Molecular Measurable Residual Disease Assessment before Hematopoietic Stem Cell Transplantation in Pediatric Acute Myeloid Leukemia Patients: A Retrospective Study by the I-BFM Study Group
Abstract
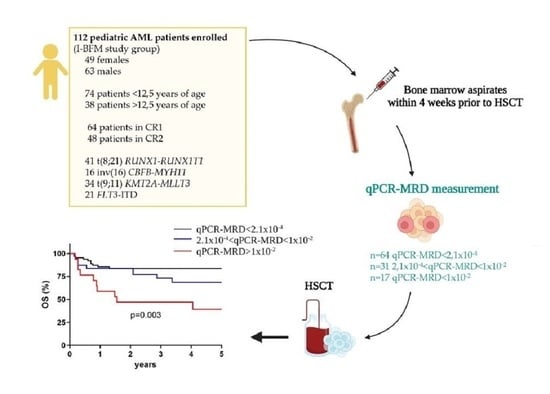
:1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. RNA Extraction and Retro-Transcription
2.3. Molecular Marker Identification by Polymerase Chain Reaction
2.4. qPCR-MRD Detection
2.5. Quality Control of Standard Operating Procedures (SOPs)
2.6. Statistical Methods
3. Results
3.1. Inter-Laboratory Quality Control (QC)
3.2. Survival According to the Type of Remission
3.3. qPCR-MRD Threshold 2.1 × 10−4 Is Associated with HSCT Outcome
3.4. qPCR-MRD Threshold 1 × 10−2 Refines HSCT Outcome Prediction
3.5. Model of Pre-HSCT Risk Stratification by qPCR-MRD
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Döhner, H.; Weisdorf, D.J.; Bloomfield, C.D. Acute Myeloid Leukemia. N. Engl. J. Med. 2015, 373, 1136–1152. [Google Scholar] [CrossRef] [Green Version]
- Rasche, M.; Zimmermann, M.; Borschel, L.; Bourquin, J.-P.; Dworzak, M.; Klingebiel, T.; Lehrnbecher, T.; Creutzig, U.; Klusmann, J.-H.; Reinhardt, D. Successes and Challenges in the Treatment of Pediatric Acute Myeloid Leukemia: A Retrospective Analysis of the AML-BFM Trials from 1987 to 2012. Leukemia 2018, 32, 2167–2177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pession, A.; Masetti, R.; Rizzari, C.; Putti, M.C.; Casale, F.; Fagioli, F.; Luciani, M.; Lo Nigro, L.; Menna, G.; Micalizzi, C.; et al. Results of the AIEOP AML 2002/01 Multicenter Prospective Trial for the Treatment of Children with Acute Myeloid Leukemia. Blood 2013, 122, 170–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaspers, G.J.L.; Zimmermann, M.; Reinhardt, D.; Gibson, B.E.S.; Tamminga, R.Y.J.; Aleinikova, O.; Armendariz, H.; Dworzak, M.; Ha, S.Y.; Hasle, H.; et al. Improved Outcome in Pediatric Relapsed Acute Myeloid Leukemia: Results of a Randomized Trial on Liposomal Daunorubicin by the International BFM Study Group. J. Clin. Oncol. 2013, 31, 599–607. [Google Scholar] [CrossRef] [PubMed]
- O’Hare, P.; Lucchini, G.; Cummins, M.; Veys, P.; Potter, M.; Lawson, S.; Vora, A.; Wynn, R.; Peniket, A.; Kirkland, K.; et al. Allogeneic Stem Cell Transplantation for Refractory Acute Myeloid Leukemia in Pediatric Patients: The UK Experience. Bone Marrow Transpl. 2017, 52, 825–831. [Google Scholar] [CrossRef] [Green Version]
- Lucchini, G.; Labopin, M.; Beohou, E.; Dalissier, A.; Dalle, J.H.; Cornish, J.; Zecca, M.; Samarasinghe, S.; Gibson, B.; Locatelli, F.; et al. Impact of Conditioning Regimen on Outcomes for Children with Acute Myeloid Leukemia Undergoing Transplantation in First Complete Remission. An Analysis on Behalf of the Pediatric Disease Working Party of the European Group for Blood and Marrow Transplanta. Biol. Blood Marrow Transpl. 2017, 23, 467–474. [Google Scholar] [CrossRef] [Green Version]
- Locatelli, F.; Masetti, R.; Rondelli, R.; Zecca, M.; Fagioli, F.; Rovelli, A.; Messina, C.; Lanino, E.; Bertaina, A.; Favre, C.; et al. Outcome of Children with High-Risk Acute Myeloid Leukemia given Autologous or Allogeneic Hematopoietic Cell Transplantation in the Aieop AML-2002/01 Study. Bone Marrow Transpl. 2015, 50, 181–188. [Google Scholar] [CrossRef] [Green Version]
- Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, G.; Hoadley, K.; Triche, T.J.; Laird, P.W.; Baty, J.D.; et al. Genomic and Epigenomic Landscapes of Adult de Novo Acute Myeloid Leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef] [Green Version]
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef]
- Tyner, J.W.; Tognon, C.E.; Bottomly, D.; Wilmot, B.; Kurtz, S.E.; Savage, S.L.; Long, N.; Schultz, A.R.; Traer, E.; Abel, M.; et al. Functional Genomic Landscape of Acute Myeloid Leukaemia. Nature 2018, 562, 526–531. [Google Scholar] [CrossRef]
- Leung, W.; Pui, C.H.; Coustan-Smith, E.; Yang, J.; Pei, D.; Gan, K.; Srinivasan, A.; Hartford, C.; Triplett, B.M.; Dallas, M.; et al. Detectable Minimal Residual Disease before Hematopoietic Cell Transplantation Is Prognostic but Does Not Preclude Cure for Children with Very-High-Risk Leukemia. Blood 2012, 120, 468–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grimwade, D.; Freeman, S.D. Defining Minimal Residual Disease in Acute Myeloid Leukemia: Which Platforms Are Ready for “Prime Time”? Blood 2014, 124, 3345–3355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dillon, R.; Hills, R.; Freeman, S.; Potter, N.; Jovanovic, J.; Ivey, A.; Kanda, A.S.; Runglall, M.; Foot, N.; Valganon, M.; et al. Molecular MRD Status and Outcome after Transplantation in NPM1-Mutated AML. Blood 2020, 135, 680–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pigazzi, M.; Manara, E.; Buldini, B.; Beqiri, V.; Bisio, V.; Tregnago, C.; Rondelli, R.; Masetti, R.; Caterina Putti, M.; Fagioli, F.; et al. Minimal Residual Disease Monitored after Induction Therapy by Rq-Pcr Can Contribute to Tailor Treatment of Patients with t(8;21) Runx1-Runx1t1 Rearrangement. Haematologica 2015, 100, e99–e101. [Google Scholar] [CrossRef]
- Buldini, B.; Rizzati, F.; Masetti, R.; Fagioli, F.; Menna, G.; Micalizzi, C.; Putti, M.C.; Rizzari, C.; Santoro, N.; Zecca, M.; et al. Prognostic Significance of Flow-Cytometry Evaluation of Minimal Residual Disease in Children with Acute Myeloid Leukaemia Treated according to the AIEOP-AML 2002/01 Study Protocol. Br. J. Haematol. 2017, 177, 116–126. [Google Scholar] [CrossRef]
- Rubnitz, J.E.; Inaba, H.; Dahl, G.; Ribeiro, R.C.; Bowman, W.P.; Taub, J.; Pounds, S.; Razzouk, B.I.; Lacayo, N.J.; Cao, X.; et al. Minimal Residual Disease-Directed Therapy for Childhood Acute Myeloid Leukaemia: Results of the AML02 Multicentre Trial. Lancet Oncol. 2010, 11, 543–552. [Google Scholar] [CrossRef] [Green Version]
- Van Der Velden, V.H.J.; Van Der Sluijs-Geling, A.; Gibson, B.E.S.; Te Marvelde, J.G.; Hoogeveen, P.G.; Hop, W.C.J.; Wheatley, K.; Bierings, M.B.; Schuurhuis, G.J.; De Graaf, S.S.N.; et al. Clinical Significance of Flowcytometric Minimal Residual Disease Detection in Pediatric Acute Myeloid Leukemia Patients Treated according to the DCOG ANLL97/MRC AML12 Protocol. Leukemia 2010, 24, 1599–1606. [Google Scholar] [CrossRef] [Green Version]
- Bolouri, H.; Farrar, J.E.; Triche, T.; Ries, R.E.; Lim, E.L.; Alonzo, T.A.; Ma, Y.; Moore, R.; Mungall, A.J.; Marra, M.A.; et al. The Molecular Landscape of Pediatric Acute Myeloid Leukemia Reveals Recurrent Structural Alterations and Age-Specific Mutational Interactions. Nat. Med. 2018, 24, 103–112. [Google Scholar] [CrossRef] [Green Version]
