Characterization and Protective Properties of Lactic Acid Bacteria Intended to Be Used in Probiotic Preparation for Honeybees (Apis mellifera L.)—An In Vitro Study
Abstract
:Simple Summary
Abstract
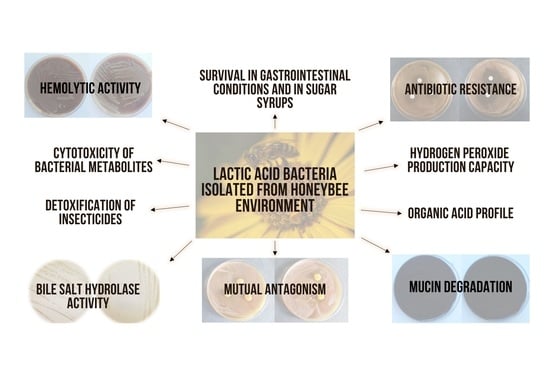
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. High-Performance Liquid Chromatography (HPLC)
2.3. Survival in the Simulated Digestive Tract
2.4. Survival in the Sugar Syrups
2.5. Cytotoxicity of CFS
2.5.1. Caco-2 Cell Culture
2.5.2. Neutral Red Uptake (NRU) Assay
2.5.3. Protective Activity of CFS
2.6. Antibiotics Susceptibility Testing
2.7. Hydrogen Peroxide Production
2.8. Hemolysis
2.9. Mucin Degradation
2.10. Bile Salts Hydrolase (BSH) Activity
2.11. Mutual Antagonistic Activity with Agar Slab Method
2.12. Statistical Analysis
3. Results and Discussion
3.1. Organic Acids Profile
3.2. Ability of LAB Strains to Survive in the Simulated Digestive Tract
3.3. Ability of LAB Strains to Survive in the Sugar Syrups
3.4. Cytotoxic Activity of CFS
3.5. Protective Activity of CFS of P. pentosaceus 14/1 against Insecticides
3.6. Susceptibility to Antibiotics
3.7. Mutual Antagonistic Activity between LAB Strains
3.8. Hydrogen Peroxide Production, Hemolytic and BSH Activity, and Mucin Degradation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Leska, A.; Nowak, A.; Nowak, I.; Górczyńska, A. Effects of Insecticides and Microbiological Contaminants on Apis mellifera Health. Molecules 2021, 26, 5080. [Google Scholar] [CrossRef] [PubMed]
- Hung, K.-L.J.; Kingston, J.M.; Albrecht, M.; Holway, D.A.; Kohn, J.R. The worldwide importance of honey bees as pollinators in natural habitats. Proc. R. Soc. B Biol. Sci. 2018, 285, 20172140. [Google Scholar] [CrossRef] [PubMed]
- Stanley, D.A.; Msweli, S.M.; Johnson, S.D. Native honeybees as flower visitors and pollinators in wild plant communities in a biodiversity hotspot. Ecosphere 2020, 11, e02957. [Google Scholar] [CrossRef]
- Ortiz-Alvarado, Y.; Clark, D.R.; Vega-Melendez, C.J.; Flores-Cruz, Z.; Domingez-Bello, M.G.; Giray, T. Antibiotics in hives and their effects on Honey Bee Physiology and behavioral development. Biol. Open 2020, 9, bio053884. [Google Scholar] [CrossRef]
- Bulson, L.; Becher, M.A.; McKinley, T.J.; Wilfert, L. Long-term effects of antibiotic treatments on honeybee colony fitness: A modelling approach. J. Appl. Ecol. 2020, 58, 70–79. [Google Scholar] [CrossRef]
- Borges, D.; Guzman-Novoa, E.; Goodwin, P.H. Effects of prebiotics and probiotics on honey bees (Apis mellifera) infected with the microsporidian parasite Nosema ceranae. Microorganisms 2021, 9, 481. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Lye, H.S.; Balakrishnan, K.; Thiagarajah, K.; Mohd Ismail, N.I.; Ooi, S.Y. Beneficial properties of probiotics. Trop. Life Sci. Res. 2016, 27, 73–90. [Google Scholar] [CrossRef]
- Salminen, S.; Nybom, S.; Meriluoto, J.; Collado, M.C.; Vesterlund, S.; El-Nezami, H. Interaction of probiotics and pathogens—Benefits to human health? Curr. Opin. Biotechnol. 2010, 21, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, J.; Lv, M.; Shao, Z.; Hungwe, M.; Wang, J.; Bai, X.; Xie, J.; Wang, Y.; Geng, W. Metabolism characteristics of lactic acid bacteria and the expanding applications in food industry. Front. Bioeng. Biotechnol. 2021, 9, 612285. [Google Scholar] [CrossRef]
- Campana, R.; van Hemert, S.; Baffone, W. Strain-specific probiotic properties of lactic acid bacteria and their interference with human intestinal pathogens invasion. Gut Pathog. 2017, 9, 12. [Google Scholar] [CrossRef]
- Agriopoulou, S.; Stamatelopoulou, E.; Sachadyn-Król, M.; Varzakas, T. Lactic Acid Bacteria as Antibacterial Agents to Extend the Shelf Life of Fresh and Minimally Processed Fruits and Vegetables: Quality and Safety Aspects. Microorganisms 2020, 8, 952. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; HuangFu, H.P.; Wang, X.; Zhao, S.S.; Liu, Y.; Lv, H.; Qin, G.Y.; Tan, Z. Antibacterial activity of lactic acid producing Leuconostoc mesenteroides QZ1178 against pathogenic Gallibacterium anatis. Front. Vet. Sci. 2021, 8, 628258. [Google Scholar] [CrossRef]
