 66, 1556–1564. [Google Scholar] [CrossRef]
- Akira, S.; Takeda, K. Toll-like receptor signaling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef]
- Sun, J.; Turner, A.; Xu, J.; Grönberg, H.; Isaacs, W. Genetic variability in inflammation pathways and prostate cancer risk. Urol. Oncol. 2007, 25, 250–259. [Google Scholar] [CrossRef]
- Iwasaki, A.; Medzhitov, R. Regulation of adaptive immunity by the innate immune system. Science 2010, 327, 291–295. [Google Scholar] [CrossRef]
- Nickel, J.C.; Moon, T. Chronic bacterial prostatitis: an evolving clinical enigma. Urology 2005, 66, 2–8. [Google Scholar] [CrossRef]
- Tanner, M.A.; Shoskes, D.; Shahed, A.; Pace, N.R. Prevalence of corynebacterial 16S rRNA sequences in patients with bacterial and "nonbacterial" prostatitis. J. Clin. Microbiol. 1999, 37, 1863–1870. [Google Scholar]
- Boldogh, I.; Baskar, J.F.; Mar, E.C.; Huang, E.S. Human cytomegalovirus and herpes simplex type 2 virus in normal and adenocarcinomatous prostate glands. J. Natl. Cancer Inst. 1983, 70, 819–826. [Google Scholar]
- Kuczyk, M.; Serth, J.; Machtens, S.; Jonas, U. Detection of viral HPV 16 DNA in prostate cancer and benign prostatic hyperplasia by quantitative PCR-directed analysis. Prostate Cancer Prostatic Dis. 2000, 3, S23. [Google Scholar]
- Serth, J.; Panitz, F.; Paeslack, U.; Kuczyk, M.A.; Jonas, U. Increased levels of human papillomavirus type 16 DNA in a subset of prostate cancers. Cancer Res. 1999, 59, 823–825. [Google Scholar]
- Kundu, S.D.; Lee, C.; Billips, B.K.; Habermacher, G.M.; Zhang, Q.; Liu, V.; Wong, L.Y.; Klumpp, D.J.; Thumbikat, P. The toll-like receptor pathway: a novel mechanism of infection-induced carcinogenesis of prostate epithelial cells. Prostate 2008, 68, 223–229. [Google Scholar] [CrossRef]
- El-Omar, E.M.; Ng, M.T.; Hold, G.L. Polymorphisms in Toll-like receptor genes and risk of cancer. Oncogene 2008, 27, 244–252. [Google Scholar] [CrossRef]
- Cheng, I.; Plummer, S.J.; Casey, G.; Witte, J.S. Toll-like receptor 4 genetic variation and advanced prostate cancer risk. Cancer Epidemiol. Biomarkers Prev. 2007, 16, 352–355. [Google Scholar] [CrossRef]
- Song, J.; Kim, D.Y.; Kim, C.S.; Kim, H.J.; Lee, D.H.; Lee, H.M.; Ko, W.; Lee, G. The association between Toll-like receptor 4 (TLR4) polymorphisms and the risk of prostate cancer in Korean men. Cancer Genet. Cytogenet. 2009, 190, 88–92. [Google Scholar] [CrossRef]
- Wang, M.H.; Helzlsouer, K.J.; Smith, M.W.; Hoffman-Bolton, J.A.; Clipp, S.L.; Grinberg, V.; De Marzo, A.M.; Isaacs, W.B.; Drake, C.G.; Shugart, Y.Y.; Platz, E.A. Association of IL10 and other immune response- and obesity-related genes with prostate cancer in CLUE II. Prostate 2009, 69, 874–885. [Google Scholar] [CrossRef]
- Chen, Y.C.; Giovannucci, E.; Lazarus, R.; Kraft, P.; Ketkar, S.; Hunter, D.J. Sequence variants of Toll-like receptor 4 and susceptibility to prostate cancer. Cancer Res. 2005, 65, 11771–11778. [Google Scholar] [CrossRef]
- Stevens, V.L.; Hsing, A.W.; Talbot, J.T.; Zheng, S.L.; Sun, J.; Chen, J.; Thun, M.J.; Xu, J.; Calle, E.E.; Rodriguez, C. Genetic variation in the toll-like receptor gene cluster (TLR10-TLR1-TLR6) and prostate cancer risk. Int. J. Cancer 2008, 123, 2644–2650. [Google Scholar] [CrossRef]
- Sun, J.; Wiklund, F.; Zheng, S.L.; Chang, B.; Balter, K.; Li, L.; Johansson, J.E.; Li, G.; Adami, H.O.; Liu, W.; Tolin, A.; Turner, A.R.; Meyers, D.A.; Isaacs, W.B.; Xu, J.; Gronberg, H. Sequence variants in Toll-like receptor gene cluster (TLR6-TLR1-TLR10) and prostate cancer risk. J. Natl. Cancer Inst. 2005, 97, 525–532. [Google Scholar] [CrossRef]
