Chemotaxonomic Classification Applied to the Identification of Two Closely-Related Citrus TCMs Using UPLC-Q-TOF-MS-Based Metabolomics
Abstract
:1. Introduction
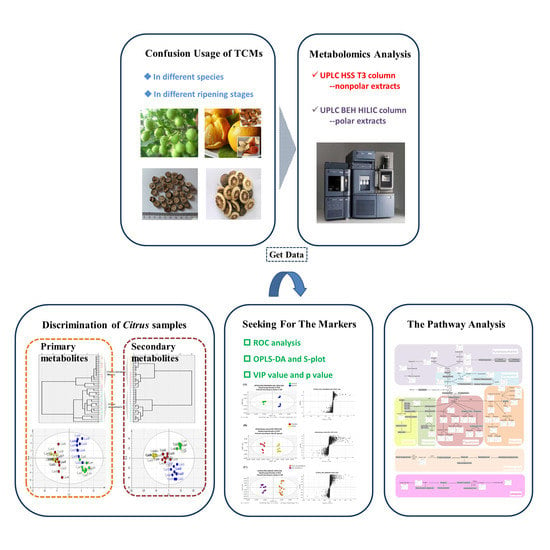
2. Results and Discussion
2.1. Optimization and Validation of the Method
2.1.1. Optimization of Chromatographic Conditions
2.1.2. Validation of the Analytical Method
2.2. Identification of Primary Metabolites
PCA Analysis of Primary Metabolites
2.3. Identification of Secondary Metabolites
Non-Targeted Analysis of Secondary Metabolite Features
2.4. Variable Selection and Annotation of Metabolites
2.4.1. Chemotaxonomy of Citrus by Primary and Secondary Metabolites
2.4.2. Potential Chemotaxonomic Markers Associated with Primary Metabolites
2.4.3. Potential Chemotaxonomic Markers Associated with Secondary Metabolites
2.5. The Pathway Analysis Associated with Chemotaxonomic Markers
2.5.1. The PB Test of the Analysis Factor
2.5.2. Analyzing a Wide Range of Secondary Metabolites to Understand the Metabolic Network
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Sample Preparation
3.2.1. Reference Standard Solution
3.2.2. Extraction of Plant Material
3.3. Chromatographic Conditions
3.4. Mass Spectrometry Condition
3.5. Data Processing and Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zhou, J.; Qi, L.; Li, P. Herbal medicine analysis by liquid chromatography/time-of-flight mass spectrometry. J. Chromgr. A 2009, 1216, 7582–7594. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Bi, K.; Han, F.; Guan, J.; Zhang, X.; Mao, X.; Zhao, L.; Li, Q.; Hou, X.; Yin, R. Identification of the absorbed components and metabolites of Zhi-Zi-Da-Huang decoction in rat plasma by ultra-high performance liquid chromatography coupled with quadrupole-time-of-flight mass spectrometry. J. Pharm. Biomed. 2015, 111, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Heidmarsson, S.; Olafsdottir, E.S.; Buonfiglio, R.; Kogej, T.; Omarsdottir, S. Secondary metabolites from cetrarioid lichens: Chemotaxonomy, biological activities and pharmaceutical potential. Phytomedicine 2016, 23, 441–459. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Li, X.; Wang, R.; Yang, Q.; Li, P.; Hu, H. Quality Traceability System of Traditional Chinese Medicine Based on Two Dimensional Barcode Using Mobile Intelligent Technology. PLoS ONE 2016, 11, e165263. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Qi, L.; Liu, E.; Zhou, J.; Wen, X. Analysis of Chinese herbal medicines with holistic approaches and integrated evaluation models. Trends Anal. Chem. 2008, 27, 66–77. [Google Scholar] [CrossRef]
- Jing, L.; Lei, Z.; Zhang, G.; Pilon, A.C.; Huhman, D.V.; Xie, R.; Xi, W.; Zhou, Z.; Sumner, L.W. Metabolite profiles of essential oils in citrus peels and their taxonomic implications. Metabolomics 2015, 11, 952–963. [Google Scholar] [CrossRef]
- Bijttebier, S.; Van der Auwera, A.; Foubert, K.; Voorspoels, S.; Pieters, L.; Apers, S. Bridging the gap between comprehensive extraction protocols in plant metabolomics studies and method validation. Anal. Chim. Acta 2016, 935, 136–150. [Google Scholar] [CrossRef] [PubMed]
- Braunberger, C.; Zehl, M.; Conrad, J.; Wawrosch, C.; Strohbach, J.; Beifuss, U.; Krenn, L. Flavonoids as chemotaxonomic markers in the genus Drosera. Phytochemistry 2015, 118, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.M.; Lee, J.; Park, S.; Lee, C.; Lee, J.W.; Lee, D.; Kim, N.; Lee, D.; Kim, H.Y.; Lee, C.H. LC–MS-based chemotaxonomic classification of wild-type Lespedeza sp. and its correlation with genotype. Plant Cell Rep. 2012, 31, 2085–2097. [Google Scholar] [CrossRef] [PubMed]