- Ibrahim, S.A.; Ayivi, R.D.; Zimmerman, T.; Siddiqui, S.A.; Altemimi, A.B.; Fidan, H.; Esatbeyoglu, T.; Bakhshayesh, R.V. Lactic Acid Bacteria as Antimicrobial Agents: Food Safety and Microbial Food Spoilage Prevention. Foods 2021, 10, 3131. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, H.; Toba, T.; Ito, A.; Kudo, S.; Sato, S.; Sato, Y. The screening of hydrogen peroxide-producing lactic acid bacteria and their application to inactivating psychrotrophic food-borne pathogens. Curr. Microbiol. 2003, 47, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Pernice, M.; Simpson, S.J.; Ponton, F. Towards an integrated understanding of gut microbiota using insects as model systems. J. Insect Physiol. 2014, 69, 12–18. [Google Scholar] [CrossRef]
- Iorizzo, M.; Letizia, F.; Ganassi, S.; Testa, B.; Petrarca, S.; Albanese, G.; Di Criscio, D.; De Cristofaro, A. Functional Properties and Antimicrobial Activity from Lactic Acid Bacteria as Resources to Improve the Health and Welfare of Honey Bees. Insects 2022, 13, 308. [Google Scholar] [CrossRef]
- Raymann, K.; Moran, N.A. The role of the gut microbiome in health and disease of Adult Honey Bee Workers. Curr. Opin. Insect. Sci. 2018, 26, 97–104. [Google Scholar] [CrossRef]
- Evans, J.D.; Lopez, D.L. Bacterial probiotics induce an immune response in the honey bee (hymenoptera: Apidae). J. Econ. Entomol. 2004, 97, 752–756. [Google Scholar] [CrossRef]
- Fanciotti, M.N.; Tejerina, M.; Benítez-Ahrendts, M.R.; Audisio, M.C. Honey yield of different commercial apiaries treated with Lactobacillus salivarius A3IOB, a new bee-probiotic strain. Benef. Microbes 2018, 9, 291–298. [Google Scholar] [CrossRef]
- Cingeľová Maruščáková, I.; Schusterová, P.; Bielik, B.; Toporčák, J.; Bíliková, K.; Mudroňová, D. Effect of application of probiotic pollen suspension on immune response and gut microbiota of honey bees (Apis mellifera). Probiotics Antimicrob. Proteins 2020, 12, 929–936. [Google Scholar] [CrossRef] [PubMed]
- Tlak Gajger, I.; Vlainić, J.; Šoštarić, P.; Prešern, J.; Bubnič, J.; Smodiš Škerl, M.I. Effects on Some Therapeutical, Biochemical, and Immunological Parameters of Honey Bee (Apis mellifera) Exposed to Probiotic Treatments, in Field and Laboratory Conditions. Insects 2020, 11, 638. [Google Scholar] [CrossRef]
- Daisley, B.A.; Pitek, A.P.; Chmiel, J.A.; Gibbons, S.; Chernyshova, A.M.; Al, K.F.; Faragalla, K.M.; Burton, J.P.; Thompson, G.J.; Reid, G. Lactobacillus spp. attenuate antibiotic-induced immune and microbiota dysregulation in honey bees. Commun. Biol. 2020, 3, 534. [Google Scholar] [CrossRef]
- Nowak, A.; Szczuka, D.; Górczyńska, A.; Motyl, I.; Kręgiel, D. Characterization of Apis mellifera Gastrointestinal Microbiota and Lactic Acid Bacteria for Honeybee Protection—A Review. Cells 2021, 10, 701. [Google Scholar] [CrossRef]
- Huang, Y.-H.; Chen, Y.-H.; Chen, J.-H.; Hsu, P.-S.; Wu, T.-H.; Lin, C.-F.; Peng, C.-C.; Wu, M.-C. A potential probiotic Leuconostoc mesenteroides TBE-8 for Honey Bee. Sci. Rep. 2021, 11, 18466. [Google Scholar] [CrossRef] [PubMed]
- Leska, A.; Nowak, A.; Czarnecka-Chrebelska, K.H. Adhesion and Anti-Adhesion Abilities of Potentially Probiotic Lactic Acid Bacteria and Biofilm Eradication of Honeybee (Apis mellifera L.) Pathogens. Molecules 2022, 27, 8945. [Google Scholar] [CrossRef]
- Leska, A.; Nowak, A.; Szulc, J.; Motyl, I.; Czarnecka-Chrebelska, K.H. Antagonistic Activity of Potentially Probiotic Lactic Acid Bacteria against Honeybee (Apis mellifera L.) Pathogens. Pathogens 2022, 11, 1367. [Google Scholar] [CrossRef] [PubMed]
- Leska, A.; Nowak, A.; Miśkiewicz, K.; Rosicka-Kaczmarek, J. Binding and Detoxification of Insecticides by Potentially Probiotic Lactic Acid Bacteria Isolated from Honeybee (Apis mellifera L.) Environment—An In Vitro Study. Cells 2022, 11, 3743. [Google Scholar] [CrossRef]
- Leska, A.; Nowak, A.; Motyl, I. Isolation and Some Basic Characteristics of Lactic Acid Bacteria from Honeybee (Apis mellifera L.) Environment—A Preliminary Study. Agriculture 2022, 12, 1562. [Google Scholar] [CrossRef]
- Chen, P.; You, Q.; Li, X.; Chang, Q.; Zhang, Y.; Zheng, B.; Hu, X.; Zeng, H. Polysaccharide fractions from Fortunella margarita affect proliferation of Bifidobacterium adolescentis ATCC 15703 and undergo structural changes following fermentation. Int. J. Biol. Macromol. 2019, 123, 1070–1078. [Google Scholar] [CrossRef]