- Chen, Y.C.; Giovannucci, E.; Kraft, P.; Lazarus, R.; Hunter, D.J. Association between Toll-like receptor gene cluster (TLR6, TLR1, and TLR10) and prostate cancer. Cancer Epidemiol. Biomarkers Prev. 2007, 16, 1982–1989. [Google Scholar] [CrossRef]
- Kormann, M.S.; Depner, M.; Hartl, D.; Klopp, N.; Illig, T.; Adamski, J.; Vogelberg, C.; Weiland, S.K.; von Mutius, E.; Kabesch, M. Toll-like receptor heterodimer variants protect from childhood asthma. J. Allergy Clin. Immunol. 2008, 122, 86–92, 92.e1–8. [Google Scholar] [CrossRef]
- Xu, J.; Lowey, J.; Wiklund, F.; Sun, J.; Lindmark, F.; Hsu, F.C.; Dimitrov, L.; Chang, B.; Turner, A.R.; Liu, W.; Adami, H.O.; Suh, E.; Moore, J.H.; Zheng, S.L.; Isaacs, W.B.; Trent, J.M.; Gronberg, H. The interaction of four genes in the inflammation pathway significantly predicts prostate cancer risk. Cancer Epidemiol. Biomarkers Prev. 2005, 14, 2563–2568. [Google Scholar] [CrossRef]
- Zheng, S.L.; Augustsson-Balter, K.; Chang, B.; Hedelin, M.; Li, L.; Adami, H.O.; Bensen, J.; Li, G.; Johnasson, J.E.; Turner, A.R.; Adams, T.S.; Meyers, D.A.; Isaacs, W.B.; Xu, J.; Gronberg, H. Sequence variants of toll-like receptor 4 are associated with prostate cancer risk: results from the CAncer Prostate in Sweden Study. Cancer Res. 2004, 64, 2918–2922. [Google Scholar] [CrossRef]
- Arbour, N.C.; Lorenz, E.; Schutte, B.C.; Zabner, J.; Kline, J.N.; Jones, M.; Frees, K.; Watt, J.L.; Schwartz, D.A. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat. Genet. 2000, 25, 187–191. [Google Scholar] [CrossRef]
- Schmitt, C.; Humeny, A.; Becker, C.M.; Brune, K.; Pahl, A. Polymorphisms of TLR4: rapid genotyping and reduced response to lipopolysaccharide of TLR4 mutant alleles. Clin. Chem. 2002, 48, 1661–1667. [Google Scholar]
- Hwang, Y.H.; Ro, H.; Choi, I.; Kim, H.; Oh, K.H.; Hwang, J.I.; Park, M.H.; Kim, S.; Yang, J.; Ahn, C. Impact of polymorphisms of TLR4/CD14 and TLR3 on acute rejection in kidney transplantation. Transplantation 2009, 88, 699–705. [Google Scholar] [CrossRef]
- Oh, D.Y.; Baumann, K.; Hamouda, O.; Eckert, J.K.; Neumann, K.; Kucherer, C.; Bartmeyer, B.; Poggensee, G.; Oh, N.; Pruss, A.; Jessen, H.; Schumann, R.R. A frequent functional toll-like receptor 7 polymorphism is associated with accelerated HIV-1 disease progression. AIDS 2009, 23, 297–307. [Google Scholar] [CrossRef]
- Shafer-Weaver, K.A.; Watkins, S.K.; Anderson, M.J.; Draper, L.J.; Malyguine, A.; Alvord, W.G.; Greenberg, N.M.; Hurwitz, A.A. Immunity to murine prostatic tumors: continuous provision of T-cell help prevents CD8 T-cell tolerance and activates tumor-infiltrating dendritic cells. Cancer Res. 2009, 69, 6256–6264. [Google Scholar] [CrossRef]
- Ebelt, K.; Babaryka, G.; Figel, A.M.; Pohla, H.; Buchner, A.; Stief, C.G.; Eisenmenger, W.; Kirchner, T.; Schendel, D.J.; Noessner, E. Dominance of CD4+ lymphocytic infiltrates with disturbed effector cell characteristics in the tumor microenvironment of prostate carcinoma. Prostate 2008, 68, 1–10. [Google Scholar] [CrossRef]
- Sfanos, K.S.; Bruno, T.C.; Maris, C.H.; Xu, L.; Thoburn, C.J.; DeMarzo, A.M.; Meeker, A.K.; Isaacs, W.B.; Drake, C.G. Phenotypic analysis of prostate-infiltrating lymphocytes reveals TH17 and Treg skewing. Clin. Cancer Res. 2008, 14, 3254–3261. [Google Scholar] [CrossRef]
- Derhovanessian, E.; Adams, V.; Hahnel, K.; Groeger, A.; Pandha, H.; Ward, S.; Pawelec, G. Pretreatment frequency of circulating IL-17+ CD4+ T-cells, but not Tregs, correlates with clinical response to whole-cell vaccination in prostate cancer patients. Int. J. Cancer 2009, 125, 1372–1379. [Google Scholar] [CrossRef]
