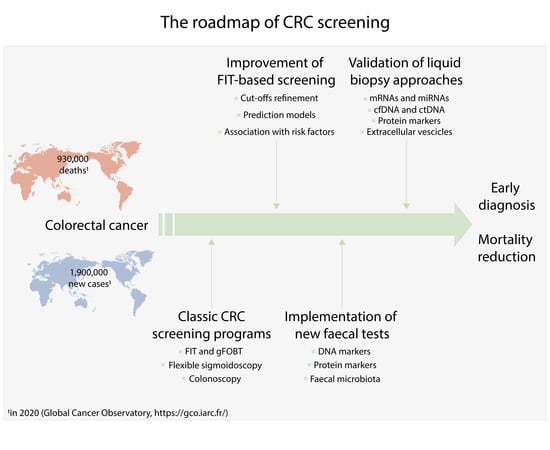
The Roadmap of Colorectal Cancer Screening
Abstract
:Simple Summary
Abstract
1. Introduction
2. Colorectal Cancer Status in Europe and in the World
3. Colorectal Cancer Screening
3.1. Advantages of Screening in Terms of Incidence and Mortality
3.2. Screening Tests
3.2.1. Stool-Based Tests
3.2.2. Imaging Tests
3.2.3. Endoscopic Tests
3.3. Screening Status in Europe and the World
4. Disadvantages of Fecal Tests (FIT/gFOBT), Room for Improvement
5. New Tests
5.1. Fecal Tests
5.2. A New Alternative: Liquid Biopsy
5.2.1. mRNA
5.2.2. miRNA
5.2.3. DNA
5.2.4. Proteins
5.2.5. Extracellular Vesicles
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Vogelstein, B.; Fearon, E.R.; Hamilton, S.R.; Kern, S.E.; Preisinger, A.C.; Leppert, M.; Smits, A.M.M.; Bos, J.L. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 1988, 319, 525–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puccini, A.; Berger, M.D.; Naseem, M.; Tokunaga, R.; Battaglin, F.; Cao, S.; Hanna, D.L.; McSkane, M.; Soni, S.; Zhang, W.; et al. Colorectal cancer: Epigenetic alterations and their clinical implications. Biochim. Biophys. Acta Rev. Cancer 2017, 1868, 439–448. [Google Scholar] [CrossRef]
- Brenner, H.; Kloor, M.; Pox, C.P. Colorectal cancer. Lancet 2014, 383, 1490–1502. [Google Scholar] [CrossRef]
- Siskova, A.; Cervena, K.; Kral, J.; Hucl, T.; Vodicka, P.; Vymetalkova, V. Colorectal adenomas—genetics and searching for new molecular screening biomarkers. Int. J. Mol. Sci. 2020, 21, 3260. [Google Scholar] [CrossRef]
- Hadjipetrou, A.; Anyfantakis, D.; Galanakis, C.G.; Kastanakis, M.; Kastanakis, S. Colorectal cancer, screening and primary care: A mini literature review. World J. Gastroenterol. 2017, 23, 6049–6058. [Google Scholar] [CrossRef]
- Global Cancer Observatory. Available online: https://gco.iarc.fr/ (accessed on 21 January 2021).
- Rosso, C.; Cabianca, L.; Gili, F.M. Non-invasive markers to detect colorectal cancer in asymptomatic population. Minerva Biotecnol. 2019, 31, 23–29. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schreuders, E.H.; Ruco, A.; Rabeneck, L.; Schoen, R.E.; Sung, J.J.Y.; Young, G.P.; Kuipers, E.J. Colorectal cancer screening: A global overview of existing programmes. Gut 2015, 64, 1637–1649. [Google Scholar] [CrossRef] [PubMed]
- Gini, A.; Jansen, E.E.L.; Zielonke, N.; Meester, R.G.S.; Senore, C.; Anttila, A.; Segnan, N.; Mlakar, D.N.; de Koning, H.J.; Lansdorp-Vogelaar, I.; et al. Impact of colorectal cancer screening on cancer-specific mortality in Europe: A systematic review. Eur. J. Cancer 2020, 127, 224–235. [Google Scholar] [CrossRef] [Green Version]
- Associazione Italiana di Oncologia Medica; Associazione Italiana dei Registri Tumori; Progressi delle Aziende Sanitarie per la Salute in Italia; Società Italiana di Anatomia Patologica e Citodiagnostica. I numeri del Cancro in Italia, 9th ed.; Intermedia Editore: Brescia, Italy, 2019. [Google Scholar]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef] [Green Version]
- Kaminski, M.F.; Robertson, D.J.; Senore, C.; Rex, D.K. Optimizing the quality of colorectal cancer screening worldwide. Gastroenterology 2020, 158, 404–417. [Google Scholar] [CrossRef] [PubMed]
- Senore, C.; Basu, P.; Anttila, A.; Ponti, A.; Tomatis, M.; Vale, D.B.; Ronco, G.; Soerjomataram, I.; Primic-Žakelj, M.; Riggi, E.; et al. Performance of colorectal cancer screening in the European Union Member States: Data from the second European screening report. Gut 2019, 68, 1232–1244. [Google Scholar] [CrossRef] [PubMed]
- Vicentini, M.; Zorzi, M.; Bovo, E.; Mancuso, P.; Zappa, M.; Manneschi, G.; Mangone, L.; Giorgi Rossi, P. Impact of screening programme using the faecal immunochemical test on stage of colorectal cancer: Results from the IMPATTO study. Int. J. Cancer 2019, 145, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Rossi, P.G.; Vicentini, M.; Sacchettini, C.; Di Felice, E.; Caroli, S.; Ferrari, F.; Mangone, L.; Pezzarossi, A.; Roncaglia, F.; Campari, C.; et al. Impact of screening program on incidence of colorectal cancer: A cohort study in Italy. Am. J. Gastroenterol. 2015, 110, 1359–1366. [Google Scholar] [CrossRef] [PubMed]
- Brenner, H.; Stock, C.; Hoffmeister, M. Effect of screening sigmoidoscopy and screening colonoscopy on colorectal cancer incidence and mortality: Systematic review and meta-analysis of randomised controlled trials and observational studies. BMJ 2014, 348, g2467. [Google Scholar] [CrossRef] [Green Version]
- Grobbee, E.J.; van der Vlugt, M.; van Vuuren, A.J.; Stroobants, A.K.; Mallant-Hent, R.C.; Lansdorp-Vogelaar, I.; Bossuyt, P.M.M.; Kuipers, E.J.; Dekker, E.; Spaander, M.C.W. Diagnostic yield of one-time colonoscopy vs one-time flexible sigmoidoscopy vs multiple rounds of mailed fecal immunohistochemical tests in colorectal cancer screening. Clin. Gastroenterol. Hepatol. 2020, 18, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Holme, Ø.; Løberg, M.; Kalager, M.; Bretthauer, M.; Hernán, M.A.; Aas, E.; Eide, T.J.; Skovlund, E.; Lekven, J.; Schneede, J.; et al. Long-term effectiveness of sigmoidoscopy screening on colorectal cancer incidence and mortality in women and men. Ann. Intern. Med. 2018, 168, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Jodal, H.C.; Helsingen, L.M.; Anderson, J.C.; Lytvyn, L.; Vandvik, P.O.; Emilsson, L. Colorectal cancer screening with faecal testing, sigmoidoscopy or colonoscopy: A systematic review and network meta-analysis. BMJ Open 2019, 9, e032773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greegor, D.H. Diagnosis of large-bowel cancer in the asymptomatic patient. J. Am. Med. Assoc. 1967, 201, 943–945. [Google Scholar] [CrossRef]