- Nowak, A.; Zakłos-Szyda, M.; Rosicka-Kaczmarek, J.; Motyl, I. Anticancer Potential of Post-Fermentation Media and Cell Extracts of Probiotic Strains: An In Vitro Study. Cancers 2022, 14, 1853. [Google Scholar] [CrossRef]
- Honey Chandran, C.; Keerthi, T.R. Probiotic potency of Lactobacillus plantarum KX519413 and KX519414 isolated from Honey Bee Gut. FEMS Microbiol. Lett. 2018, 365, fnx285. [Google Scholar] [CrossRef]
- de Oliveira Coelho, B.; Fiorda-Mello, F.; de Melo Pereira, G.V.; Thomaz-Soccol, V.; Rakshit, S.K.; de Carvalho, J.C.; Soccol, C.R. In Vitro Probiotic Properties and DNA Protection Activity of Yeast and Lactic Acid Bacteria Isolated from A Honey-Based Kefir Beverage. Foods 2019, 8, 485. [Google Scholar] [CrossRef]
- Bamidele, J.A.; Idowu, A.B.; Ademolu, K.O.; Osipitan, A.A.; Atayese, A.O. Gut digestive enzymes and bacterial and fungal diversity of Apis mellifera adansonii (hymenoptera: Apidae) from three ecological zones of Nigeria. Egypt. J. Basic Appl. Sci. 2021, 8, 54–63. [Google Scholar] [CrossRef]
- Hurtado-Romero, A.; Del Toro-Barbosa, M.; Gradilla-Hernández, M.S.; Garcia-Amezquita, L.E.; García-Cayuela, T. Probiotic Properties, Prebiotic Fermentability, and GABA-Producing Capacity of Microorganisms Isolated from Mexican Milk Kefir Grains: A Clustering Evaluation for Functional Dairy Food Applications. Foods 2021, 10, 2275. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; Lombardi, S.J.; Ganassi, S.; Testa, B.; Ianiro, M.; Letizia, F.; Succi, M.; Tremonte, P.; Vergalito, F.; Cozzolino, A.; et al. Antagonistic Activity against Ascosphaera apis and Functional Properties of Lactobacillus kunkeei Strains. Antibiotics 2020, 9, 262. [Google Scholar] [CrossRef] [PubMed]
- Peghaire, E.; Moné, A.; Delbac, F.; Debroas, D.; Chaucheyras-Durand, F.; El Alaoui, H. A Pediococcus strain to rescue honeybees by decreasing Nosema ceranae- and pesticide-induced adverse effects. Pestic. Biochem. Physiol. 2020, 163, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Pachla, A.; Ptaszyńska, A.A.; Wicha, M.; Kunat, M.; Wydrych, J.; Oleńska, E.; Małek, W. Insight into probiotic properties of lactic acid bacterial endosymbionts of Apis mellifera L. derived from the Polish apiary. Saudi J. Biol. Sci. 2021, 28, 1890–1899. [Google Scholar] [CrossRef]
- Okrasa, M.; Leszczyńska, M.; Sałasińska, K.; Szczepkowski, L.; Kozikowski, P.; Nowak, A.; Szulc, J.; Adamus-Włodarczyk, A.; Gloc, M.; Majchrzycka, K.; et al. Viscoelastic Polyurethane Foams with Reduced Flammability and Cytotoxicity. Materials 2022, 15, 151. [Google Scholar] [CrossRef]
- Pisano, M.B.; Viale, S.; Conti, S.; Fadda, M.E.; Deplano, M.; Melis, M.P.; Deiana, M.; Cosentino, S. Preliminary evaluation of probiotic properties of Lactobacillus strains isolated from Sardinian dairy products. BioMed Res. Int. 2014, 2014, 286390. [Google Scholar] [CrossRef]
- Raghavan, K.T. Antibiotic susceptibility profile of lactic acid bacteria with probiotic potential isolated from humans. Biomed. J. Sci. Technol. Res. 2019, 17, 12964–12966. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement; CLSI Document M100-S22; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015. [Google Scholar]
- Cebeci, A.; Gürakan, C. Properties of potential probiotic Lactobacillus plantarum strains. Food Microbiol. 2003, 20, 511–518. [Google Scholar] [CrossRef]
- Charteris, W.P.; Kelly, P.M.; Morelli, L.; Collins, J.K. Antibiotic susceptibility of potentially probiotic Lactobacillus species. J. Food. Prot. 1998, 61, 1636–1643. [Google Scholar] [CrossRef] [PubMed]
- Turchi, B.; Mancini, S.; Fratini, F.; Pedonese, F.; Nuvoloni, R.; Bertelloni, F.; Ebani, V.V.; Cerri, D. Preliminary evaluation of probiotic potential of Lactobacillus plantarum strains isolated from Italian food products. World J. Microbiol. Biotechnol. 2013, 29, 1913–1922. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.; Puniya, A.K.; Shukla, P. Probiotic properties of lactobacillus plantarum rypr1 from an indigenous fermented beverage Raabadi. Front. Microbiol. 2016, 7, 1683. [Google Scholar] [CrossRef] [PubMed]
- Verdenelli, M.C.; Ghelfi, F.; Silvi, S.; Orpianesi, C.; Cecchini, C.; Cresci, A. Probiotic properties of Lactobacillus rhamnosus and Lactobacillus paracasei isolated from human faeces. Eur. J. Nutr. 2009, 48, 355–363. [Google Scholar] [CrossRef]