- Zabaleta, J.; Su, L.J.; Lin, H.Y.; Sierra, R.A.; Hall, M.C.; Sartor, A.O.; Clark, P.E.; Hu, J.J.; Ochoa, A.C. Cytokine Genetic Polymorphisms and Prostate Cancer Aggressiveness. Carcinogenesis 2009, 30, 1358–1362. [Google Scholar] [CrossRef]
- Wu, H.C.; Chang, C.H.; Wan, L.; Wu, C.I.; Tsai, F.J.; Chen, W.C. IL-2 gene C/T polymorphism is associated with prostate cancer. J. Clin. Lab. Anal. 2006, 20, 245–249. [Google Scholar] [CrossRef]
- Kesarwani, P.; Ahirwar, D.K.; Mandhani, A.; Mittal, R.D. Association between -174 G/C promoter polymorphism of the interleukin-6 gene and progression of prostate cancer in North Indian population. DNA Cell Biol. 2008, 27, 505–510. [Google Scholar] [CrossRef]
- Moore, S.C.; Leitzmann, M.F.; Albanes, D.; Weinstein, S.J.; Snyder, K.; Virtamo, J.; Ahn, J.; Mayne, S.T.; Yu, H.; Peters, U.; Gunter, M.J. Adipokine genes and prostate cancer risk. Int. J. Cancer 2009, 124, 869–876. [Google Scholar] [CrossRef]
- Tan, D.; Wu, X.; Hou, M.; Lee, S.; Ok, LW.; Wang, J.; Janarthan, B.; Nallapareddy, S.; Trump, D.L.; Gao, A.C. Interleukin-6 polymorphism is associated with more aggressive prostate cancer. J. Urol. 2005, 174, 753–756. [Google Scholar] [CrossRef]
- Faupel-Badger, J.M.; Kidd, L.C.; Albanes, D.; Virtamo, J.; Woodson, K.; Tangrea, J.A. Association of IL-10 polymorphisms with prostate cancer risk and grade of disease. Cancer Causes Contr. 2008, 19, 119–124. [Google Scholar] [CrossRef]
- Kesarwani, P.; Ahirwar, D.K.; Mandhani, A.; Singh, A.N.; Dalela, D.; Srivastava, A.N.; Mittal, R.D. IL-10 -1082 G>A: a risk for prostate cancer but may be protective against progression of prostate cancer in North Indian cohort. World J. Urol. 2009, 27, 389–396. [Google Scholar] [CrossRef]
- McCarron, S.L.; Edwards, S.; Evans, P.R.; Gibbs, R.; Dearnaley, D.P.; Dowe, A.; Southgate, C.; The Cancer Research Campaign/British Prostate Group United Kingdom Familial Prostate Cancer Study Collaborators; Easton, D.F.; Eeles, R.A.; Howell, W. Martin Influence of Cytokine Gene Polymorphisms on the Development of Prostate Cancer. Cancer Res. 2002, 62, 3369–3372. [Google Scholar]
- Zabaleta, J.; Lin, H.Y.; Sierra, R.A.; Hall, M.C.; Clark, P.E.; Sartor, O.A.; Hu, J.J.; Ochoa, A.C. Interactions of cytokine gene polymorphisms in prostate cancer risk. Carcinogenesis 2008, 29, 573–578. [Google Scholar]
- Liu, Y.; Lin, N.; Huang, L.; Xu, Q.; Pang, G. Genetic polymorphisms of the interleukin-18 gene and risk of prostate cancer. DNA Cell Biol. 2007, 26, 613–618. [Google Scholar] [CrossRef]
- Brand, T.C.; Bermejo, C.; Canby-Hagino, E.; Troyer, D.A.; Baillargeon, J.; Thompson, I.M.; Leach, R.J.; Naylor, S.L. Association of polymorphisms in TGFB1 and prostate cancer prognosis. J. Urol. 2008, 179, 754–758. [Google Scholar] [CrossRef]
- Ewart-Toland, A.; Chan, J.M.; Yuan, J.; Balmain, A.; Ma, J. A gain of function TGFB1 polymorphism may be associated with late stage prostate cancer. Cancer Epidemiol. Biomarkers Prev. 2004, 13, 759–764. [Google Scholar]
- Li, Z.; Habuchi, T.; Tsuchiya, N.; Mitsumori, K.; Wang, L.; Ohyama, C.; Sato, K.; Kamoto, T.; Ogawa, O.; Kato, T. Increased risk of prostate cancer and benign prostatic hyperplasia associated with transforming growth factor-beta 1 gene polymorphism at codon10. Carcinogenesis 2004, 25, 237–240. [Google Scholar]
- Wei, B.B.; Xi, B.; Wang, R.; Bai, J.M.; Chang, J.K.; Zhang, Y.Y.; Yoneda, R.; Su, J.T.; Hua, L.X. TGFbeta1 T29C polymorphism and cancer risk: a meta-analysis based on 40 case-control studies. Cancer Genet. Cytogenet. 2010, 196, 68–75. [Google Scholar] [CrossRef]