- Lee, J.K.; Liles, E.G.; Bent, S.; Levin, T.R.; Corley, D.A. Accuracy of fecal immunochemical tests for colorectal cancer. Ann. Intern. Med. 2014, 160, 171–181. [Google Scholar] [CrossRef]
- Selby, K.; Levine, E.H.; Doan, C.; Gies, A.; Brenner, H.; Quesenberry, C.; Lee, J.K.; Corley, D.A. Effect of sex, age, and positivity threshold on fecal immunochemical test accuracy: A systematic review and meta-analysis. Gastroenterology 2019, 157, 1494–1505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stracci, F.; Zorzi, M.; Grazzini, G. Colorectal cancer screening: Tests, strategies, and perspectives. Front. Public Health 2014, 2, 210. [Google Scholar] [CrossRef] [Green Version]
- Cusumano, V.T.; May, F.P. Making FIT count: Maximizing appropriate use of the fecal immunochemical test for colorectal cancer screening programs. J. Gen. Intern. Med. 2020, 35, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Amitay, E.L.; Cuk, K.; Niedermaier, T.; Weigl, K.; Brenner, H. Factors associated with false-positive fecal immunochemical tests in a large German colorectal cancer screening study. Int. J. Cancer 2019, 144, 2419–2427. [Google Scholar] [CrossRef]
- Spada, C.; Hassan, C.; Bellini, D.; Burling, D.; Cappello, G.; Carretero, C.; Dekker, E.; Eliakim, R.; de Haan, M.; Kaminski, M.F.; et al. Imaging alternatives to colonoscopy: CT colonography and colon capsule. European Society of Gastrointestinal Endoscopy (ESGE) and European Society of Gastrointestinal and Abdominal Radiology (ESGAR) Guideline—Update 2020. Endoscopy 2020, 52, 1127–1141. [Google Scholar]
- Malila, N.; Senore, C.; Armaroli, P. European guidelines for quality assurance in colorectal cancer screening and diagnosis. Endoscopy 2012, 44, SE31–SE48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ONS (Osservatorio Nazionale Screening). Rapporto 2019; ONS: Firenze, Italy, 2019; pp. 1–45. [Google Scholar]
- Venturelli, F.; Sampaolo, L.; Carrozzi, G.; Zappa, M.; Giorgi Rossi, P. Associations between cervical, breast and colorectal cancer screening uptake, chronic diseases and health-related behaviours: Data from the Italian PASSI nationwide surveillance. Prev. Med. 2019, 120, 60–70. [Google Scholar] [CrossRef]
- Mancini, S.; Bucchi, L.; Giuliani, O.; Ravaioli, A.; Vattiato, R.; Baldacchini, F.; Ferretti, S.; Sassoli de Bianchi, P.; Mezzetti, F.; Triossi, O.; et al. Proportional incidence of interval colorectal cancer in a large population-based faecal immunochemical test screening programme. Dig. Liver Dis. 2020, 52, 452–456. [Google Scholar] [CrossRef]
- Basu, P.; Ponti, A.; Anttila, A.; Ronco, G.; Senore, C.; Vale, D.B.; Segnan, N.; Tomatis, M.; Soerjomataram, I.; Primic Žakelj, M.; et al. Status of implementation and organization of cancer screening in The European Union Member States—Summary results from the second European screening report. Int. J. Cancer 2018, 142, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Vale, D.B.; Anttila, A.; Ponti, A.; Senore, C.; Sankaranaryanan, R.; Ronco, G.; Segnan, N.; Tomatis, M.; Žakelj, M.P.; Elfström, K.M.; et al. Invitation strategies and coverage in the population-based cancer screening programmes in the European Union. Eur. J. Cancer Prev. 2019, 28, 131–140. [Google Scholar] [CrossRef]
- Ponti, A.; Anttila, A.; Ronco, G.; Senore, C.; Basu, P.; Segnan, N.; Tomatis, M.; Primic-Žakelj, M.; Dillner, J.; Fernan, M.; et al. Cancer Screening in the European Union. Report on the Implementation of the Council Recommendation on Cancer Screening; International Agency for Research on Cancer: Lyon, France, 2017; pp. 1–313. [Google Scholar]
- Navarro, M.; Nicolas, A.; Ferrandez, A.; Lanas, A. Colorectal cancer population screening programs worldwide in 2016: An update. World J. Gastroenterol. 2017, 23, 3632–3642. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, R.; Guo, F.; Heisser, T.; Hoffmeister, M.; Brenner, H. Utilisation of colorectal cancer screening tests in european countries by type of screening offer: Results from the european health interview survey. Cancers 2020, 12, 1409. [Google Scholar] [CrossRef] [PubMed]
- Altobelli, E.; D’Aloisio, F.; Angeletti, P.M. Colorectal cancer screening in countries of European Council outside of the EU-28. World J. Gastroenterol. 2016, 22, 4946–4957. [Google Scholar] [CrossRef]
- Altobelli, E.; Rapacchietta, L.; Marziliano, C.; Campagna, G.; Profeta, V.; Fagnano, R. Differences in colorectal cancer surveillance epidemiology and screening in the WHO European Region. Oncol. Lett. 2019, 17, 2531–2542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- United States Preventive Services Taskforce Recommendation: Colorectal Cancer Screening. Available online: https://www.uspreventiveservicestaskforce.org/uspstf/draft-update-summary/colorectal-cancer-screening3 (accessed on 21 January 2021).