- Song, Y.-L.; Kato, N.; Matsumiya, Y.; Liu, C.-X.; Kato, H.; Watanabe, K. Identification of and hydrogen peroxide production by fecal and vaginal lactobacilli isolated from Japanese women and newborn infants. J. Clin. Microbiol. 1999, 37, 3062–3064. [Google Scholar] [CrossRef]
- Yasmin, I.; Saeed, M.; Khan, W.A.; Khaliq, A.; Chughtai, M.F.J.; Iqbal, R.; Tehseen, S.; Naz, S.; Liaqat, A.; Mehmood, T.; et al. In Vitro Probiotic Potential and Safety Evaluation (Hemolytic, Cytotoxic Activity) of Bifidobacterium Strains Isolated from Raw Camel Milk. Microorganisms 2020, 8, 354. [Google Scholar] [CrossRef]
- de Albuquerque, T.M.; Garcia, E.F.; de Oliveira Araújo, A.; Magnani, M.; Saarela, M.; de Souza, E.L. In vitro characterization of Lactobacillus strains isolated from fruit processing by-products as potential probiotics. Probiotics Antimicrob. Proteins 2017, 10, 704–716. [Google Scholar] [CrossRef]
- Wang, L.-J.; Yang, X.; Qian, S.-Y.; Liu, Y.-C.; Yao, K.-H.; Dong, F.; Song, W.-Q. Identification of hemolytic activity and hemolytic genes of methicillin-resistant Staphylococcus aureus isolated from Chinese children. Chin. Med. J. 2019, 133, 88–90. [Google Scholar] [CrossRef]
- Rakhmanova, A.; Khan, Z.A.; Shah, K. A mini review fermentation and preservation: Role of lactic acid bacteria. MOJ Food Process. Technol. 2018, 6, 414–417. [Google Scholar] [CrossRef]
- Wieërs, G.; Belkhir, L.; Enaud, R.; Leclercq, S.; Philippart de Foy, J.-M.; Dequenne, I.; de Timary, P.; Cani, P.D. How probiotics affect the microbiota. Front. Cell. Infect. Microbiol. 2020, 9, 454. [Google Scholar] [CrossRef] [PubMed]
- Quigley, E.M.M. Prebiotics and probiotics in Digestive Health. Clin. Gastroenterol. Hepatol. 2019, 17, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Shah, N.P. Concomitant ingestion of lactic acid bacteria and black tea synergistically enhances flavonoid bioavailability and attenuates D-galactose-induced oxidative stress in mice via modulating glutathione Antioxidant System. J. Nutr. Biochem. 2016, 38, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Ruiz Rodríguez, L.G.; Mohamed, F.; Bleckwedel, J.; Medina, R.; De Vuyst, L.; Hebert, E.M.; Mozzi, F. Diversity and functional properties of lactic acid bacteria isolated from wild fruits and flowers present in northern Argentina. Front. Microbiol. 2019, 10, 1091. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Nguyen, D.-T.; Park, Y.-S.; Hwang, K.-Y.; Cho, Y.-S.; Kang, K.-D.; Yoon, J.-H.; Yu, J.-D.; Yee, S.-T.; Ahn, Y.-H.; et al. Organic acid profiling analysis in culture media of lactic acid bacteria by gas chromatography-mass spectrometry. Mass Spectrom. Lett. 2012, 3, 74–77. [Google Scholar] [CrossRef] [Green Version]
- Pachla, A.; Ptaszyńska, A.A.; Wicha, M.; Oleńska, E.; Małek, W. Fascinating fructophilic lactic acid bacteria associated with various fructose-rich niches. Ann. Univ. Mariae Curie-Skłodowska C Biol. 2019, 72, 41. [Google Scholar] [CrossRef]
- Wang, L.; Cen, S.; Wang, G.; Lee, Y.-K.; Zhao, J.; Zhang, H.; Chen, W. Acetic acid and butyric acid released in large intestine play different roles in the alleviation of constipation. J. Funct. Foods 2020, 69, 103953. [Google Scholar] [CrossRef]
- Filannino, P.; Di Cagno, R.; Addante, R.; Pontonio, E.; Gobbetti, M. Metabolism of fructophilic lactic acid bacteria isolated from the Apis mellifera L. Bee Gut: Phenolic acids as external electron acceptors. Appl. Environ. Microbiol. 2016, 82, 6899–6911. [Google Scholar] [CrossRef]
- Foglietta, F.; Serpe, L.; Canaparo, R.; Vivenza, N.; Riccio, G.; Imbalzano, E.; Gasco, P.; Zara, G.P. Modulation of butyrate anticancer activity by solid lipid nanoparticle delivery: An in vitro investigation on human breast cancer and leukemia cell lines. J. Pharm. Pharm. Sci. 2014, 17, 231. [Google Scholar] [CrossRef]
- Pattayil, L.; Balakrishnan-Saraswathi, H.-T. In vitro evaluation of apoptotic induction of butyric acid derivatives in colorectal carcinoma cells. Anticancer Res. 2019, 39, 3795–3801. [Google Scholar] [CrossRef] [PubMed]
- Al-Lahham, S.H.; Peppelenbosch, M.P.; Roelofsen, H.; Vonk, R.J.; Venema, K. Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2010, 1801, 1175–1183. [Google Scholar] [CrossRef] [PubMed]
- Załęski, A.; Banaszkiewicz, A.; Walkowiak, J. Butyric acid in irritable bowel syndrome. Prz. Gastroenterol. 2013, 6, 350–353. [Google Scholar] [CrossRef]