- Sandasi, M.; Kamatou, G.P.P.; Viljoen, A.M. An untargeted metabolomic approach in the chemotaxonomic assessment of two Salvia species as a potential source of α-bisabolol. Phytochemistry 2012, 84, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, Y.; Liu, C.; Song, Z.; Li, Q.; Zha, Q.; Lu, C.; Wang, C.; Ning, Z.; Zhang, Y.; et al. The chemotaxonomic classification of Rhodiola plants and its correlation with morphological characteristics and genetic taxonomy. Chem. Cent. J. 2013, 7, 118. [Google Scholar] [CrossRef] [PubMed]
- Ning, Z.; Lu, C.; Zhang, Y.; Zhao, S.; Liu, B.; Xu, X.; Liu, Y. Application of plant metabonomics in quality assessment for large-scale production of traditional Chinese medicine. Planta Med. 2013, 79, 897–908. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Gao, W.; Gao, Y.; Man, S.; Huang, L.; Liu, C. Identification of chemical constituents in extracts and rat plasma from Fructus Aurantii by UPLC-PDA-Q-TOF/MS. Phytochem. Anal. 2011, 22, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Zhao, S.; Ning, Z.; Zeng, H.; Shu, Y.; Tao, O.; Xiao, C.; Lu, C.; Liu, Y. Citrus fruits as a treasure trove of active natural metabolites that potentially provide benefits for human health. Chem. Cent. J. 2015, 9, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Chinese Pharmacopoeia Commission (CPC). Pharmacopoeia of the People’s Republic of China; Chinese Medical Science and Technology Press: Beijing, China, 2015; Volume 1, pp. 191, 197, 246–247. [Google Scholar]
- Chen, H.; Zhang, W.; Yuan, J.; Li, Y.; Yang, S.; Yang, W. Simultaneous quantification of polymethoxylated flavones and coumarins in Fructus aurantii and Fructus aurantii immaturus using HPLC-ESI-MS/MS. J. Pharm. Biomed. 2012, 59, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Pan, Y.; Fan, G.; Chai, Y.; Wu, Y. Application of an efficient strategy based on MAE, HPLC-DAD-MS/MS and HSCCC for the rapid extraction, identification, separation and purification of flavonoids from Fructus Aurantii Immaturus. Biomed. Chromatogr. 2010, 24, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Barreca, D.; Bellocco, E.; Caristi, C.; Leuzzi, U.; Gattuso, G. Distribution of C- and O-glycosyl flavonoids, (3-hydroxy-3-methylglutaryl)glycosyl flavanones and furocoumarins in Citrus aurantium L. juice. Food Chem. 2011, 124, 576–582. [Google Scholar] [CrossRef]
- Zeng, H.; Liu, Z.; Zhao, S.; Shu, Y.; Song, Z.; Wang, C.; Dong, Y.; Ning, Z.; He, D.; Wang, M.; et al. Preparation and quantification of the total phenolic products in Citrus fruit using solid-phase extraction coupled with high-performance liquid chromatography with diode array and UV detection. J. Sep. Sci. 2016, 39, 3806–3817. [Google Scholar] [CrossRef] [PubMed]
- Fernie, A.R.; Schauer, N. Metabolomics-assisted breeding: A viable option for crop improvement? Trends Genet. 2009, 25, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Li, K.; Alseekh, S.; Omranian, N.; Zhao, L.; Zhou, Y.; Xiao, Y.; Jin, M.; Yang, N.; Liu, H.; et al. Genetic Determinants of the Network of Primary Metabolism and Their Relationships to Plant Performance in a Maize Recombinant Inbred Line Population. Plant Cell 2015, 27, 1839–1856. [Google Scholar] [CrossRef] [PubMed]
- Lisec, J.; Meyer, R.C.; Steinfath, M.; Redestig, H.; Becher, M.; Witucka-Wall, H.; Fiehn, O.; Torjek, O.; Selbig, J.; Altmann, T.; et al. Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. Plant J. 2008, 53, 960–972. [Google Scholar] [CrossRef] [PubMed]