- Sung, J.J.Y.; Ng, S.C.; Chan, F.K.L.; Chiu, H.M.; Kim, H.S.; Matsuda, T.; Ng, S.S.M.; Lau, J.Y.W.; Zheng, S.; Adler, S.; et al. An updated Asia Pacific Consensus Recommendations on colorectal cancer screening. Gut 2015, 64, 121–132. [Google Scholar] [CrossRef]
- The Asia Cohort Consortium. Available online: https://www.asiacohort.org/index.html (accessed on 22 February 2021).
- Kooyker, A.I.; Toes-Zoutendijk, E.; Opstal-van Winden, A.W.J.; Spaander, M.C.W.; Buskermolen, M.; Vuuren, H.J.; Kuipers, E.J.; Kemenade, F.J.; Ramakers, C.; Thomeer, M.G.J.; et al. The second round of the Dutch colorectal cancer screening program: Impact of an increased fecal immunochemical test cut-off level on yield of screening. Int. J. Cancer 2020, 147, 1098–1106. [Google Scholar] [CrossRef]
- Rutka, M.; Bor, R.; Molnár, T.; Farkas, K.; Pigniczki, D.; Fábián, A.; Győrffy, M.; Bálint, A.; Milassin, Á.; Szűcs, M.; et al. Efficacy of the population-based pilot colorectal cancer screening, Csongrád county, Hungary, 2015. Turk. J. Med. Sci. 2020, 50, 756–763. [Google Scholar] [CrossRef]
- Nielson, C.M.; Petrik, A.F.; Jacob, L.; Vollmer, W.M.; Keast, E.M.; Schneider, J.L.; Rivelli, J.S.; Kapka, T.J.; Meenan, R.T.; Mummadi, R.R.; et al. Positive predictive values of fecal immunochemical tests used in the STOP CRC pragmatic trial. Cancer Med. 2018, 7, 4781–4790. [Google Scholar] [CrossRef] [PubMed]
- Berry, E.; Miller, S.; Koch, M.; Balasubramanian, B.; Argenbright, K.; Gupta, S. Lower abnormal fecal immunochemical test cut-off values improve detection of colorectal cancer in system-level screens. Clin. Gastroenterol. Hepatol. 2020, 18, 647–653. [Google Scholar] [CrossRef]
- de Klerk, C.M.; Vendrig, L.M.; Bossuyt, P.M.; Dekker, E. Participant-related risk factors for false-positive and false-negative fecal immunochemical tests in colorectal cancer screening: Systematic review and meta-analysis. Am. J. Gastroenterol. 2018, 113, 1778–1787. [Google Scholar] [CrossRef]
- Stegeman, I.; De Wijkerslooth, T.R.; Stoop, E.M.; Van Leerdam, M.; Van Ballegooijen, M.; Kraaijenhagen, R.A.; Fockens, P.; Kuipers, E.J.; Dekker, E.; Bossuyt, P.M. Risk factors for false positive and for false negative test results in screening with fecal occult blood testing. Int. J. Cancer 2013, 133, 2408–2414. [Google Scholar] [CrossRef] [Green Version]
- Wong, M.C.S.; Ching, J.Y.L.; Chan, V.C.W.; Lam, T.Y.T.; Luk, A.K.C.; Ng, S.S.M.; Sung, J.J.Y. Factors associated with false-positive and false-negative fecal immunochemical test results for colorectal cancer screening. Gastrointest. Endosc. 2015, 81, 596–607. [Google Scholar] [CrossRef]
- Gies, A.; Cuk, K.; Schrotz-King, P.; Brenner, H. Direct comparison of diagnostic performance of 9 quantitative fecal immunochemical tests for colorectal cancer screening. Gastroenterology 2018, 154, 93–104. [Google Scholar] [CrossRef] [Green Version]
- Imperiale, T.F.; Gruber, R.N.; Stump, T.E.; Emmett, T.W.; Monahan, P.O. Performance characteristics of fecal immunochemical tests for colorectal cancer and advanced adenomatous polyps: A systematic review and meta-analysis. Ann. Intern. Med. 2019, 170, 319–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibañez-Sanz, G.; Garcia, M.; Mila, N.; Hubbard, R.A.; Vidal, C.; Binefa, G.; Benito, L.; Moreno, V. False-positive results in a population-based colorectal screening program: Cumulative risk from 2000 to 2017 with biennial screening. Cancer Epidemiol. Biomarkers Prev. 2019, 28, 1909–1916. [Google Scholar] [CrossRef] [Green Version]
- van de Veerdonk, W.; Hoeck, S.; Peeters, M.; Van Hal, G. Towards risk-stratified colorectal cancer screening. Adding risk factors to the fecal immunochemical test: Evidence, evolution and expectations. Prev. Med. 2019, 126, 105746. [Google Scholar] [CrossRef]
- Senore, C.; Zappa, M.; Campari, C.; Crotta, S.; Armaroli, P.; Arrigoni, A.; Cassoni, P.; Colla, R.; Fracchia, M.; Gili, F.; et al. Faecal haemoglobin concentration among subjects with negative FIT results is associated with the detection rate of neoplasia at subsequent rounds: A prospective study in the context of population based screening programmes in Italy. Gut 2020, 69, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Blom, J.; Löwbeer, C.; Elfström, K.M.; Sventelius, M.; Öhman, D.; Saraste, D.; Törnberg, S. Gender-specific cut-offs in colorectal cancer screening with FIT: Increased compliance and equal positivity rate. J. Med. Screen. 2019, 26, 92–97. [Google Scholar] [CrossRef]
- Mannucci, A.; Zuppardo, R.A.; Rosati, R.; Di Leo, M.; Perea, J.; Cavestro, G.M. Colorectal cancer screening from 45 years of age: Thesis, antithesis and synthesis. World J. Gastroenterol. 2019, 25, 2565–2580. [Google Scholar] [CrossRef]
- Sidransky, D.; Tokino, T.; Hamilton, S.R.; Kinzler, K.W.; Levin, B.; Frost, P.; Vogelstein, B. Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science 1992, 256, 102–105. [Google Scholar] [CrossRef]
- Niedermaier, T.; Weigl, K.; Hoffmeister, M.; Brenner, H. Fecal immunochemical tests combined with other stool tests for colorectal cancer and advanced adenoma detection: A systematic review. Clin. Transl. Gastroenterol. 2016, 7, e175. [Google Scholar] [CrossRef] [PubMed]
- Gies, A.; Cuk, K.; Schrotz-King, P.; Brenner, H. Fecal immunochemical test for hemoglobin in combination with fecal transferrin in colorectal cancer screening. United Eur. Gastroenterol. J. 2018, 6, 1223–1231. [Google Scholar] [CrossRef] [Green Version]
- Widlak, M.M.; Neal, M.; Daulton, E.; Thomas, C.L.; Tomkins, C.; Singh, B.; Harmston, C.; Wicaksono, A.; Evans, C.; Smith, S.; et al. Risk stratification of symptomatic patients suspected of colorectal cancer using faecal and urinary markers. Color. Dis. 2018, 20, 335–342. [Google Scholar] [CrossRef]