- Khalil, E.S.; Abd Manap, M.Y.; Mustafa, S.; Alhelli, A.M.; Shokryazdan, P. Probiotic Properties of Exopolysaccharide-Producing Lactobacillus Strains Isolated from Tempoyak. Molecules 2018, 23, 398. [Google Scholar] [CrossRef]
- Park, C.; Park, J.; Kim, W.-J.; Kim, W.; Cheong, H.; Kim, S.-J. Malonic Acid Isolated from Pinus densiflora Inhibits UVB-Induced Oxidative Stress and Inflammation in HaCaT Keratinocytes. Polymers 2021, 13, 816. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.J.; Park, A.R.; Kim, S.; Yeon, J.; Yu, N.H.; Ha, S.; Chang, J.Y.; Park, H.W.; Kim, J.-C. Biological control of root-knot nematodes by organic acid-producing Lactobacillus brevis wikim0069 isolated from kimchi. Plant Pathol. J. 2019, 35, 662–673. [Google Scholar] [CrossRef]
- Binda, S.; Hill, C.; Johansen, E.; Obis, D.; Pot, B.; Sanders, M.E.; Tremblay, A.; Ouwehand, A.C. Criteria to qualify microorganisms as “probiotic” in foods and dietary supplements. Front Microbiol. 2020, 11, 01662. [Google Scholar] [CrossRef]
- Huys, G.; Botteldoorn, N.; Delvigne, F.; De Vuyst, L.; Heyndrickx, M.; Pot, B.; Dubois, J.J.; Daube, G. Microbial characterization of probiotics–Advisory report of the Working Group “8651 Probiotics” of the Belgian Superior Health Council (SHC). Mol. Nutr. Food Res. 2013, 57, 1479–1504. [Google Scholar] [CrossRef]
- Usman; Hosono, A. Viability of lactobacillus gasseri and its cholesterol-binding and antimutagenic activities during subsequent refrigerated storage in nonfermented milk. Saudi J. Biol. Sci. 1999, 82, 2536–2542. [Google Scholar] [CrossRef]
- Tokatlı, M.; Gülgör, G.; Bağder Elmacı, S.; Arslankoz İşleyen, N.; Özçelik, F. In vitro properties of potential probiotic indigenous lactic acid bacteria originating from traditional pickles. BioMed Res. Int. 2015, 2015, 315819. [Google Scholar] [CrossRef]
- Farhangfar, A.; Gandomi, H.; Akhondzadeh Basti, A.; Misaghi, A.; Noori, N. Study of growth kinetic and gastrointestinal stability of acid-bile resistant Lactobacillus plantarum strains isolated from Siahmazgi traditional cheese. Vet. Res. Forum 2021, 12, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Sharafi, H.; Derakhshan, V.; Paknejad, M.; Alidoust, L.; Tohidi, A.; Pornour, M.; Hajfarajollah, H.; Zahiri, H.S.; Noghabi, K.A. Lactobacillus crustorum kh: Novel prospective probiotic strain isolated from Iranian traditional dairy products. Appl. Biochem. Biotechnol. 2014, 175, 2178–2194. [Google Scholar] [CrossRef] [PubMed]
- Mantzourani, I.; Chondrou, P.; Bontsidis, C.; Karolidou, K.; Terpou, A.; Alexopoulos, A.; Bezirtzoglou, E.; Galanis, A.; Plessas, S. Assessment of the probiotic potential of lactic acid bacteria isolated from kefir grains: Evaluation of adhesion and antiproliferative properties in in vitro experimental systems. Ann. Microbiol. 2019, 69, 751–763. [Google Scholar] [CrossRef]
- Feng, Y.; Qiao, L.; Liu, R.; Yao, H.; Gao, C. Potential probiotic properties of lactic acid bacteria isolated from the intestinal mucosa of healthy piglets. Ann. Microbiol. 2017, 67, 239–253. [Google Scholar] [CrossRef]
- Masco, L.; Crockaert, C.; Van Hoorde, K.; Swings, J.; Huys, G. In vitro assessment of the gastrointestinal transit tolerance of taxonomic reference strains from human origin and probiotic product isolates of Bifidobacterium. J. Dairy Sci. 2007, 90, 3572–3578. [Google Scholar] [CrossRef] [PubMed]
- Benavides, A.B.; Ulcuango, M.; Yépez, L.; Tenea, G.N. Assessment of the in vitro bioactive properties of lactic acid bacteria isolated from native ecological niches of Ecuador. Rev. Argent. Microbiol. 2016, 48, 236–244. [Google Scholar] [CrossRef]
- Čeksteryte, V. The quality of syrups used for bee feeding before winter and their suitability for bee wintering. J. Apic. Sci. 2006, 5, 5–14. [Google Scholar]
- Ptaszyńska, A.A.; Borsuk, G.; Zdybicka-Barabas, A.; Cytryńska, M.; Małek, W. Are commercial probiotics and prebiotics effective in the treatment and prevention of Honeybee Nosemosis C? Parasitol. Res. 2015, 115, 397–406. [Google Scholar] [CrossRef]
- Sambuy, Y.; De Angelis, I.; Ranaldi, G.; Scarino, M.L.; Stammati, A.; Zucco, F. The Caco-2 cell line as a model of the intestinal barrier: Influence of cell and culture-related factors on Caco-2 cell functional characteristics. Cell Biol. Toxicol. 2005, 21, 1–26. [Google Scholar] [CrossRef]