- Grimwade, D. The Changing Paradigm of Prognostic Factors in Acute Myeloid Leukaemia. Best Pract. Res. Clin. Haematol. 2012, 25, 419–425. [Google Scholar] [CrossRef]
- Cheson, B.D.; Bennett, J.M.; Kopecky, K.J.; Büchner, T.; Willman, C.L.; Estey, E.H.; Schiffer, C.A.; Doehner, H.; Tallman, M.S.; Lister, T.A.; et al. Revised Recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. J. Clin. Oncol. 2003, 21, 4642–4649. [Google Scholar] [CrossRef]
- Czyz, A.; Nagler, A. The Role of Measurable Residual Disease (MRD) in Hematopoietic Stem Cell Transplantation for Hematological Malignancies Focusing on Acute Leukemia. Int. J. Mol. Sci. 2019, 20, 5362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buckley, S.A.; Wood, B.L.; Othus, M.; Hourigan, C.S.; Ustun, C.; Linden, M.A.; Defor, T.E.; Malagola, M.; Anthias, C.; Valkova, V.; et al. Minimal Residual Disease Prior to Allogeneic Hematopoietic Cell Transplantation in Acute Myeloid Leukemia: A Meta-Analysis. Haematologica 2017, 102, 865–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walter, R.B.; Gooley, T.A.; Wood, B.L.; Milano, F.; Fang, M.; Sorror, M.L.; Estey, E.H.; Salter, A.I.; Lansverk, E.; Chien, J.W.; et al. Impact of Pretransplantation Minimal Residual Disease, as Detected by Multiparametric Flow Cytometry, on Outcome of Myeloablative Hematopoietic Cell Transplantation for Acute Myeloid Leukemia. J. Clin. Oncol. 2011, 29, 1190–1197. [Google Scholar] [CrossRef] [Green Version]
- Kayser, S.; Benner, A.; Thiede, C.; Martens, U.; Huber, J.; Stadtherr, P.; Janssen, J.W.G.; Röllig, C.; Uppenkamp, M.J.; Bochtler, T.; et al. Pretransplant NPM1 MRD Levels Predict Outcome after Allogeneic Hematopoietic Stem Cell Transplantation in Patients with Acute Myeloid Leukemia. Blood Cancer J. 2016, 6, e449. [Google Scholar] [CrossRef] [PubMed]
- Juul-Dam, K.L.; Ommen, H.B.; Nyvold, C.G.; Walter, C.; Vålerhaugen, H.; Kairisto, V.; Abrahamsson, J.; Alm, S.J.; Jahnukainen, K.; Lausen, B.; et al. Measurable Residual Disease Assessment by QPCR in Peripheral Blood Is an Informative Tool for Disease Surveillance in Childhood Acute Myeloid Leukaemia. Br. J. Haematol. 2020, 190, 198–208. [Google Scholar] [CrossRef] [PubMed]
- Van Dongen, J.J.M.; Macintyre, E.A.; Gabert, J.A.; Delabesse, E.; Rossi, V.; Saglio, G.; Gottardi, E.; Rambaldi, A.; Dotti, G.; Griesinger, F.; et al. Standardized RT-PCR Analysis of Fusion Gene Transcripts from Chromosome Aberrations in Acute Leukemia for Detection of Minimal Residual Disease. Report of the BIOMED-1 Concerted Action: Investigation of Minimal Residual Disease in Acute Leukemia. Leukemia 1999, 13, 1901–1928. [Google Scholar] [CrossRef]
- Pigazzi, M.; Manara, E.; Bisio, V.; Aveic, S.; Masetti, R.; Menna, G.; Zecca, M.; Pession, A.; Locatelli, F.; Basso, G. Screening of Novel Genetic Aberrations in Pediatric Acute Myeloid Leukemia: A Report from the AIEOP AML-2002 Study Group. Blood 2012, 120, 3860–3862. [Google Scholar] [CrossRef] [Green Version]
- Pigazzi, M.; Masetti, R.; Bresolin, S.; Beghin, A.; Di Meglio, A.; Gelain, S.; Trentin, L.; Baron, E.; Giordan, M.; Zangrando, A.; et al. MLL Partner Genes Drive Distinct Gene Expression Profiles and Genomic Alterations in Pediatric Acute Myeloid Leukemia: An AIEOP Study. Leukemia 2011, 25, 560–563. [Google Scholar] [CrossRef] [Green Version]
