Phosphatase of Regenerating Liver-1 (PRL-1)-Overexpressing Placenta-Derived Mesenchymal Stem Cells Enhance Antioxidant Effects via Peroxiredoxin 3 in TAA-Injured Rat Livers
Abstract
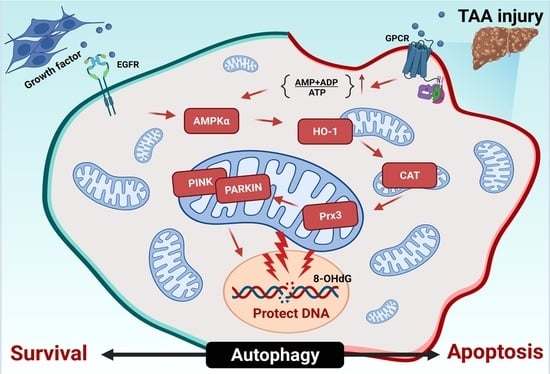
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Cell Culture
2.3. Blood Chemistry
2.4. Quantitative Reverse Transcription–Polymerase Chain Reaction (qRT–PCR)
2.5. Western Blotting
2.6. Enzyme-Linked Immunosorbent Assay (ELISA)
2.7. Analysis of mtDNA Copy Number
2.8. Histological Analysis
2.9. Immunohistochemistry
2.10. Immunofluorescence
2.11. TUNEL Assay
2.12. Statistical Analysis
3. Results
3.1. Attenuation of Cell Death in Naïve and PRL-1(+) Cells Transplanted into TAA-Injured Rat Livers
3.2. The Effect of Naïve and PRL-1(+) Cells Transplanted into TAA-Injured Rat Livers on DNA Repair
3.3. Regulation of Reactive Oxygen Species (ROS) in Naïve and PRL-1(+) Cells Transplanted into TAA-Injured Rat Livers
3.4. Effect of the Mitochondrial Peroxiredoxin Family in Naïve-and PRL-1(+)-Cell-Transplanted, TAA-Injured Rat Livers
3.5. The Effect of Naïve and PRL-1(+) Cell Transplantation on Mitophagy in TAA-Injured Rat Livers
3.6. Analysis of Mitochondrial Biogenesis Efficacy through Naïve and PRL-1(+) Transplantation in TAA-Injured Rat Livers
3.7. Expression of Antioxidant Factors in TAA-Treated WB-F344s
3.8. The Effect of Naïve and PRL-1(+) Cell Transplantation on Cell Proliferation in TAA-Injured Rat Livers
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zargar, S.; Alonazi, M.; Rizwana, H.; Wani, T.A. Resveratrol Reverses Thioacetamide-Induced Renal Assault with respect to Oxidative Stress, Renal Function, DNA Damage, and Cytokine Release in Wistar Rats. Oxid. Med. Cell. Longev. 2019, 2019, 1702959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geng, A.; Tang, H.; Huang, J.; Qian, Z.; Qin, N.; Yao, Y.; Xu, Z.; Chen, H.; Lan, L.; Xie, H.; et al. The deacetylase SIRT6 promotes the repair of UV-induced DNA damage by targeting DDB2. Nucleic Acids Res. 2020, 48, 9181–9194. [Google Scholar] [CrossRef] [PubMed]
- Cichoż-Lach, H.; Michalak, A. Oxidative stress as a crucial factor in liver diseases. World J. Gastroenterol. 2014, 20, 8082–8091. [Google Scholar] [CrossRef]
- Jain, I.H.; Calvo, S.E.; Markhard, A.L.; Skinner, O.S.; To, T.L.; Ast, T.; Mootha, V.K. Genetic Screen for Cell Fitness in High or Low Oxygen Highlights Mitochondrial and Lipid Metabolism. Cell 2020, 181, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Tudek, B.; Zdżalik-Bielecka, D.; Tudek, A.; Kosicki, K.; Fabisiewicz, A.; Speina, E. Lipid peroxidation in face of DNA damage, DNA repair and other cellular processes. Free Radic. Biol. Med. 2017, 107, 77–89. [Google Scholar] [CrossRef]
- Santos, J.D.B.; Mendonça, A.A.S.; Sousa, R.C.; Silva, T.G.S.; Bigonha, S.M.; Santos, E.C.; Gonçalves, R.V.; Novaes, R.D. Food-drug interaction: Anabolic steroids aggravate hepatic lipotoxicity and nonalcoholic fatty liver disease induced by trans fatty acids. Food Chem. Toxicol. 2018, 116, 360–368. [Google Scholar] [CrossRef]