- Vinderola, G.; Sanders, M.E.; Salminen, S. The Concept of Postbiotics. Foods 2022, 11, 1077. [Google Scholar] [CrossRef]
- Thorakkattu, P.; Khanashyam, A.C.; Shah, K.; Babu, K.S.; Mundanat, A.S.; Deliephan, A.; Deokar, G.S.; Santivarangkna, C.; Nirmal, N.P. Postbiotics: Current Trends in Food and Pharmaceutical Industry. Foods 2022, 11, 3094. [Google Scholar] [CrossRef]
- Avand, A.; Akbari, V.; Shafizadegan, S. In Vitro Cytotoxic Activity of a Lactococcus lactis Antimicrobial Peptide Against Breast Cancer Cells. Iran J. Biotechnol. 2018, 16, e1867. [Google Scholar] [CrossRef] [PubMed]
- Chuah, L.-O.; Foo, H.L.; Loh, T.C.; Mohammed Alitheen, N.B.; Yeap, S.K.; Abdul Mutalib, N.E.; Abdul Rahim, R.; Yusoff, K. Postbiotic metabolites produced by Lactobacillus plantarum strains exert selective cytotoxicity effects on cancer cells. BMC Complement. Altern. Med. 2019, 19, 114. [Google Scholar] [CrossRef] [PubMed]
- Stafford, K.; Coles, G. Drug Resistance in Ectoparasites of Medical and Veterinary Importance. In Antimicrobial Drug Resistance. Infectious Disease; Mayers, D.L., Ed.; Springer Nature Switzerland AG: Cham, Switzerland, 2017. [Google Scholar]
- Berry, J.A.; Hood, W.M.; Pietravalle, S.; Delaplane, K.S. Field-level sublethal effects of approved bee hive chemicals on Honey Bees (Apis mellifera L). PLoS ONE 2013, 8, e76536. [Google Scholar] [CrossRef] [PubMed]
- Cutler, G.C.; Purdy, J.; Giesy, J.P.; Solomon, K.R. Risk to pollinators from the use of chlorpyrifos in the United States. Ecol. Risk Assess. Chlorpyrifos Terr. Aquat. Syst. USA 2014, 231, 219–265. [Google Scholar]
- Li, Z.; Li, M.; Huang, J.; Ma, C.; Xiao, L.; Huang, Q.; Zhao, Y.; Nie, H.; Su, S. Effects of Sublethal Concentrations of Chlorpyrifos on Olfactory Learning and Memory Performances in Two Bee Species, Apis mellifera and Apis cerana. Sociobiology 2017, 64, 174. [Google Scholar] [CrossRef]
- Zhou, C.; Li, X. Cytotoxicity of chlorpyrifos to human liver hepatocellular carcinoma cells: Effects on mitochondrial membrane potential and intracellular free Ca2+. Toxin Rev. 2017, 37, 259–268. [Google Scholar] [CrossRef]
- Oostingh, G.J.; Wichmann, G.; Schmittner, M.; Lehmann, I.; Duschl, A. The cytotoxic effects of the organophosphates chlorpyrifos and Diazinon differ from their immunomodulating effects. J. Immunotoxicol. 2009, 6, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Salazar Mercado, S.A.; Maldonado Bayona, H.A. Evaluation of cytotoxic potential of chlorpyrifos using Lens culinaris med as efficient bioindicator. Ecotoxicol. Environ. Saf. 2019, 183, 109528. [Google Scholar] [CrossRef]
- Gultekin, F.; Patat, S.; Akca, H.; Akdogan, M.; Altuntas, I. Melatonin can suppress the cytotoxic effects of chlorpyrifos on human hepg2 cell lines. Hum. Exp. Toxicol. 2006, 25, 47–55. [Google Scholar] [CrossRef]
- Cho, K.M.; Math, R.K.; Islam, S.M.; Lim, W.J.; Hong, S.Y.; Kim, J.M.; Yun, M.G.; Cho, J.J.; Yun, H.D. Biodegradation of chlorpyrifos by lactic acid bacteria during kimchi fermentation. J. Agric. Food Chem. 2009, 57, 1882–1889. [Google Scholar] [CrossRef] [PubMed]
- Bagherpour Shamloo, H.; Golkari, S.; Faghfoori, Z.; Movassaghpour, A.A.; Lotfi, H.; Barzegari, A.; Yari Khosroushahi, A. Lactobacillus casei decreases organophosphorus pesticide DIAZINON cytotoxicity in human HUVEC cell line. Adv. Pharm. Bull. 2016, 6, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Cuesta-Maté, A.; Renelies-Hamilton, J.; Kryger, P.; Jensen, A.B.; Sinotte, V.M.; Poulsen, M. Resistance and vulnerability of honeybee (Apis mellifera) gut bacteria to commonly used pesticides. Front. Microbiol. 2021, 12, 2428. [Google Scholar] [CrossRef]
- Elzeini, H.M.; Ali, A.R.; Nasr, N.F.; Hassan, M.; Hassan, A.A.; Elenany, Y.E. Probiotic capability of novel lactic acid bacteria isolated from worker honey bees gut microbiota. FEMS Microbiol. Lett. 2021, 368, fnab030. [Google Scholar] [CrossRef] [PubMed]
- Ovung, A.; Bhattacharyya, J. Sulfonamide drugs: Structure, antibacterial property, toxicity, and biophysical interactions. Biophys. Rev. 2021, 13, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Erginkaya, Z.; Turhan, E.U.; Tatlı, D. Determination of antibiotic resistance of lactic acid bacteria isolated from traditional Turkish fermented dairy products. Iran J. Vet. Res. 2018, 19, 53–56. [Google Scholar] [CrossRef]