- Ahlquist, D.A.; Skoletsky, J.E.; Boynton, K.A.; Harrington, J.J.; Mahoney, D.W.; Pierceall, W.E.; Thibodeau, S.N.; Shuber, A.P. Colorectal cancer screening by detection of altered human DNA in stool: Feasibility of a multitarget assay panel. Gastroenterology 2000, 119, 1219–1227. [Google Scholar] [CrossRef] [PubMed]
- Imperiale, T.F.; Levin, T.R.; Lidgard, G.P.; Itzkowitz, S.H.; Ahlquist, D.A.; Ransohoff, D.F.; Lavin, P.; Berger, B.M. Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 2014, 370, 1287–1297. [Google Scholar] [CrossRef] [Green Version]
- Bosch, L.J.W.; Melotte, V.; Mongera, S.; Daenen, K.L.J.; Coupé, V.M.H.; Van Turenhout, S.T.; Stoop, E.M.; De Wijkerslooth, T.R.; Mulder, C.J.J.; Rausch, C.; et al. Multitarget stool DNA test performance in an average-risk colorectal cancer screening population. Am. J. Gastroenterol. 2019, 114, 1909–1918. [Google Scholar] [CrossRef] [Green Version]
- Mu, J.; Huang, Y.; Cai, S.; Li, Q.; Song, Y.; Yuan, Y.; Zhang, S.; Zheng, S. Plausibility of an extensive use of stool DNA test for screening advanced colorectal neoplasia. Clin. Chim. Acta 2020, 501, 42–47. [Google Scholar] [CrossRef]
- Rengucci, C.; De Maio, G.; Menghi, M.; Scarpi, E.; Guglielmo, S.; Fusaroli, P.; Caletti, G.; Saragoni, L.; Casadei Gardini, A.; Zoli, W.; et al. Improved stool DNA integrity method for early colorectal cancer diagnosis. Cancer Epidemiol. Biomarkers Prev. 2014, 23, 2553–2560. [Google Scholar] [CrossRef] [Green Version]
- Calistri, D.; Rengucci, C.; Casadei Gardini, A.; Frassineti, G.L.; Scarpi, E.; Zoli, W.; Falcini, F.; Silvestrini, R.; Amadori, D. Fecal DNA for noninvasive diagnosis of colorectal cancer in immunochemical fecal occult blood test-positive individuals. Cancer Epidemiol. Biomarkers Prev. 2010, 19, 2647–2654. [Google Scholar] [CrossRef] [Green Version]
- Grobbee, E.J.; Lam, S.Y.; Fuhler, G.M.; Blakaj, B.; Konstantinov, S.R.; Bruno, M.J.; Peppelenbosch, M.P.; Kuipers, E.J.; Spaander, M.C.W. First steps towards combining faecal immunochemical testing with the gut microbiome in colorectal cancer screening. United Eur. Gastroenterol. J. 2020, 8, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Malagón, M.; Ramió-Pujol, S.; Serrano, M.; Serra-Pagès, M.; Amoedo, J.; Oliver, L.; Bahí, A.; Mas–de–Xaxars, T.; Torrealba, L.; Gilabert, P.; et al. Reduction of faecal immunochemical test false-positive results using a signature based on faecal bacterial markers. Aliment. Pharmacol. Ther. 2019, 49, 1410–1420. [Google Scholar] [CrossRef]
- Komor, M.A.; Bosch, L.J.W.; Coupé, V.M.H.; Rausch, C.; Pham, T.V.; Piersma, S.R.; Mongera, S.; Mulder, C.J.J.; Dekker, E.; Kuipers, E.J.; et al. Proteins in stool as biomarkers for non-invasive detection of colorectal adenomas with high risk of progression. J. Pathol. 2020, 250, 288–298. [Google Scholar] [CrossRef] [Green Version]
- Redwood, D.G.; Blake, I.D.; Provost, E.M.; Kisiel, J.B.; Sacco, F.D.; Ahlquist, D.A. Alaska native patient and provider perspectives on the multitarget stool DNA test compared with colonoscopy for colorectal cancer screening. J. Prim. Care Community Health 2019, 10, 215013271988429. [Google Scholar] [CrossRef]
- Symonds, E.L.; Cock, C.; Meng, R.; Cole, S.R.; Fraser, R.J.L.; Young, G.P. Uptake of a colorectal cancer screening blood test in people with elevated risk for cancer who cannot or will not complete a faecal occult blood test. Eur. J. Cancer Prev. 2018, 27, 425–432. [Google Scholar] [CrossRef]
- Normanno, N.; Cervantes, A.; Ciardiello, F.; De Luca, A.; Pinto, C. The liquid biopsy in the management of colorectal cancer patients: Current applications and future scenarios. Cancer Treat. Rev. 2018, 70, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Benning, T.M.; Dellaert, B.G.C.; Dirksen, C.D.; Severens, J.L. Preferences for potential innovations in non-invasive colorectal cancer screening: A labeled discrete choice experiment for a Dutch screening campaign. Acta Oncol. 2014, 53, 898–908. [Google Scholar] [CrossRef] [Green Version]
- Osborne, J.M.; Flight, I.; Wilson, C.J.; Chen, G.; Ratcliffe, J.; Young, G.P. The impact of sample type and procedural attributes on relative acceptability of different colorectal cancer screening regimens. Patient Prefer. Adherence 2018, 12, 1825–1836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcuello, M.; Vymetalkova, V.; Neves, R.P.L.; Duran-Sanchon, S.; Vedeld, H.M.; Tham, E.; van Dalum, G.; Flügen, G.; Garcia-Barberan, V.; Fijneman, R.J.; et al. Circulating biomarkers for early detection and clinical management of colorectal cancer. Mol. Asp. Med. 2019, 69, 107–122. [Google Scholar] [CrossRef]
- Niedermaier, T.; Weigl, K.; Hoffmeister, M.; Brenner, H. Fecal immunochemical tests in combination with blood tests for colorectal cancer and advanced adenoma detection—Systematic review. United Eur. Gastroenterol. J. 2018, 6, 13–21. [Google Scholar] [CrossRef] [Green Version]
- Byron, S.A.; Van Keuren-Jensen, K.R.; Engelthaler, D.M.; Carpten, J.D.; Craig, D.W. Translating RNA sequencing into clinical diagnostics: Opportunities and challenges. Nat. Rev. Genet. 2016, 17, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Rodia, M.T.; Ugolini, G.; Mattei, G.; Montroni, I.; Zattoni, D.; Ghignone, F.; Veronese, G.; Marisi, G.; Lauriola, M.; Strippoli, P.; et al. Systematic large-scale meta-analysis identifies a panel of two mRNAs as blood biomarkers for colorectal cancer detection. Oncotarget 2016, 7, 30295–30306. [Google Scholar] [CrossRef]
- Rodia, M.T.; Solmi, R.; Pasini, F.; Nardi, E.; Mattei, G.; Ugolini, G.; Ricciardiello, L.; Strippoli, P.; Miglio, R.; Lauriola, M. LGALS4, CEACAM6, TSPAN8, and COL1A2: Blood markers for colorectal cancer—validation in a cohort of subjects with positive fecal immunochemical test result. Clin. Colorectal Cancer 2018, 17, e217–e228. [Google Scholar] [CrossRef]