- Field, C.S.; Baixauli, F.; Kyle, R.L.; Puleston, D.J.; Cameron, A.M.; Sanin, D.E.; Hippen, K.L.; Loschi, M.; Thangavelu, G.; Corrado, M.; et al. Mitochondrial Integrity Regulated by Lipid Metabolism Is a Cell-Intrinsic Checkpoint for Treg Suppressive Function. Cell Metab. 2020, 31, 422–437.e425. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.B.; Louie, S.M.; Daniele, J.R.; Tran, Q.; Dillin, A.; Zoncu, R.; Nomura, D.K.; Olzmann, J.A. DGAT1-Dependent Lipid Droplet Biogenesis Protects Mitochondrial Function during Starvation-Induced Autophagy. Dev. Cell 2017, 42, 9–21.e5. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef]
- Scully, R.; Panday, A.; Elango, R.; Willis, N.A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell. Biol. 2019, 20, 698–714. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Kang, T.H. DNA Oxidation and Excision Repair Pathways. Int. J. Mol. Sci. 2019, 20, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.; Wei, J.; Cui, X.; Yu, C.; Ni, W.; Bungert, J.; Wu, L.; He, C.; Qian, Z. Post-translational modification of RNA m6A demethylase ALKBH5 regulates ROS-induced DNA damage response. Nucleic Acids Res. 2021, 49, 5779–5797. [Google Scholar] [CrossRef] [PubMed]
- Karplus, P.A. A primer on peroxiredoxin biochemistry. Free Radic. Biol. Med. 2015, 80, 183–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smirnoff, N.; Arnaud, D. Hydrogen peroxide metabolism and functions in plants. New Phytol. 2019, 221, 1197–1214. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Li, X.Y.; Liu, Y.J.; Feng, J.; Wu, Y.; Shen, H.M.; Lu, G.D. Full-coverage regulations of autophagy by ROS: From induction to maturation. Autophagy 2021, 8, 1240–1255. [Google Scholar] [CrossRef]
- Rhee, S.G. Overview on Peroxiredoxin. Mol. Cells 2016, 39, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Arkat, S.; Umbarkar, P.; Singh, S.; Sitasawad, S.L. Mitochondrial Peroxiredoxin-3 protects against hyperglycemia induced myocardial damage in Diabetic cardiomyopathy. Free Radic. Biol. Med. 2016, 97, 489–500. [Google Scholar] [CrossRef]
- Zhang, Y.; Park, J.; Han, S.J.; Park, I.; Huu, T.N.; Kim, J.S.; Woo, H.A.; Lee, S.R. The critical role of redox regulation of PTEN and peroxiredoxin III in alcoholic fatty liver. Free Radic. Biol. Med. 2021, 162, 141–148. [Google Scholar] [CrossRef]
- Na, J.; Song, J.; Kim, H.H.; Seok, J.; Kim, J.Y.; Jun, J.H.; Kim, G.J. Human placenta-derived mesenchymal stem cells trigger repair system in TAA-injured rat model via antioxidant effect. Aging 2020, 13, 61–76. [Google Scholar] [CrossRef]
- Park, M.; Banga, J.P.; Kim, G.J.; Kim, M.; Lew, H. Human placenta-derived mesenchymal stem cells ameliorate orbital adipogenesis in female mice models of Graves’ ophthalmopathy. Stem Cell Res. Ther. 2019, 10, 246. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Du, K.; Ramirez, S.; Diamond, R.H.; Taub, R. Mitogenic up-regulation of the PRL-1 protein-tyrosine phosphatase gene by Egr-1. Egr-1 activation is an early event in liver regeneration. J. Biol. Chem. 1999, 274, 4513–4520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, Y.; Ye, D.Z.; Li, Z.; Teta-Bissett, M.; Peng, Y.; Taub, R.; Greenbaum, L.E.; Kaestner, K.H. Protein tyrosine phosphatase of liver regeneration-1 is required for normal timing of cell cycle progression during liver regeneration. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 308, G85–G91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.Y.; Choi, J.H.; Jun, J.H.; Park, S.; Jung, J.; Bae, S.H.; Kim, G.J. Enhanced PRL-1 expression in placenta-derived mesenchymal stem cells accelerates hepatic function via mitochondrial dynamics in a cirrhotic rat model. Stem Cell Res. Ther. 2020, 11, 512. [Google Scholar] [CrossRef] [PubMed]