- Amiranashvili, L.L.; Gagelidze, N.A.; Varsimashvili, K.I.; Tinikashvili, L.M.; Tolordava, L.L.; Gamkrelidze, M.D.; Amashukeli, N.V.; Makaradze, L.A. Antimicrobial susceptibility and antibiotic resistance profiles of cultivable lactic acid bacteria from intestinal tract of domestic chickens collected in Adjara. Ann. Agrar. Sci. 2016, 14, 182–186. [Google Scholar] [CrossRef]
- Shazali, N.; Foo, H.L.; Loh, T.C.; Choe, D.W.; Abdul Rahim, R. Prevalence of antibiotic resistance in lactic acid bacteria isolated from the faeces of Broiler Chicken in Malaysia. Gut Pathog. 2014, 6, 1. [Google Scholar] [CrossRef]
- Somashekaraiah, R.; Shruthi, B.; Deepthi, B.V.; Sreenivasa, M.Y. Probiotic properties of lactic acid bacteria isolated from Neera: A naturally fermenting coconut palm nectar. Front. Microbiol. 2019, 10, 1382. [Google Scholar] [CrossRef]
- Tian, B.; Fadhil, N.H.; Powell, J.E.; Kwong, W.K.; Moran, N.A. Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of Honeybees. mBio 2012, 3, e00377-12. [Google Scholar] [CrossRef]
- Raymann, K.; Shaffer, Z.; Moran, N.A. Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol. 2017, 15, e2001861. [Google Scholar] [CrossRef]
- Aljedani, D.M. Antibiotic treatment (tetracycline) effect on bio-efficiency of the larvae honey bee (Apis mellifera jemenatica). Saudi J. Biol. Sci. 2022, 29, 1477–1486. [Google Scholar] [CrossRef]
- Lindgren, S.E.; Dobrogosz, W.J. Antagonistic activities of lactic acid bacteria in food and feed fermentations. FEMS Microbiol. Lett. 1990, 87, 149–164. [Google Scholar] [CrossRef] [PubMed]
- Olofsson, T.C.; Butler, È.; Markowicz, P.; Lindholm, C.; Larsson, L.; Vásquez, A. Lactic acid bacterial symbionts in honeybees—An unknown key to Honey’s antimicrobial and therapeutic activities. Int. Wound J. 2014, 13, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Wilks, M.; Wiggins, R.; Whiley, A.; Hennessy, E.; Warwick, S.; Porter, H.; Corfield, A.; Millar, M. Identification and H2O2 production of vaginal lactobacilli from pregnant women at high risk of preterm birth and relation with outcome. J. Clin. Microbiol. 2004, 42, 713–717. [Google Scholar] [CrossRef] [PubMed]
- Mosbah, A.; Mesbah, M.R. A study of the role of hydrogen peroxide production by lactobacilli in preterm labor. Int. J. Med. Sci. 2009, 1, 388–395. [Google Scholar] [CrossRef]
- Rabe, L.K.; Hillier, S.L. Optimization of media for detection of hydrogen peroxide production by Lactobacillus species. J. Clin. Microbiol. 2003, 41, 3260–3264. [Google Scholar] [CrossRef]
- Nizet, V. Streptococcal β-hemolysins: Genetics and role in disease pathogenesis. Trends Microbiol. 2002, 10, 575–580. [Google Scholar] [CrossRef]
- Zhang, H.; Zheng, Y.; Gao, H.; Xu, P.; Wang, M.; Li, A.; Miao, M.; Xie, X.; Deng, Y.; Zhou, H.; et al. Identification and characterization of Staphylococcus aureus strains with an incomplete hemolytic phenotype. Front. Cell. Infect. Microbiol. 2016, 6, 146. [Google Scholar] [CrossRef]
- Halder, D.; Mandal, M.; Chatterjee, S.S.; Pal, N.K.; Mandal, S. Indigenous Probiotic Lactobacillus Isolates Presenting Antibiotic like Activity against Human Pathogenic Bacteria. Biomedicines 2017, 5, 31. [Google Scholar] [CrossRef]
- Li, M.; Wang, Y.; Cui, H.; Li, Y.; Sun, Y.; Qiu, H.-J. Characterization of lactic acid bacteria isolated from the gastrointestinal tract of a wild boar as potential probiotics. Front. Vet. Sci. 2020, 7, 49. [Google Scholar] [CrossRef] [PubMed]
- Benítez-Cabello, A.; Calero-Delgado, B.; Rodríguez-Gómez, F.; Garrido-Fernández, A.; Jiménez-Díaz, R.; Arroyo-López, F.N. Biodiversity and multifunctional features of lactic acid bacteria isolated from table olive biofilms. Front. Microbiol. 2019, 10, 836. [Google Scholar] [CrossRef] [PubMed]