- Kesarwani, P.; Mandhani, A.; Mittal, R.D. Polymorphisms in tumor necrosis factor-A gene and prostate cancer risk in North Indian cohort. J. Urol. 2009, 182, 2938–2943. [Google Scholar] [CrossRef]
- Saenz-Lopez, P.; Carretero, R.; Cozar, J.M.; Romero, J.M.; Canton, J.; Vilchez, J.R.; Tallada, M.; Garrido, F.; Ruiz-Cabello, F. Genetic polymorphisms of RANTES, IL1-A, MCP-1 and TNF-A genes in patients with prostate cancer. BMC Cancer 2008, 8, 382. [Google Scholar] [CrossRef]
- Turner, D.M.; Williams, D.M.; Sankaran, D.; Lazarus, M.; Sinnott, P.J.; Hutchinson, I.V. An investigation of polymorphism in the interleukin-10 gene promoter. Eur. J. Immunogenet. 1997, 24, 1–8. [Google Scholar] [CrossRef]
- Giedraitis, V.; He, B.; Huang, W.X.; Hillert, J. Cloning and mutation analysis of the human IL-18 promoter: a possible role of polymorphisms in expression regulation. J. Neuroimmunol. 2001, 112, 146–152. [Google Scholar] [CrossRef]
- Bennermo, M.; Held, C.; Stemme, S.; Ericsson, C.G.; Silveira, A.; Green, F.; Tornvall, P. Genetic Predisposition of the Interleukin-6 Response to Inflammation: Implications for a Variety of Major Diseases? Clin. Chem. 2004, 50, 2136–2140. [Google Scholar] [CrossRef]
- Boiardi, L.; Casali, B.; Farnetti, E.; Pipitone, N.; Nicoli, D.; Cantini, F.; Macchioni, P.; Bajocchi, G.; Catanoso, M.G.; Pulsatelli, L.; Consonni, D.; Salvarani, C. Relationship between interleukin 6 promoter polymorphism at position -174, IL-6 serum levels, and the risk of relapse/recurrence in polymyalgia rheumatica. J. Rheumatol. 2006, 33, 703–708. [Google Scholar]
- Brull, D.J.; Montgomery, H.E.; Sanders, J.; Dhamrait, S.; Luong, L.; Rumley, A.; Lowe, G.D.; Humphries, S.E. Interleukin-6 gene -174g>c and -572g>c promoter polymorphisms are strong predictors of plasma interleukin-6 levels after coronary artery bypass surgery. Arterioscler. Thromb. Vasc. Biol. 2001, 21, 1458–1463. [Google Scholar] [CrossRef]
- Fishman, D.; Faulds, G.; Jeffery, R.; Mohamed-Ali, V.; Yudkin, J.S.; Humphries, S.; Woo, P. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J. Clin. Invest. 1998, 102, 1369–1376. [Google Scholar] [CrossRef]
- Ravaglia, G.; Forti, P.; Maioli, F.; Chiappelli, M.; Dolzani, P.; Martelli, M.; Bianchin, M.; Mariani, E.; Bolondi, L.; Licastro, F. Associations of the -174 G/C interleukin-6 gene promoter polymorphism with serum interleukin 6 and mortality in the elderly. Biogerontology 2005, 6, 415–423. [Google Scholar] [CrossRef]
- Grainger, D.J.; Heathcote, K.; Chiano, M.; Snieder, H.; Kemp, P.R.; Metcalfe, J.C.; Carter, N.D.; Spector, T.D. Genetic control of the circulating concentration of transforming growth factor type beta1. Hum. Mol. Genet. 1999, 8, 93–97. [Google Scholar] [CrossRef]
- Meng, J.; Thongngarm, T.; Nakajima, M.; Yamashita, N.; Ohta, K.; Bates, C.A.; Grunwald, G.K.; Rosenwasser, L.J. Association of transforming growth factor-beta1 single nucleotide polymorphism C-509T with allergy and immunological activities. Int. Arch. Allergy Immunol. 2005, 138, 151–160. [Google Scholar] [CrossRef]
- Silverman, E.S.; Palmer, L.J.; Subramaniam, V.; Hallock, A.; Mathew, S.; Vallone, J.; Faffe, D.S.; Shikanai, T.; Raby, B.A.; Weiss, S.T.; Shore, S.A. Transforming growth factor-beta1 promoter polymorphism C-509T is associated with asthma. Am. J. Respir. Crit. Care Med. 2004, 169, 214–219. [Google Scholar] [CrossRef]
- Dunning, A.M.; Ellis, P.D.; McBride, S.; Kirschenlohr, H.L.; Healey, C.S.; Kemp, P.R.; Luben, R.N.; Chang-Claude, J.; Mannermaa, A.; Kataja, V.; Pharoah, P.D.; Easton, D.F.; Ponder, B.A.; Metcalfe, J.C. A transforming growth factorbeta1 signal peptide variant increases secretion in vitro and is associated with increased incidence of invasive breast cancer. Cancer Res. 2003, 63, 2610–2615. [Google Scholar]