- Ferlizza, E.; Solmi, R.; Miglio, R.; Nardi, E.; Mattei, G.; Sgarzi, M.; Lauriola, M. Colorectal cancer screening: Assessment of CEACAM6, LGALS4, TSPAN8 and COL1A2 as blood markers in faecal immunochemical test negative subjects. J. Adv. Res. 2020, 24, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Chao, S.; Ying, J.; Liew, G.; Marshall, W.; Liew, C.-C.; Burakoff, R. Blood RNA biomarker panel detects both left- and right-sided colorectal neoplasms: A case-control study. J. Exp. Clin. Cancer Res. 2013, 32, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marshall, K.W.; Mohr, S.; El Khettabi, F.; Nossova, N.; Chao, S.; Bao, W.; Ma, J.; Li, X.J.; Liew, C.C. A blood-based biomarker panel for stratifying current risk for colorectal cancer. Int. J. Cancer 2010, 126, 1177–1186. [Google Scholar] [CrossRef] [PubMed]
- Ciarloni, L.; Ehrensberger, S.H.; Imaizumi, N.; Monnier-Benoit, S.; Nichita, C.; Myung, S.J.; Kim, J.S.; Song, S.Y.; Kim, T.; Van Der Weg, B.; et al. Development and clinical validation of a blood test based on 29-gene expression for early detection of colorectal cancer. Clin. Cancer Res. 2016, 22, 4604–4611. [Google Scholar] [CrossRef] [Green Version]
- Ciarloni, L.; Hosseinian, S.; Monnier-Benoit, S.; Imaizumi, N.; Dorta, G.; Ruegg, C. Discovery of a 29-gene panel in peripheral blood mononuclear cells for the detection of colorectal cancer and adenomas using high throughput real-time PCR. PLoS ONE 2015, 10, e0123904. [Google Scholar] [CrossRef] [Green Version]
- Alamro, R.; Mustafa, M.; Al-Asmari, A. Inflammatory gene mRNA expression in human peripheral blood and its association with colorectal cancer. J. Inflamm. Res. 2018, 11, 351–357. [Google Scholar] [CrossRef] [Green Version]
- Sahengbieke, S.; Wang, J.; Li, X.; Wang, Y.; Lai, M.; Wu, J. Circulating cell-free high mobility group AT-hook 2 mRNA as a detection marker in the serum of colorectal cancer patients. J. Clin. Lab. Anal. 2018, 32, e22332. [Google Scholar] [CrossRef] [Green Version]
- Hamm, A.; Prenen, H.; Van Delm, W.; Di Matteo, M.; Wenes, M.; Delamarre, E.; Schmidt, T.; Weitz, J.; Sarmiento, R.; Dezi, A.; et al. Tumour-educated circulating monocytes are powerful candidate biomarkers for diagnosis and disease follow-up of colorectal cancer. Gut 2016, 65, 990–1000. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Murphy, J. RNA biomarkers in colorectal cancer. Methods 2013, 59, 116–125. [Google Scholar] [CrossRef]
- Moridikia, A.; Mirzaei, H.; Sahebkar, A.; Salimian, J. MicroRNAs: Potential candidates for diagnosis and treatment of colorectal cancer. J. Cell. Physiol. 2018, 233, 901–913. [Google Scholar] [CrossRef]
- Nikolaou, S.; Qiu, S.; Fiorentino, F.; Rasheed, S.; Tekkis, P.; Kontovounisios, C. Systematic review of blood diagnostic markers in colorectal cancer. Tech. Coloproctol. 2018, 22, 481–498. [Google Scholar] [CrossRef] [Green Version]
- Thomas, J.; Ohtsuka, M.; Pichler, M.; Ling, H. MicroRNAs: Clinical relevance in colorectal cancer. Int. J. Mol. Sci. 2015, 16, 28063–28076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganepola, G.A.; Nizin, J.; Rutledge, J.R.; Chang, D.H. Use of blood-based biomarkers for early diagnosis and surveillance of colorectal cancer. World J. Gastrointest. Oncol. 2014, 6, 83–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikolouzakis, T.K.; Vassilopoulou, L.; Fragkiadaki, P.; Sapsakos, T.M.; Papadakis, G.Z.; Spandidos, D.A.; Tsatsakis, A.M.; Tsiaoussis, J. Improving diagnosis, prognosis and prediction by using biomarkers in CRC patients. Oncol. Rep. 2018, 39, 2455–2472. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.; Wang, Z.; Shen, L.; Wei, Q. Circulating microRNA-21 as a potential diagnostic marker for colorectal cancer: A meta-analysis. Mol. Clin. Oncol. 2016, 4, 237–244. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.; Huang, D.; Ni, S.; Peng, Z.; Sheng, W.; Du, X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int. J. Cancer 2010, 127, 118–126. [Google Scholar] [CrossRef]
- Herreros-Villanueva, M.; Duran-Sanchon, S.; Martín, A.C.; Pérez-Palacios, R.; Vila-Navarro, E.; Marcuello, M.; Diaz-Centeno, M.; Cubiella, J.; Diez, M.S.; Bujanda, L.; et al. Plasma microRNA signature validation for early detection of colorectal cancer. Clin. Transl. Gastroenterol. 2019, 10, e00003. [Google Scholar] [CrossRef]
- Yamada, A.; Horimatsu, T.; Okugawa, Y.; Nishida, N.; Honjo, H.; Ida, H.; Kou, T.; Kusaka, T.; Sasaki, Y.; Yagi, M.; et al. Serum MIR-21, MIR-29a, and MIR-125b are promising biomarkers for the early detection of colorectal neoplasia. Clin. Cancer Res. 2015, 21, 4234–4242. [Google Scholar] [CrossRef] [Green Version]
- Zanutto, S.; Ciniselli, C.M.; Belfiore, A.; Lecchi, M.; Masci, E.; Delconte, G.; Primignani, M.; Tosetti, G.; Dal Fante, M.; Fazzini, L.; et al. Plasma miRNA-based signatures in CRC screening programs. Int. J. Cancer 2020, 146, 1164–1173. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zhu, M.; Shan, X.; Zhou, X.; Wang, T.; Zhang, J.; Tao, J.; Cheng, W.; Chen, G.; Li, J.; et al. A panel of seven-miRNA signature in plasma as potential biomarker for colorectal cancer diagnosis. Gene 2019, 687, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Khakoo, S.; Georgiou, A.; Gerlinger, M.; Cunningham, D.; Starling, N. Circulating tumour DNA, a promising biomarker for the management of colorectal cancer. Crit. Rev. Oncol. Hematol. 2018, 122, 72–82. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas. Available online: https://portal.gdc.cancer.gov/ (accessed on 22 February 2021).