- Manara, E.; Basso, G.; Zampini, M.; Buldini, B.; Tregnago, C.; Rondelli, R.; Masetti, R.; Bisio, V.; Frison, M.; Polato, K.; et al. Characterization of Children with FLT3-ITD Acute Myeloid Leukemia: A Report from the AIEOP AML-2002 Study Group. Leukemia 2017, 31, 18–25. [Google Scholar] [CrossRef] [Green Version]
- Testi, A.M.; Pession, A.; Diverio, D.; Grimwade, D.; Gibson, B.; Cardoso de Azevedo, A.; Moran, L.; Leverger, G.; Elitzur, S.; Hasle, H.; et al. Risk-Adapted Treatment of Acute Promyelocytic Leukemia: Results from the International Consortium for Childhood APL. Blood 2018, 132, 405–412. [Google Scholar] [CrossRef]
- Grimwade, D.; Vyas, P.; Freeman, S. Assessment of Minimal Residual Disease in Acute Myeloid Leukemia. Curr. Opin. Oncol. 2010, 22, 656–663. [Google Scholar] [CrossRef] [PubMed]
- Kayser, S.; Schlenk, R.F.; Grimwade, D.; Yosuico, V.E.D.; Walter, R.B. Evidence-Based Focused Review Minimal Residual Disease—Directed Therapy in Acute Myeloid Leukemia. Blood 2015, 125, 2331–2336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabert, J.; Beillard, E.; van der Velden, V.H.J.; Bi, W.; Grimwade, D.; Pallisgaard, N.; Barbany, G.; Cazzaniga, G.; Cayuela, J.M.; Cavé, H.; et al. Standardization and Quality Control Studies of “real Time” Quantitative Reverse Transcriptase Polymerase Chain Reaction of Fusion Gene Transcripts for Residual Disease Detection in Leukemia—A Europe Against Cancer Program. Leukemia 2003, 17, 2318–2357. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; Levine, R.L.; Lo-coco, F.; et al. Diagnosis and Management of AML in Adults: 2017 ELN Recommendations from an International Expert Panel. Blood 2017, 129, 424–448. [Google Scholar] [CrossRef] [Green Version]
- Heuser, M.; Freeman, S.D.; Ossenkoppele, G.J.; Buccisano, F.; Hourigan, C.S.; Ngai, L.L.; Tettero, J.M.; Bachas, C.; Baer, C.; Béné, M.C.; et al. 2021 Update on MRD in Acute Myeloid Leukemia: A Consensus Document from the European LeukemiaNet MRD Working Party. Blood 2021, 138, 2753–2767. [Google Scholar] [CrossRef]
- Buccisano, F.; Maurillo, L.; Del Principe, M.I.; Del Poeta, G.; Sconocchia, G.; Lo-Coco, F.; Arcese, W.; Amadori, S.; Venditti, A. Prognostic and Therapeutic Implications of Minimal Residual Disease Detection in Acute Myeloid Leukemia. Blood 2012, 119, 332–341. [Google Scholar] [CrossRef] [Green Version]
- Sison, E.A.R.; Brown, P. Does Hematopoietic Stem Cell Transplantation Benefit Infants with Acute Leukemia? Hematol. Am. Soc. Hematol. Educ. Program 2013, 2013, 601–604. [Google Scholar] [CrossRef] [Green Version]
- Fagioli, F.; Zecca, M.; Locatelli, F.; Lanino, E.; Uderzo, C.; Di Bartolomeo, P.; Berger, J.M.; Favre, C.; Rondelli, R.; Pession, A.; et al. Allogeneic Stem Cell Transplantation for Children with Acute Myeloid Leukemia in Second Complete Remission. J. Pediatr. Hematol. Oncol. 2008, 30, 575–583. [Google Scholar] [CrossRef] [Green Version]
- Sander, A.; Zimmermann, M.; Dworzak, M.; Fleischhack, G.; Von Neuhoff, C.; Reinhardt, D.; Kaspers, G.J.L.; Creutzig, U. Consequent and Intensified Relapse Therapy Improved Survival in Pediatric AML: Results of Relapse Treatment in 379 Patients of Three Consecutive AML-BFM Trials. Leukemia 2010, 24, 1422–1428. [Google Scholar] [CrossRef] [Green Version]