- Corfield, A.P. The Interaction of the Gut Microbiota with the Mucus Barrier in Health and Disease in Human. Microorganisms 2018, 6, 78. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; van Passel, M.W.J.; van de Bovenkamp, J.H.B.; Schipper, R.; de Vos, W.; Dekker, J. Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes 2010, 1, 254–268. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.S.; Gopal, P.K.; Gill, H.S. Potential probiotic lactic acid bacteria Lactobacillus rhamnosus (HN001), Lactobacillus acidophilus (HN017) and Bifidobacterium lactis (HN019) do not degrade gastric mucin in vitro. Int. J. Food Microbiol. 2001, 63, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Fernández, M.F.; Boris, S.; Barbés, C. Safety evaluation of Lactobacillus delbrueckii subsp. lactis UO 004, a probiotic bacterium. Res. Microbiol. 2005, 156, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Wickström, C.; Chávez de Paz, L.; Davies, J.R.; Svensäter, G. Surface-associated MUC5B mucins promote protease activity in Lactobacillus fermentum biofilms. BMC Oral Health 2013, 13, 43. [Google Scholar] [CrossRef]
- Zarepour, M.; Bhullar, K.; Montero, M.; Ma, C.; Huang, T.; Velcich, A.; Xia, L.; Vallance, B.A. The mucin muc2 limits pathogen burdens and epithelial barrier dysfunction during Salmonella enterica serovar typhimurium colitis. Infect. Immun. 2013, 81, 3672–3683. [Google Scholar] [CrossRef]
- Hernández-Gómez, J.G.; López-Bonilla, A.; Trejo-Tapia, G.; Ávila-Reyes, S.V.; Jiménez-Aparicio, A.R.; Hernández-Sánchez, H. In Vitro Bile Salt Hydrolase (BSH) Activity Screening of Different Probiotic Microorganisms. Foods 2021, 10, 674. [Google Scholar] [CrossRef]
- Tanaka, H.; Doesburg, K.; Iwasaki, T.; Mierau, I. Screening of lactic acid bacteria for bile salt hydrolase activity. J. Dairy Sci. 1999, 82, 2530–2535. [Google Scholar] [CrossRef]
- Shehata, M.G.; El Sohaimy, S.A.; El-Sahn, M.A.; Youssef, M.M. Screening of isolated potential probiotic lactic acid bacteria for cholesterol lowering property and bile salt hydrolase activity. Ann. Agric. Sci. 2016, 61, 65–75. [Google Scholar] [CrossRef]
- Begley, M.; Hill, C.; Gahan, C.G. Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 2006, 72, 1729–1738. [Google Scholar] [CrossRef] [PubMed]







| Antibiotic (Concentration) | Antibiotic Susceptibility Profile of LAB * | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A. kunkeei DSM 12361 12361 | P. acidilactici 4/1 | P. pentosaceus 5/2 | P. pentosaceus 7/1 | P. acidilactici 11/3 | P. pentosaceus 14/1 | P. acidilactici 18/1 | P. acidilactici 21/1 | P. pentosaceus 25/1 | P. pentosaceus OK-S | L. casei 12AN | |
| Ampicillin (10 µg) | S | S | MS | S | S | MS | S | S | S | S | S |
| Chloramphenicol (30 µg) | S | S | S | S | S | S | S | S | S | S | S |
| Chloramphenicol (50 µg) | S | S | S | S | S | S | S | S | S | S | S |
| Enrofloxacin (5 µg) | R | R | R | R | R | R | R | R | R | R | R |
| Erythromycin (30 µg) | S | S | S | S | S | S | S | S | S | S | S |
| Gentamicin (30 µg) | R | R | R | R | R | R | R | R | R | R | R |
| Kanamycin (30 µg) | R | R | R | R | R | R | R | R | R | R | R |
| Lincomycin (2 µg) | S | R | MS | MS | MS | MS | MS | MS | MS | MS | S |
| Lincomycin (15 µg) | S | R | S | S | S | MS | MS | S | S | MS | S |
| Oxytetracycline (30 µg) | MS | S | S | S | S | S | S | S | S | S | MS |
| Streptomycin (25 µg) | R | R | R | R | R | R | R | R | R | R | R |
| Sulfonamides (300 µg) | R | R | R | R | R | R | R | R | R | R | R |
| Tetracycline (30 µg) | MS | MS | MS | S | S | S | S | S | MS | S | MS |
| Tylosin (30 µg) | R | R | R | R | R | R | R | R | R | R | R |
| Vancomycin (30 µg) | R | R | R | R | R | R | R | R | R | R | R |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leska, A.; Nowak, A.; Rosicka-Kaczmarek, J.; Ryngajłło, M.; Czarnecka-Chrebelska, K.H. Characterization and Protective Properties of Lactic Acid Bacteria Intended to Be Used in Probiotic Preparation for Honeybees (Apis mellifera L.)—An In Vitro Study. Animals 2023, 13, 1059. https://doi.org/10.3390/ani13061059
Leska A, Nowak A, Rosicka-Kaczmarek J, Ryngajłło M, Czarnecka-Chrebelska KH. Characterization and Protective Properties of Lactic Acid Bacteria Intended to Be Used in Probiotic Preparation for Honeybees (Apis mellifera L.)—An In Vitro Study. Animals. 2023; 13(6):1059. https://doi.org/10.3390/ani13061059
Chicago/Turabian StyleLeska, Aleksandra, Adriana Nowak, Justyna Rosicka-Kaczmarek, Małgorzata Ryngajłło, and Karolina Henryka Czarnecka-Chrebelska. 2023. "Characterization and Protective Properties of Lactic Acid Bacteria Intended to Be Used in Probiotic Preparation for Honeybees (Apis mellifera L.)—An In Vitro Study" Animals 13, no. 6: 1059. https://doi.org/10.3390/ani13061059