- Kaur, V.; Kumar, M.; Kaur, P.; Kaur, S.; Singh, A.P.; Kaur, S. Hepatoprotective activity of Butea monosperma bark against thioacetamide-induced liver injury in rats. Biomed. Pharmacother. 2017, 89, 332–341. [Google Scholar] [CrossRef]
- Vokálová, L.; Lauková, L.; Čonka, J.; Melišková, V.; Borbélyová, V.; Bábíčková, J.; Tóthová, L.; Hodosy, J.; Vlková, B.; Celec, P. Deoxyribonuclease partially ameliorates thioacetamide-induced hepatorenal injury. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 312, G457–G463. [Google Scholar] [CrossRef] [Green Version]
- Nicolussi, A.; D’Inzeo, S.; Capalbo, C.; Giannini, G.; Coppa, A. The role of peroxiredoxins in cancer. Mol. Clin. Oncol. 2017, 6, 139–153. [Google Scholar] [CrossRef] [Green Version]
- Yoo, S.M.; Jung, Y.K. A Molecular Approach to Mitophagy and Mitochondrial Dynamics. Mol. Cells 2018, 41, 18–26. [Google Scholar] [CrossRef]
- Zhan, T.; Chen, Q.; Zhang, C.; Bi, C.; Zhang, X. Constructing a Novel Biosynthetic Pathway for the Production of Glycolate from Glycerol in Escherichia coli. ACS Synth. Biol. 2020, 9, 2600–2609. [Google Scholar] [CrossRef]
- Singh, G.; Pachouri, U.C.; Khaidem, D.C.; Kundu, A.; Chopra, C.; Singh, P. Mitochondrial DNA Damage and Diseases. F1000Research 2015, 4, 176. [Google Scholar] [CrossRef]
- Quan, Y.; Xin, Y.; Tian, G.; Zhou, J.; Liu, X. Mitochondrial ROS-Modulated mtDNA: A Potential Target for Cardiac Aging. Oxid. Med. Cell. Longev. 2020, 2020, 9423593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chao, T.; Shih, H.T.; Hsu, S.C.; Chen, P.J.; Fan, Y.S.; Jeng, Y.M.; Shen, Z.Q.; Tsai, T.F.; Chang, Z.F. Autophagy restricts mitochondrial DNA damage-induced release of ENDOG (endonuclease G) to regulate genome stability. Autophagy 2021, 17, 3444–3460. [Google Scholar] [CrossRef] [PubMed]
- Forman, H.J.; Zhang, H. Targeting oxidative stress in disease: Promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 2021, 20, 689–709. [Google Scholar] [CrossRef]
- Sies, H. Oxidative stress: A concept in redox biology and medicine. Redox Biol. 2015, 4, 180–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, P.; Wei, L.L.; Zhao, S.; Sverdlov, D.Y.; Vaid, K.A.; Miyamoto, M.; Kuramitsu, K.; Lai, M.; Popov, Y.V. Hepatocyte mitochondria-derived danger signals directly activate hepatic stellate cells and drive progression of liver fibrosis. Nat. Commun. 2020, 11, 2362. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, M.; Wang, X.; Bu, Q.; Wang, Q.; Su, W.; Li, L.; Zhou, H.; Lu, L. XBP1 deficiency promotes hepatocyte pyroptosis by impairing mitophagy to activate mtDNA-cGAS-STING signaling in macrophages during acute liver injury. Redox Biol. 2022, 52, 102305. [Google Scholar] [CrossRef] [PubMed]
- Łuczaj, W.; Skrzydlewska, E. DNA damage caused by lipid peroxidation products. Cell. Mol. Biol. Lett. 2003, 8, 391–413. [Google Scholar]
- Liu, Z.; Hu, Y.; Gong, Y.; Zhang, W.; Liu, C.; Wang, Q.; Deng, H. Hydrogen peroxide mediated mitochondrial UNG1-PRDX3 interaction and UNG1 degradation. Free Radic. Biol. Med. 2016, 99, 54–62. [Google Scholar] [CrossRef]
- Lee, M.J.; Jung, J.; Na, K.H.; Moon, J.S.; Lee, H.J.; Kim, J.H.; Kim, G.I.; Kwon, S.W.; Hwang, S.G.; Kim, G.J. Anti-fibrotic effect of chorionic plate-derived mesenchymal stem cells isolated from human placenta in a rat model of CCl(4)-injured liver: Potential application to the treatment of hepatic diseases. J. Cell. Biochem. 2010, 111, 1453–1463. [Google Scholar] [CrossRef]