- Karlsson, L.; Forestier, E.; Hasle, H.; Jahnukainen, K.; Jónsson, Ó.G.; Lausen, B.; Norén Nyström, U.; Palle, J.; Tierens, A.; Zeller, B.; et al. Outcome after Intensive Reinduction Therapy and Allogeneic Stem Cell Transplant in Paediatric Relapsed Acute Myeloid Leukaemia. Br. J. Haematol. 2017, 178, 592–602. [Google Scholar] [CrossRef] [Green Version]
- Creutzig, U.; Zimmermann, M.; Ritter, J.; Reinhardt, D.; Hermann, J.; Henze, G.; Jürgens, H.; Kabisch, H.; Reiter, A.; Riehm, H.; et al. Treatment Strategies and Long-Term Results in Paediatric Patients Treated in Four Consecutive AML-BFM Trials. Leukemia 2005, 19, 2030–2042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, B.E.S.; Webb, D.K.H.; Howman, A.J.; de Graaf, S.S.N.; Harrison, C.J.; Wheatley, K. Results of a Randomized Trial in Children with Acute Myeloid Leukaemia: Medical Research Council AML12 Trial. Br. J. Haematol. 2011, 155, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, H.; Tabuchi, K.; Tawa, A.; Tsukimoto, I.; Tsuchida, M.; Morimoto, A.; Yabe, H.; Horibe, K.; Hanada, R.; Imaizumi, M.; et al. Outcome of Children with Relapsed Acute Myeloid Leukemia Following Initial Therapy under the AML99 Protocol. Int. J. Hematol. 2014, 100, 171–179. [Google Scholar] [CrossRef]
- Viehmann, S.; Teigler-Schlegel, A.; Bruch, J.; Langebrake, C.; Reinhardt, D.; Harbott, J. Monitoring of Minimal Residual Disease (MRD) by Real-Time Quantitative Reverse Transcription PCR (RQ-RT-PCR) in Childhood Acute Myeloid Leukemia with AML1/ETO Rearrangement. Leukemia 2003, 17, 1130–1136. [Google Scholar] [CrossRef] [Green Version]
- Walter, R.B.; Gyurkocza, B.; Storer, B.E.; Godwin, C.D.; Pagel, J.M.; Buckley, S.A.; Sorror, M.L.; Wood, B.L.; Storb, R.; Appelbaum, F.R.; et al. Comparison of Minimal Residual Disease as Outcome Predictor for AML Patients in First Complete Remission Undergoing Myeloablative or Nonmyeloablative Allogeneic Hematopoietic Cell Transplantation. Leukemia 2015, 29, 137–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ustun, C.; Courville, E.L.; DeFor, T.; Dolan, M.; Randall, N.; Yohe, S.; Bejanyan, N.; Warlick, E.; Brunstein, C.; Weisdorf, D.J.; et al. Myeloablative, but Not Reduced-Intensity, Conditioning Overcomes the Negative Effect of Flow-Cytometric Evidence of Leukemia in Acute Myeloid Leukemia. Biol. Blood Marrow Transpl. 2016, 22, 669–675. [Google Scholar] [CrossRef] [Green Version]
- Hourigan, C.S.; Dillon, L.W.; Gui, G.; Logan, B.R.; Fei, M.; Ghannam, J.; Li, Y.; Licon, A.; Alyea, E.P.; Bashey, A.; et al. Impact of Conditioning Intensity of Allogeneic Transplantation for Acute Myeloid Leukemia with Genomic Evidence of Residual Disease. J. Clin. Oncol. 2020, 38, 1273–1283. [Google Scholar] [CrossRef]
- Pastore, F.; Dufour, A.; Benthaus, T.; Metzeler, K.H.; Maharry, K.S.; Schneider, S.; Ksienzyk, B.; Mellert, G.; Zellmeier, E.; Kakadia, P.M.; et al. Combined Molecular and Clinical Prognostic Index for Relapse and Survival in Cytogenetically Normal Acute Myeloid Leukemia. J. Clin. Oncol. 2014, 32, 1586–1594. [Google Scholar] [CrossRef] [Green Version]
- Hayes, D.N.; Kim, W.Y.; Hayes, D.N.; Kim, W.Y. The next Steps in Next-Gen Sequencing of Cancer Genomes Find the Latest Version: The next Steps in next-Gen Sequencing of Cancer Genomes. J. Clin. Invest. 2015, 125, 462–468. [Google Scholar] [CrossRef] [Green Version]