- Hinke, V.; Seck, T.; Clanget, C.; Scheidt-Nave, C.; Ziegler, R.; Pfeilschifter, J. Association of transforming growth factor-beta1 (TGFbeta1) T29 --> C gene polymorphism with bone mineral density (BMD), changes in BMD, and serum concentrations of TGF-beta1 in a population-based sample of postmenopausal german women. Calcif. Tissue Int. 2001, 69, 315–320. [Google Scholar] [CrossRef]
- Yokota, M.; Ichihara, S.; Lin, T.L.; Nakashima, N.; Yamada, Y. Association of a T29-->C polymorphism of the transforming growth factor-beta1 gene with genetic susceptibility to myocardial infarction in Japanese. Circulation 2000, 101, 2783–2787. [Google Scholar] [CrossRef]
- Lv, K.; Chen, R.; Cai, Q.; Fang, M.; Sun, S. Effects of a single nucleotide polymorphism on the expression of human tumor necrosis factor-alpha. Scand. J. Immunol. 2006, 64, 164–169. [Google Scholar] [CrossRef]
- Kroeger, K.M.; Carville, K.S.; Abraham, L.J. The -308 tumor necrosis factor-alpha promoter polymorphism effects transcription. Mol. Immunol. 1997, 34, 391–399. [Google Scholar] [CrossRef]
- Louis, E.; Franchimont, D.; Piron, A.; Gevaert, Y.; Schaaf-Lafontaine, N.; Roland, S.; Mahieu, P.; Malaise, M.; De Groote, D.; Louis, R.; Belaiche, J. Tumour necrosis factor (TNF) gene polymorphism influences TNF-alpha production in lipopolysaccharide (LPS)-stimulated whole blood cell culture in healthy humans. Clin. Exp. Immunol. 1998, 113, 401–406. [Google Scholar] [CrossRef]
- Dominici, R.; Cattaneo, M.; Malferrari, G.; Archi, D.; Mariani, C.; Grimaldi, L.M.; Biunno, I. Cloning and functional analysis of the allelic polymorphism in the transcription regulatory region of interleukin-1 alpha. Immunogenetics 2002, 54, 82–86. [Google Scholar] [CrossRef]
- Chakravorty, M.; Datta De, D.; Choudhury, A.; Roychoudhury, S. IL1B promoter polymorphism regulates the expression of gastric acid stimulating hormone gastrin. Int. J. Biochem. Cell Biol. 2009, 41, 1502–1510. [Google Scholar] [CrossRef]
- Lind, H.; Haugen, A.; Zienolddiny, S. Differential binding of proteins to the IL1B -31 T/C polymorphism in lung epithelial cells. Cytokine 2007, 38, 43–48. [Google Scholar] [CrossRef]
- Michaud, D.S.; Daugherty, S.E.; Berndt, S.I.; Platz, E.A.; Yeager, M.; Crawford, E.D.; Hsing, A.; Huang, W.Y.; Hayes, R.B. Genetic polymorphisms of interleukin-1B (IL-1B), IL-6, IL-8, and IL-10 and risk of prostate cancer. Cancer Res. 2006, 66, 4525–4530. [Google Scholar] [CrossRef]
- Hwang, I.R.; Kodama, T.; Kikuchi, S.; Sakai, K.; Peterson, L.E.; Graham, D.Y.; Yamaoka, Y. Effect of interleukin 1 polymorphisms on gastric mucosal interleukin 1beta production in Helicobacter pylori infection. Gastroenterology 2002, 123, 1793–1803. [Google Scholar] [CrossRef]
- Xuan, J.; Deguchi, R.; Watanabe, S.; Ozawa, H.; Urano, T.; Ogawa, Y.; Fukuda, R.; Kijima, H.; Koga, Y.; Takagi, A. Relationship between IL-1beta gene polymorphism and gastric mucosal IL-1beta levels in patients with Helicobacter pylori infection. J. Gastroenterol. 2005, 40, 796–801. [Google Scholar] [CrossRef]
- Lindmark, F.; Zheng, S.L.; Wiklund, F.; Bälter, K.A.; Sun, J.; Chang, B.; Hedelin, M.; Clark, J.; Johansson, J.E.; Meyers, D.A.; Adami, H.O.; Isaacs, W.; Grönberg, H.; Xu, J. Interleukin-1 receptor antagonist haplotype associated with prostate cancer risk. Br. J. Cancer 2005, 93, 493–497. [Google Scholar] [CrossRef]