- Wu, W.B.; Menon, R.; Xu, Y.Y.; Zhao, J.R.; Wang, Y.L.; Liu, Y.; Zhang, H.J. Downregulation of peroxiredoxin-3 by hydrophobic bile acid induces mitochondrial dysfunction and cellular senescence in human trophoblasts. Sci. Rep. 2016, 6, 38946. [Google Scholar] [CrossRef] [Green Version]
- Sevilla, F.; Camejo, D.; Ortiz-Espín, A.; Calderón, A.; Lázaro, J.J.; Jiménez, A. The thioredoxin/peroxiredoxin/sulfiredoxin system: Current overview on its redox function in plants and regulation by reactive oxygen and nitrogen species. J. Exp. Bot. 2015, 66, 2945–2955. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Park, S.J.; Kim, J.H.; Seong, J.B.; Kim, K.M.; Woo, H.A.; Lee, D.S. Peroxiredoxin 5 regulates adipogenesis-attenuating oxidative stress in obese mouse models induced by a high-fat diet. Free Radic. Biol. Med. 2018, 123, 27–38. [Google Scholar] [CrossRef] [PubMed]








| Gene | Primers (Rat) | Tm (℃) | NCBI Ref. | |
|---|---|---|---|---|
| qRT-PCR | PPARγ | F: 5’-GACAGACCTCAGGCAGATTG-3’ R: 5’-GTCAGCGACTGGGACTTTTC-3’ | 57 | NM_013124.3 |
| Adiponectin | F: 5’-GACTGCCACTAATTCAGAGC-3’ R: 5’-CTCATGGGGATAACACTCAG-3’ | 55 | NM_144744.3 | |
| Adipsin | F: 5’-CACGTACCATGATGGGGCAA-3’ R: 5’-TCGAGATCCCACGTAACCA-3’ | 59 | NM_000102.4 | |
| LPL | F: 5’-ACAGGTGCAATTCCAAGGAAG-3’ R: 5’-CTTTCAGCCACTGTGCCATA-3’ | 57 | M92059.1 | |
| γH2AX | F: 5’-TGGAAAGGGTCAGGGAACG-3’ R: 5’-GACTTGTGCTGGTATCTGGGTG-3’ | 58 | NM_001109291.1 | |
| PINK | F: 5’-CATGGCTTTGGATGGAGAGT-3’ R: 5’-TGGGAGTTTGCTCTTCAAGG-3’ | 56 | XM_032895606.1 | |
| PARKIN | F: 5’-CTGGCAGTCATTCTGGACAC-3’ R:5’-CTCTCCACTCATCCGGTTTG-3’ | 57 | XM_032894705.1 | |
| 8-OHdG | F: 5’-GGGCCCAAGCAGTGCTGTTC-3’ R: 5’-GATCCCTTTTTGCGCTTTTGC-3’ | 61 | XM_034511266.1 | |
| GAPDH | F: 5’-CGAGATCCCTCCAAAATCAA-3’ R: 5’-TGTGGTCATGAGTCCTTCCA-3’ | 55 | NM_001357943.2 | |
| TaqMan | Mitochondrial D-loop | F: 5’-GGTTCTTACTTCAGGGCCATCA-4’ R: 5’-GATTAGACCCGTTACCATCGAGAT-3’ | 60 | EU194676.1 |
| β-actin | F: 5’-GGGATGTTTGCTCCAACCAA-3’ R: 5’-GCGCTTTTGACTCAAGGATTTAA-3’ | 58 | XM_039089807.1 | |
| mitochondrial D-loop probe | JOE-TTGGTTCATCGTCCATACGTTCCCCTTA-3’ | |||
| β-actin probe | FAM-CGGTCGCCTTCACCGTTCCAGTT-3’ | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, H.J.; Jun, J.H.; Kim, J.Y.; Jang, H.J.; Lim, J.-Y.; Bae, S.H.; Kim, G.J. Phosphatase of Regenerating Liver-1 (PRL-1)-Overexpressing Placenta-Derived Mesenchymal Stem Cells Enhance Antioxidant Effects via Peroxiredoxin 3 in TAA-Injured Rat Livers. Antioxidants 2023, 12, 46. https://doi.org/10.3390/antiox12010046
Park HJ, Jun JH, Kim JY, Jang HJ, Lim J-Y, Bae SH, Kim GJ. Phosphatase of Regenerating Liver-1 (PRL-1)-Overexpressing Placenta-Derived Mesenchymal Stem Cells Enhance Antioxidant Effects via Peroxiredoxin 3 in TAA-Injured Rat Livers. Antioxidants. 2023; 12(1):46. https://doi.org/10.3390/antiox12010046
Chicago/Turabian StylePark, Hee Jung, Ji Hye Jun, Jae Yeon Kim, Hye Jung Jang, Ja-Yun Lim, Si Hyun Bae, and Gi Jin Kim. 2023. "Phosphatase of Regenerating Liver-1 (PRL-1)-Overexpressing Placenta-Derived Mesenchymal Stem Cells Enhance Antioxidant Effects via Peroxiredoxin 3 in TAA-Injured Rat Livers" Antioxidants 12, no. 1: 46. https://doi.org/10.3390/antiox12010046