- Delsing Malmberg, E.; Rehammar, A.; Pereira, M.B.; Abrahamsson, J.; Samuelsson, T.; Ståhlman, S.; Asp, J.; Tierens, A.; Palmqvist, L.; Kristiansson, E.; et al. Accurate and Sensitive Analysis of Minimal Residual Disease in Acute Myeloid Leukemia Using Deep Sequencing of Single Nucleotide Variations. J. Mol. Diagnostics 2019, 21, 149–162. [Google Scholar] [CrossRef] [Green Version]
- Jongen-Lavrencic, M.; Grob, T.; Hanekamp, D.; Kavelaars, F.G.; Al Hinai, A.; Zeilemaker, A.; Erpelinck-Verschueren, C.A.J.; Gradowska, P.L.; Meijer, R.; Cloos, J.; et al. Molecular Minimal Residual Disease in Acute Myeloid Leukemia. N. Engl. J. Med. 2018, 378, 1189–1199. [Google Scholar] [CrossRef] [PubMed]



| AIEOP | NOPHO | BFM | Total | |
|---|---|---|---|---|
| n° pts | 63 | 38 | 11 | 112 |
| Age (average) | 7.9 | 9.1 | 9.8 | 8.9 |
| Gender | ||||
| male | 37 | 19 | 7 | 63 |
| female | 26 | 19 | 4 | 49 |
| WBC (average) (n = 107) | 50,382 | 59,245 | 52,635 | 53,347 |
| Genetics | ||||
| Standard risk | ||||
| t(8;21)RUNX1::RUNX1T1 | 20 | 17 | 4 | 41 |
| inv(16)CBFB::MYH11 | 8 | 6 | 2 | 16 |
| High risk | ||||
| t(9;11)KMT2A::MLLT3 | 19 | 10 | 5 | 34 |
| FLT3-ITD | 16 | 5 | 0 | 21 |
| Karyotype (n = 82) | ||||
| complex | 6 | 8 | 1 | 15 |
| Type of remission | ||||
| CR1 | 53 | 7 | 4 | 64 |
| CR2 | 10 | 31 | 7 | 48 |
| pts status | ||||
| relapse post HSCT (n°) | 7 | 11 | 0 | 18 |
| death (n°) | 8 | 18 | 3 | 29 |
| Median Follow-up (years) | 3.3 | 2.3 | 1.4 | 3.1 |
| range | 0.3–14 | 0.1–16 | 0.1–4 | 0.1–16 |
| Type of HSCT (n = 108) | ||||
| MUD-ALLO | 21 | 28 | 4 | 53 |
| SIBLING-RELATED | 34 | 10 | 3 | 47 |
| AUTOLOGOUS | 8 | 0 | 0 | 8 |
| Source (n = 94) | ||||
| BM | 45 | 12 | 4 | 61 |
| PB | 15 | 14 | 1 | 30 |
| CB | 3 | 0 | 0 | 3 |
| Preparative regimen before HSCT | ||||
| BUS-based | 52 | 11 | 2 | 65 |
| TBI-based | 6 | 0 | 0 | 6 |
| other | 5 | 15 | 5 | 25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benetton, M.; Merli, P.; Walter, C.; Hansen, M.; Da Ros, A.; Polato, K.; Tregnago, C.; Abrahamsson, J.; Strocchio, L.; Sonneveld, E.; et al. Molecular Measurable Residual Disease Assessment before Hematopoietic Stem Cell Transplantation in Pediatric Acute Myeloid Leukemia Patients: A Retrospective Study by the I-BFM Study Group. Biomedicines 2022, 10, 1530. https://doi.org/10.3390/biomedicines10071530
Benetton M, Merli P, Walter C, Hansen M, Da Ros A, Polato K, Tregnago C, Abrahamsson J, Strocchio L, Sonneveld E, et al. Molecular Measurable Residual Disease Assessment before Hematopoietic Stem Cell Transplantation in Pediatric Acute Myeloid Leukemia Patients: A Retrospective Study by the I-BFM Study Group. Biomedicines. 2022; 10(7):1530. https://doi.org/10.3390/biomedicines10071530
Chicago/Turabian StyleBenetton, Maddalena, Pietro Merli, Christiane Walter, Maria Hansen, Ambra Da Ros, Katia Polato, Claudia Tregnago, Jonas Abrahamsson, Luisa Strocchio, Edwin Sonneveld, and et al. 2022. "Molecular Measurable Residual Disease Assessment before Hematopoietic Stem Cell Transplantation in Pediatric Acute Myeloid Leukemia Patients: A Retrospective Study by the I-BFM Study Group" Biomedicines 10, no. 7: 1530. https://doi.org/10.3390/biomedicines10071530