- Song, L.; Jia, J.; Peng, X.; Xiao, W.; Li, Y. The performance of the SEPT9 gene methylation assay and a comparison with other CRC screening tests: A meta-analysis. Sci. Rep. 2017, 7, 3032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.; Hu, B.; Gui, Y.C.; Tan, Z.B.; Xu, J.W. Diagnostic value and clinical significance of methylated SEPT9 for colorectal cancer: A meta-analysis. Med. Sci. Monit. 2019, 25, 5813–5822. [Google Scholar] [CrossRef]
- He, N.; Song, L.; Kang, Q.; Jin, P.; Cai, G.; Zhou, J.; Zhou, G.; Sheng, J.; Cai, S.; Wang, J.; et al. The pathological features of colorectal cancer determine the detection performance on blood ctDNA. Technol. Cancer Res. Treat. 2018, 17, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, S.K.; Symonds, E.L.; Baker, R.T.; Murray, D.H.; McEvoy, A.; Van Doorn, S.C.; Mundt, M.W.; Cole, S.R.; Gopalsamy, G.; Mangira, D.; et al. Evaluation of an assay for methylated BCAT1 and IKZF1 in plasma for detection of colorectal neoplasia. BMC Cancer 2015, 15, 654. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef] [Green Version]
- Phallen, J.; Sausen, M.; Adleff, V.; Leal, A.; Hruban, C.; White, J.; Anagnostou, V.; Fiksel, J.; Cristiano, S.; Papp, E.; et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 2017, 9, eaan2415. [Google Scholar] [CrossRef] [Green Version]
- Wan, N.; Weinberg, D.; Liu, T.-Y.; Niehaus, K.; Ariazi, E.A.; Delubac, D.; Kannan, A.; White, B.; Bailey, M.; Bertin, M.; et al. Machine learning enables detection of early-stage colorectal cancer by whole-genome sequencing of plasma cell-free DNA. BMC Cancer 2019, 19, 832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, I.S.; Kato, S.; Fanta, P.T.; Leichman, L.; Okamura, R.; Raymond, V.M.; Lanman, R.B.; Lippman, S.M.; Kurzrock, R. Genomic profiling of blood-derived circulating tumor DNA from patients with colorectal cancer: Implications for response and resistance to targeted therapeutics. Mol. Cancer Ther. 2019, 18, 1852–1862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odegaard, J.I.; Vincent, J.J.; Mortimer, S.; Vowles, J.V.; Ulrich, B.C.; Banks, K.C.; Fairclough, S.R.; Zill, O.A.; Sikora, M.; Mokhtari, R.; et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin. Cancer Res. 2018, 24, 3539–3549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zill, O.A.; Banks, K.C.; Fairclough, S.R.; Mortimer, S.A.; Vowles, J.V.; Mokhtari, R.; Gandara, D.R.; Mack, P.C.; Odegaard, J.I.; Nagy, R.J.; et al. The landscape of actionable genomic alterations in cell-free circulating tumor DNA from 21,807 advanced cancer patients. Clin. Cancer Res. 2018, 24, 3528–3538. [Google Scholar] [CrossRef] [Green Version]
- Kuespert, K.; Pils, S.; Hauck, C.R. CEACAMs: Their role in physiology and pathophysiology. Curr. Opin. Cell Biol. 2006, 18, 565–571. [Google Scholar] [CrossRef] [Green Version]
- Beauchemin, N.; Arabzadeh, A. Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) in cancer progression and metastasis. Cancer Metastasis Rev. 2013, 32, 643–671. [Google Scholar] [CrossRef]
- Jelski, W.; Mroczko, B. Biochemical markers of colorectal cancer—present and future. Cancer Manag. Res. 2020, 12, 4789–4797. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, L.; Nielsen, H.J.; Christensen, I.J. Early detection and recurrence of colorectal adenomas by combination of eight cancer-associated biomarkers in plasma. Clin. Exp. Gastroenterol. 2020, 13, 273–284. [Google Scholar] [CrossRef]
- Hall, C.; Clarke, L.; Pal, A.; Buchwald, P.; Eglinton, T.; Wakeman, C.; Frizelle, F. A review of the role of carcinoembryonic antigen in clinical practice. Ann. Coloproctol. 2019, 35, 294–305. [Google Scholar] [CrossRef]
- Chen, H.; Zucknick, M.; Werner, S.; Knebel, P.; Brenner, H. Head-to-head comparison and evaluation of 92 plasma protein biomarkers for early detection of colorectal cancer in a true screening setting. Clin. Cancer Res. 2015, 21, 3318–3326. [Google Scholar] [CrossRef] [Green Version]
- Halilovic, E.; Rasic, I.; Sofic, A.; Mujic, A.; Rovcanin, A.; Hodzic, E.; Kulovic, E. The importance of determining preoperative serum concentration of carbohydrate antigen 19-9 and carcinoembryonic antigen in assessing the progression of colorectal cancer. Med. Arch. 2020, 74, 346–349. [Google Scholar] [CrossRef]
- Baqar, A.R.; Wilkins, S.; Staples, M.; Angus Lee, C.H.; Oliva, K.; McMurrick, P. The role of preoperative CEA in the management of colorectal cancer: A cohort study from two cancer centres. Int. J. Surg. 2019, 64, 10–15. [Google Scholar] [CrossRef]
- Koulis, C.; Yap, R.; Engel, R.; Jardé, T.; Wilkins, S.; Solon, G.; Shapiro, J.D.; Abud, H.; McMurrick, P. Personalized medicine—current and emerging predictive and prognostic biomarkers in colorectal cancer. Cancers 2020, 12, 812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ushigome, M.; Nabeya, Y.; Soda, H.; Takiguchi, N.; Kuwajima, A.; Tagawa, M.; Matsushita, K.; Koike, J.; Funahashi, K.; Shimada, H. Multi-panel assay of serum autoantibodies in colorectal cancer. Int. J. Clin. Oncol. 2018, 23, 917–923. [Google Scholar] [CrossRef]