- Cheng, I.; Krumroy, L.M.; Plummer, S.J.; Casey, G.; Witte, J.S. MIC1 and IL1RN Genetic Variation and Advanced Prostate Cancer Risk. Cancer Epidemiol. Biomarkers Prev. 2007, 16, 1309–1311. [Google Scholar] [CrossRef]
- Tindall, E.A.; Hoang, H.N.; Southey, M.C.; English, D.R.; Hopper, J.L.; Giles, G.G.; Severi, G.; Hayes, V.M. The 4q27 locus and familial prostate cancer risk. BMC Cancer 2010, 10, 69. [Google Scholar] [CrossRef]
- Hoffmann, S.C.; Stanley, E.M.; Darrin Cox, E.; Craighead, N.; DiMercurio, B.S.; Koziol, D.E.; Harlan, D.M.; Kirk, A.D.; Blair, P.J. Association of cytokine polymorphic inheritance and in vitro cytokine production in anti-CD3/CD28-stimulated peripheral blood lymphocytes. Transplantation 2001, 72, 1444–1450. [Google Scholar] [CrossRef]
- Matesanz, F.; Fedetz, M.; Leyva, L.; Delgado, C.; Fernandez, O.; Alcina, A. Effects of the multiple sclerosis associated -330 promoter polymorphism in IL2 allelic expression. J. Neuroimmunol. 2004, 148, 212–217. [Google Scholar] [CrossRef]
- Nakashima, H.; Miyake, K.; Inoue, Y.; Shimizu, S.; Akahoshi, M.; Tanaka, Y.; Otsuka, T.; Harada, M. Association between IL-4 genotype and IL-4 production in the Japanese population. Genes Immun. 2002, 3, 107–109. [Google Scholar] [CrossRef]
- Sun, J.; Hedelin, M.; Zheng, S.L.; Adami, H.O.; Bensen, J.; Augustsson-Bälter, K.; Chang, B.; Adolfsson, J.; Adams, T.; Turner, A.; Meyers, D.A.; Isaacs, W.B.; Xu, J.; Grönberg, H. Interleukin-6 Sequence Variants Are not Associated with Prostate Cancer Risk Cancer Epidemiol. Biomarkers Prev. 2004, 13, 1677–1679. [Google Scholar]
- Pierce, B.L.; Biggs, M.L.; Decambre, M.; Reiner, A.P.; Li, C.; Fitzpatrick, A.; Carlson, C.S.; Stanford, J.L.; Austin, M.A. C-reactive protein, interleukin-6, and prostate cancer risk in men aged 65 years and older. Cancer Causes Cont. 2009, 20, 1193–1203. [Google Scholar] [CrossRef]
- Malarstig, A.; Wallentin, L.; Siegbahn, A. Genetic variation in the interleukin-6 gene in relation to risk and outcomes in acute coronary syndrome. Thromb. Res. 2007, 119, 467–473. [Google Scholar] [CrossRef]
- Eder, T.; Mayer, R.; Langsenlehner, U.; Renner, W.; Krippl, P.; Wascher, T.C.; Pummer, K.; Kapp, K.S. Interleukin-10 [ATA] promoter haplotype and prostate cancer risk: a population-based study. Eur. J. Cancer 2007, 43, 472–475. [Google Scholar] [CrossRef]
- Kang, D.; Lee, K.M.; Park, S.K.; Berndt, S.I.; Reding, D.; Chatterjee, N.; Welch, R.; Chanock, S.; Huang, W.Y.; Hayes, R.B. Lack of association of transforming growth factor-beta1 polymorphisms and haplotypes with prostate cancer risk in the prostate, lung, colorectal, and ovarian trial. Cancer Epidemiol. Biomarkers Prev. 2007, 16, 1303–1305. [Google Scholar] [CrossRef]
- Meyer, A.; Dork, T.; Bogdanova, N.; Brinkhaus, M.J.; Wiese, B.; Hagemann, J.; Serth, J.; Bremer, M.; Baumann, R.; Karstens, J.H.; Machtens, S. TGFB1 gene polymorphism Leu10Pro (c.29T>C), prostate cancer incidence and quality of life in patients treated with brachytherapy. World J. Urol. 2009, 27, 371–377. [Google Scholar] [CrossRef]
- Awad, M.R.; El-Gamel, A.; Hasleton, P.; Turner, D.M.; Sinnott, P.J.; Hutchinson, I.V. Genotypic variation in the transforming growth factor-beta1 gene: association with transforming growth factor-beta1 production, fibrotic lung disease, and graft fibrosis after lung transplantation. Transplantation 1998, 66, 1014–1020. [Google Scholar] [CrossRef]
- Danforth, K.N.; Rodriguez, C.; Hayes, R.B.; Sakoda, L.C.; Huang, W.Y.; Yu, K.; Calle, E.E.; Jacobs, E.J.; Chen, B.E.; Andriole, G.L.; Figueroa, J.D.; Yeager, M.; Platz, E.A.; Michaud, D.S.; Chanock, S.J.; Thun, M.J.; Hsing, A.W. TNF polymorphisms and prostate cancer risk. Prostate 2008, 68, 400–407. [Google Scholar] [CrossRef]