- Chauvin, A.; Boisvert, F.-M. Clinical proteomics in colorectal cancer, a promising tool for improving personalised medicine. Proteomes 2018, 6, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, S.B.; Sharma, S.; Mohamedali, A.; Mahboob, S.; Redmond, W.J.; Pascovici, D.; Wu, J.X.; Zaw, T.; Adhikari, S.; Vaibhav, V.; et al. Potential early clinical stage colorectal cancer diagnosis using a proteomics blood test panel. Clin. Proteomics 2019, 16, 34. [Google Scholar] [CrossRef]
- Ivancic, M.M.; Megna, B.W.; Sverchkov, Y.; Craven, M.; Reichelderfer, M.; Pickhardt, P.J.; Sussman, M.R.; Kennedy, G.D. Noninvasive detection of colorectal carcinomas using serum protein biomarkers. J. Surg. Res. 2020, 246, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Wilhelmsen, M.; Christensen, I.J.; Rasmussen, L.; Jørgensen, L.N.; Madsen, M.R.; Vilandt, J.; Hillig, T.; Klærke, M.; Nielsen, K.T.; Laurberg, S.; et al. Detection of colorectal neoplasia: Combination of eight blood-based, cancer-associated protein biomarkers. Int. J. Cancer 2017, 140, 1436–1446. [Google Scholar] [CrossRef]
- Desmond, B.J.; Dennett, E.R.; Danielson, K.M. Circulating extracellular vesicle microRNA as diagnostic biomarkers in early colorectal cancer—a review. Cancers 2019, 12, 52. [Google Scholar] [CrossRef] [Green Version]
- González-Masiá, J.A.; García-Olmo, D.; García-Olmo, D.C. Circulating nucleic acids in plasma and serum (CNAPS): Applications in oncology. Onco. Targets. Ther. 2013, 6, 819–832. [Google Scholar]
- Mousavi, S.; Moallem, R.; Hassanian, S.M.; Sadeghzade, M.; Mardani, R.; Ferns, G.A.; Khazaei, M.; Avan, A. Tumor-derived exosomes: Potential biomarkers and therapeutic target in the treatment of colorectal cancer. J. Cell. Physiol. 2019, 234, 12422–12432. [Google Scholar] [CrossRef] [PubMed]
- Rashed, M.H.; Bayraktar, E.; Helal, G.K.; Abd-Ellah, M.; Amero, P.; Chavez-Reyes, A.; Rodriguez-Aguayo, C. Exosomes: From garbage bins to promising therapeutic targets. Int. J. Mol. Sci. 2017, 18, 538. [Google Scholar] [CrossRef] [Green Version]
- Shao, Y.; Shen, Y.; Chen, T.; Xu, F.; Chen, X.; Zheng, S. The functions and clinical applications of tumor-derived exosomes. Oncotarget 2016, 7, 60736–60751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menck, K.; Bleckmann, A.; Wachter, A.; Hennies, B.; Ries, L.; Schulz, M.; Balkenhol, M.; Pukrop, T.; Schatlo, B.; Rost, U.; et al. Characterisation of tumour-derived microvesicles in cancer patients’ blood and correlation with clinical outcome. J. Extracell. Vesicles 2017, 6, 1340745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tugutova, E.A.; Tamkovich, S.N.; Patysheva, M.R.; Afanas’ev, S.G.; Tsydenova, A.A.; Grigor’eva, A.E.; Kolegova, E.S.; Kondakova, I.V.; Yunusova, N.V. Relation between tetraspanin-associated and tetraspanin-non-associated exosomal proteases and metabolic syndrome in colorectal cancer patients. Asian Pac. J. Cancer Prev. 2019, 20, 809–815. [Google Scholar] [CrossRef] [Green Version]
- Tamkovich, S.N.; Yunusova, N.V.; Stakheeva, M.N.; Somov, A.K.; Frolova, A.E.; Kiryushina, N.A.; Afanasyev, S.G.; Grigor’eva, A.E.; Laktionov, P.P.; Kondakova, I.V. Isolation and characterization of exosomes from blood plasma of breast cancer and colorectal cancer patients. Biochem. Suppl. Ser. B Biomed. Chem. 2017, 11, 291–295. [Google Scholar]
- Menck, K.; Bleckmann, A.; Schulz, M.; Ries, L.; Binder, C. Isolation and characterization of microvesicles from peripheral blood. J. Vis. Exp. 2017, 119, e55057. [Google Scholar] [CrossRef] [Green Version]
- Coumans, F.A.W.; Brisson, A.R.; Buzas, E.I.; Dignat-George, F.; Drees, E.E.E.; El-Andaloussi, S.; Emanueli, C.; Gasecka, A.; Hendrix, A.; Hill, A.F.; et al. Methodological guidelines to study extracellular vesicles. Circ. Res. 2017, 120, 1632–1648. [Google Scholar] [CrossRef]
- Mateescu, B.; Kowal, E.J.K.; van Balkom, B.W.M.; Bartel, S.; Bhattacharyya, S.N.; Buzás, E.I.; Buck, A.H.; de Candia, P.; Chow, F.W.N.; Das, S.; et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA—An ISEV position paper. J. Extracell. Vesicles 2017, 6, 1286095. [Google Scholar] [CrossRef] [Green Version]
- Belov, L.; Matic, K.J.; Hallal, S.; Best, O.G.; Mulligan, S.P.; Christopherson, R.I. Extensive surface protein profiles of extracellular vesicles from cancer cells may provide diagnostic signatures from blood samples. J. Extracell. Vesicles 2016, 5, 25355. [Google Scholar] [CrossRef]
- Ogata-Kawata, H.; Izumiya, M.; Kurioka, D.; Honma, Y.; Yamada, Y.; Furuta, K.; Gunji, T.; Ohta, H.; Okamoto, H.; Sonoda, H.; et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 2014, 9, e92921. [Google Scholar] [CrossRef]
- Yan, S.; Han, B.; Gao, S.; Wang, X.; Wang, Z.; Wang, F.; Zhang, J.; Xu, D.; Sun, B. Exosome-encapsulated microRNAs as circulating biomarkers for colorectal cancer. Oncotarget 2017, 8, 60149–60158. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Eng, C.; Shen, J.; Lu, Y.; Yoko, T.; Mehdizadeh, A.; Chang, G.J.; Rodriguez-Bigas, M.A.; Li, Y.; Chang, P.; et al. Serum exosomal miR-4772-3p is a predictor of tumor recurrence in stage II and III colon cancer. Oncotarget 2016, 7, 76250–76260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Z.Y.; Gu, R.H.; Yan, B. Downregulation of exosome-encapsulated miR-548c-5p is associated with poor prognosis in colorectal cancer. J. Cell. Biochem. 2019, 120, 1457–1463. [Google Scholar] [CrossRef] [PubMed]
- Min, L.; Zhu, S.; Chen, L.; Liu, X.; Wei, R.; Zhao, L.; Yang, Y.; Zhang, Z.; Kong, G.; Li, P.; et al. Evaluation of circulating small extracellular vesicles derived miRNAs as biomarkers of early colon cancer: A comparison with plasma total miRNAs. J. Extracell. Vesicles 2019, 8, 1643670. [Google Scholar] [CrossRef] [Green Version]
- Cha, B.S.; Park, K.S.; Park, J.S. Signature mRNA markers in extracellular vesicles for the accurate diagnosis of colorectal cancer. J. Biol. Eng. 2020, 14, 4. [Google Scholar] [CrossRef]
- Dong, L.; Lin, W.; Qi, P.; Xu, M.D.; Wu, X.; Ni, S.; Huang, D.; Weng, W.W.; Tan, C.; Sheng, W.; et al. Circulating long RNAs in serum extracellular vesicles: Their characterization and potential application as biomarkers for diagnosis of colorectal cancer. Cancer Epidemiol. Biomarkers Prev. 2016, 25, 1158–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Country | Program | Test 1 | Cut-Off 2 | Target Age 3 | Interval 3 (years) | Invited 4 (%) | Participation 5 (%) |
|---|---|---|---|---|---|---|---|
| Albania | NA | NA | NA | NA | NA | NA | NA |
| Austria | Regional/Opportunistic | FIT/gFOBT/CS | NA | 40–80 | 1 | NA | NA |
| Belarus | NA | NA | NA | NA | NA | NA | NA |
| Belgium | Regional | FIT/CS | 15 | 50–74 | 2 | 99.2 | 27.7 |
| Bosnia and Herzegovina | Regional/Opportunistic | gFOBT | NA | >50 | NA | NA | NA |
| Bulgaria | No/Opportunistic | gFOBT | NA | NA | NA | NA | NA |
| Croatia | National | gFOBT | NA | 50–74 | 2 | 100 | 15.3 |
| Cyprus | Pilot/Planned | FIT | NA | 50–69 | 2 | NA | NA |
| Czech Republic | Regional/Opportunistic | FIT/gFOBT/CS | 15 | 50–79 | 2 | NA | 22.7 |
| Denmark | National | FIT | 20 | 50–74 | 2 | 25 | 64 |
| Estonia | Pilot/Planned | FIT | NA | 60–69 | 2 | ||
| Finland | Pilot/Planned | gFOBT | NA | 60–69 | 2 | 23.9 | 66.4 |
| France | Regional | FIT/gFOBT | 30 | 50–74 | 2 | 99.1 | 26.5 |
| Germany | Opportunistic/Pilot/planned | FIT/gFOBT/CS | NA | 50–74 | 2–10 | NA | NA |
| Greece | No/Opportunistic | gFOBT/CS | NA | 50–74 | NA | NA | NA |
| Hungary | Pilot/Planned | FIT | 20 | 50–70 | 2 | 21.1 | 36.7 |
| Iceland | Opportunistic/Planned | gFOBT/CS | NA | 55–75 | 2–10 | NA | 30 |
| Ireland | National | FIT | 20 | 60–69 | 2 | 10.9 | 39.6 |
| Italy | National | FIT/FS 6 | 20 | 50–74 6 | 2 | 75 | 42 |
| Latvia | No/Opportunistic | gFOBT | NA | 50–74 | NA | NA | 11.1 |
| Lithuania | Opportunistic/Pilot/Planned | FIT | NA | 50–74 | 2 | NA | 53.1 |
| Luxembourg | Opportunistic/Planned | FIT/gFOBT/CS | NA | 55–74 | 2 | NA | NA |
| Macedonia | No/Opportunistic | FIT | NA | NA | NA | NA | NA |
| Malta | National | FIT | 16–20 | 55–66 | 2 | 100 | 45.4 |
| Montenegro | Regional | FIT | NA | 50–74 | 2 | NA | 33.3 |
| The Netherlands | National | FIT | 47 | 55–75 | 2 | 38.5 | 71.2 |
| Norway | Regional/Pilot | FIT | NA | 55–64 | 2 | NA | 64.8 |
| Poland | National | CS | NA | 55–64 | 10 | 12.5 | 16.7 |
| Portugal | Regional | FIT/gFOBT | 20 | 50–70 | 2 | 1.6 | 62 |
| Romania | No/Opportunistic | NA | NA | NA | NA | NA | NA |
| Russian Federation | Opportunistic/ Pilot | FIT/CS | NA | 48–75 | NA | NA | NA |
| Serbia | National | FIT | NA | 50–74 | 2 | NA | 58.4 |
| Slovakian Republic | No/Opportunistic | FIT/gFOBT/CS | NA | NA | NA | NA | NA |
| Slovenia | National | FIT | 20 | 50–69 | 2 | 93 | 47.1 |
| Spain | National | FIT/gFOBT | 20 | 50–69 | 2 | 14.2 | 50.2 |
| Sweden | Regional | gFOBT | NA | 60–69 | 2 | 100 | 62.7 |
| Switzerland | No/Opportunistic | FIT/CS | NA | 50–69 | 2–10 | NA | 22 |
| Ukraine | NA | NA | NA | NA | NA | NA | NA |
| United Kingdom | National | FIT/gFOBT/FS | NA | 50–74 6 | 2 | 100 | 56.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferlizza, E.; Solmi, R.; Sgarzi, M.; Ricciardiello, L.; Lauriola, M. The Roadmap of Colorectal Cancer Screening. Cancers 2021, 13, 1101. https://doi.org/10.3390/cancers13051101
Ferlizza E, Solmi R, Sgarzi M, Ricciardiello L, Lauriola M. The Roadmap of Colorectal Cancer Screening. Cancers. 2021; 13(5):1101. https://doi.org/10.3390/cancers13051101
Chicago/Turabian StyleFerlizza, Enea, Rossella Solmi, Michela Sgarzi, Luigi Ricciardiello, and Mattia Lauriola. 2021. "The Roadmap of Colorectal Cancer Screening" Cancers 13, no. 5: 1101. https://doi.org/10.3390/cancers13051101