- Wilson, A.G.; Symons, J.A.; McDowell, T.L.; McDevitt, H.O.; Duff, G.W. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc. Natl. Acad. Sci. USA 1997, 94, 3195–3199. [Google Scholar] [CrossRef]
- Huizinga, T.W.; Westendorp, R.G.; Bollen, E.L.; Keijsers, V.; Brinkman, B.M.; Langermans, J.A.; Breedveld, F.C.; Verweij, C.L.; van de Gaer, L.; Dams, L.; Crusius, J.B.; Garcia-Gonzalez, A.; van Oosten, B.W.; Polman, C.H.; Pena, A.S. TNF-alpha promoter polymorphisms, production and susceptibility to multiple sclerosis in different groups of patients. J. Neuroimmunol. 1997, 72, 149–153. [Google Scholar] [CrossRef]
- Crawley, E.; Kay, R.; Sillibourne, J.; Patel, P.; Hutchinson, I.; Woo, P. Polymorphic haplotypes of the interleukin-10 5' flanking region determine variable interleukin-10 transcription and are associated with particular phenotypes of juvenile rheumatoid arthritis. Arthritis. Rheum. 1999, 42, 1101–1108. [Google Scholar] [CrossRef]
- Rad, R.; Dossumbekova, A.; Neu, B.; Lang, R.; Bauer, S.; Saur, D.; Gerhard, M.; Prinz, C. Cytokine gene polymorphisms influence mucosal cytokine expression, gastric inflammation, and host specific colonisation during Helicobacter pylori infection. Gut 2004, 53, 1082–1089. [Google Scholar] [CrossRef]
- Balkwill, F. Cancer and the chemokine network. Nat. Rev. Cancer 2004, 4, 540–550. [Google Scholar]
- Gerber, P.A.; Hippe, A.; Buhren, B.A.; Muller, A.; Homey, B. Chemokines in tumor-associated angiogenesis. Biol. Chem. 2009, 390, 1213–1223. [Google Scholar]
- Vandercappellen, J.; Van Damme, J.; Struyf, S. The role of CXC chemokines and their receptors in cancer. Cancer Lett. 2008, 267, 226–244. [Google Scholar] [CrossRef]
- Shen, H.; Lentsch, A.B. Progressive dysregulation of transcription factors NF-kappa B and STAT1 in prostate cancer cells causes proangiogenic production of CXC chemokines. Am. J. Physiol. Cell Physiol. 2004, 286, C840–C847. [Google Scholar] [CrossRef]
- Vindrieux, D.; Escobar, P.; Lazennec, G. Emerging roles of chemokines in prostate cancer. Endocr. Relat. Cancer 2009, 16, 663–673. [Google Scholar] [CrossRef]
- Waugh, D.J.; Wilson, C.; Seaton, A.; Maxwell, P.J. Multi-faceted roles for CXC-chemokines in prostate cancer progression. Front Biosci. 2008, 13, 4595–4604. [Google Scholar]
- Haverkamp, J.; Charbonneau, B.; Ratliff, T.L. Prostate inflammation and its potential impact on prostate cancer: a current review. J. Cell Biochem. 2008, 103, 1344–1353. [Google Scholar] [CrossRef]
- Yang, H.P.; Woodson, K.; Taylor, P.R.; Pietinen, P.; Albanes, D.; Virtamo, J.; Tangrea, J.A. Genetic variation in interleukin 8 and its receptor genes and its influence on the risk and prognosis of prostate cancer among Finnish men in a large cancer prevention trial. Eur. J. Cancer Prev. 2006, 15, 249–253. [Google Scholar] [CrossRef]
- Hirata, H.; Hinoda, Y.; Kikuno, N.; Kawamoto, K.; Dahiya, A.V.; Suehiro, Y.; Tanaka, Y.; Dahiya, R. CXCL12 G801A polymorphism is a risk factor for sporadic prostate cancer susceptibility. Clin. Cancer Res. 2007, 13, 5056–5062. [Google Scholar] [CrossRef]
- Petersen, D.C.; Severi, G.; Hoang, H.N.; Padilla, E.J.; Southey, M.C.; English, D.R.; Hopper, J.L.; Giles, G.G.; Hayes, V.M. No association between common chemokine and chemokine receptor gene variants and prostate cancer risk. Cancer Epidemiol. Biomarkers Prev. 2008, 17, 3615–3617. [Google Scholar] [CrossRef]
- Balistreri, C.R.; Carruba, G.; Calabro, M.; Campisi, I.; Di Carlo, D.; Lio, D.; Colonna-Romano, G.; Candore, G.; Caruso, C. CCR5 proinflammatory allele in prostate cancer risk: a pilot study in patients and centenarians from Sicily. Ann. N. Y. Acad. Sci. 2009, 1155, 289–292. [Google Scholar]
- Rovin, B.H.; Lu, L.; Saxena, R. A novel polymorphism in the MCP-1 gene regulatory region that influences MCP-1 expression. Biochem. Biophys. Res. Commun. 1999, 259, 344–348. [Google Scholar] [CrossRef]
- Nickel, R.G.; Casolaro, V.; Wahn, U.; Beyer, K.; Barnes, K.C.; Plunkett, B.S.; Freidhoff, L.R.; Sengler, C.; Plitt, J.R.; Schleimer, R.P.; Caraballo, L.; Naidu, R.P.; Levett, P.N.; Beaty, T.H.; Huang, S.K. Atopic dermatitis is associated with a functional mutation in the promoter of the C-C chemokine RANTES. J. Immunol. 2000, 164, 1612–1616. [Google Scholar]
- Liu, R.; Paxton, W.A.; Choe, S.; Ceradini, D.; Martin, S.R.; Horuk, R.; MacDonald, M.E.; Stuhlmann, H.; Koup, R.A.; Landau, N.R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 1996, 86, 367–377. [Google Scholar] [CrossRef]
- Nakayama, E.E.; Tanaka, Y.; Nagai, Y.; Iwamoto, A.; Shioda, T. A CCR2-V64I polymorphism affects stability of CCR2A isoform. AIDS 2004, 18, 729–738. [Google Scholar] [CrossRef]
- Hull, J.; Thomson, A.; Kwiatkowski, D. Association of respiratory syncytial virus bronchiolitis with the interleukin 8 gene region in UK families. Thorax 2000, 55, 1023–1027. [Google Scholar] [CrossRef]
- Winkler, C.; Modi, W.; Smith, M.W.; Nelson, G.W.; Wu, X.; Carrington, M.; Dean, M.; Honjo, T.; Tashiro, K.; Yabe, D.; Buchbinder, S.; Vittinghoff, E.; Goedert, J.J.; O'Brien, T.R; Jacobson, L.P.; Detels, R.; Donfield, S.; Willoughby, A.; Gomperts, E.; Vlahov, D.; Phair, J.; O'Brien, S.J. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. ALIVE Study, Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC). Science 1998, 279, 389–393. [Google Scholar] [CrossRef]
- Faure, S.; Meyer, L.; Costagliola, D.; Vaneensberghe, C.; Genin, E.; Autran, B.; Delfraissy, J.F.; McDermott, D.H.; Murphy, P.M.; Debre, P.; Theodorou, I.; Combadiere, C. Rapid progression to AIDS in HIV+ individuals with a structural variant of the chemokine receptor CX3CR1. Science 2000, 287, 2274–2277. [Google Scholar] [CrossRef]
- Hattersley, A.T.; McCarthy, M.I. What makes a good genetic association study? Lancet 2005, 366, 1315–1323. [Google Scholar] [CrossRef]
- Williams, H.; Powell, I.J. Epidemiology, pathology, and genetics of prostate cancer among African Americans compared with other ethnicities. Methods Mol. Biol. 2009, 472, 439–453. [Google Scholar] [CrossRef]
- Yu, K.; Wang, Z.; Li, Q.; Wacholder, S.; Hunter, D.J.; Hoover, R.N.; Chanock, S.; Thomas, G. Population substructure and control selection in genome-wide association studies. PLoS ONE 2008, 3, e2551. [Google Scholar]
- Varghese, J.S.; Easton, D.F. Genome-wide association studies in common cancers-what have we learnt? Curr. Opin. Genet. Dev. 2010. [Google Scholar] [CrossRef]
- Schork, N.J.; Murray, S.S.; Frazer, K.A.; Topol, E.J. Common vs.. rare allele hypotheses for complex diseases. Curr. Opin. Genet. Dev. 2009, 19, 212–219. [Google Scholar] [CrossRef]
© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Tindall, E.A.; Hayes, V.M.; Petersen, D.C. Inflammatory Genetic Markers of Prostate Cancer Risk. Cancers 2010, 2, 1198-1220. https://doi.org/10.3390/cancers2021198
Tindall EA, Hayes VM, Petersen DC. Inflammatory Genetic Markers of Prostate Cancer Risk. Cancers. 2010; 2(2):1198-1220. https://doi.org/10.3390/cancers2021198
Chicago/Turabian StyleTindall, Elizabeth A., Vanessa M. Hayes, and Desiree C. Petersen. 2010. "Inflammatory Genetic Markers of Prostate Cancer Risk" Cancers 2, no. 2: 1198-1220. https://doi.org/10.3390/cancers2021198