- Fahy, E.; Sud, M.; Cotter, D.; Subramaniam, S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007, 35, W606–W612. [Google Scholar] [CrossRef] [PubMed]
- Hurlock, A.K.; Roston, R.L.; Wang, K.; Benning, C. Lipid Trafficking in Plant Cells. Traffic 2014, 15, 915–932. [Google Scholar] [CrossRef] [PubMed]
- Michaud, M.; Prinz, W.A.; Jouhet, J. Glycerolipid synthesis and lipid trafficking in plant mitochondria. FEBS J. 2016, 284, 376–390. [Google Scholar] [CrossRef] [PubMed]
- Schmid, K.M. Lipid Metabolism in Plants. In Biochemistry of Lipids, Lipoproteins and Membranes, 6th ed.; Elsevier: Amsterdam, The Netherlands, 2015; Chapter 4; pp. 113–147. [Google Scholar]
- Kennedy, E.P. Biosynthesis of complex lipids. Fed. Proc. 1961, 20, 934–940. [Google Scholar] [PubMed]
- Guo, S.; Duan, J.; Qian, D.; Tang, Y.; Qian, Y.; Wu, D.; Su, S.; Shang, E. Rapid Determination of Amino Acids in Fruits of Ziziphus jujubaby Hydrophilic Interaction Ultra-High-Performance Liquid Chromatography Coupled with Triple-Quadrupole Mass Spectrometry. J. Agric. Food Chem. 2013, 61, 2709–2719. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Duan, J.; Qian, D.; Wang, H.; Tang, Y.; Qian, Y.; Wu, D.; Su, S.; Shang, E. Hydrophilic interaction ultra-high performance liquid chromatography coupled with triple quadrupole mass spectrometry for determination of nucleotides, nucleosides and nucleobases in Ziziphus plants. J. Chromatogr. A 2013, 1301, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Durand-Hulak, M.; Dugrand, A.; Duval, T.; Bidel, L.P.R.; Jay-Allemand, C.; Froelicher, Y.; Bourgaud, F.; Fanciullino, A.L. Mapping the genetic and tissular diversity of 64 phenolic compounds in Citrus species using a UPLC-MS approach. Ann. Bot-Lond. 2015, 115, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Agati, G.; Brunetti, C.; Di Ferdinando, M.; Ferrini, F.; Pollastri, S.; Tattini, M. Functional roles of flavonoids in photoprotection: New evidence, lessons from the past. Plant Physiol. Biochem. 2013, 72, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Cheynier, V. Polyphenols in foods are more complex than often thought. Am. J. Clin. Nutr. 2005, 81, 223S–229S. [Google Scholar] [PubMed]
- Da Costa, D.; E Silva, C.; Pinheiro, A.; Frommenwiler, D.; Arruda, M.; Guilhon, G.; Alves, C.; Arruda, A.; Da Silva, M. Using LC and Hierarchical Cluster Analysis as Tools to Distinguish Timbó Collections into Two Deguelia Species: A Contribution to Chemotaxonomy. Molecules 2016, 21, 569. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Li, Q.; Zhou, C.; Yu, M.; Yang, Y.; Zhang, H.; Ding, G.; Shang, H.; Zou, Z. Chronic unpredictive mild stress leads to altered hepatic metabolic profile and gene expression. Sci. Rep. 2016, 6, 23441. [Google Scholar] [CrossRef] [PubMed]
- Sarangowa, O.; Kanazawa, T.; Nishizawa, M.; Myoda, T.; Bai, C.; Yamagishi, T. Flavonol glycosides in the petal of Rosa species as chemotaxonomic markers. Phytochemistry 2014, 107, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Alcalde-Eon, C.; García-Estévez, I.; Rivas-Gonzalo, J.C.; De La Cruz, D.R.; Escribano-Bailón, M.T. Anthocyanins of the anthers as chemotaxonomic markers in the genus Populus L. Differentiation between Populus nigra, Populus alba and Populus tremula. Phytochemistry 2016, 128, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef] [PubMed]
- Plackett, P.L.; Burman, J.P. The design of optimum multifactorial experiments. Biometrike 1946, 33, 305–325. [Google Scholar] [CrossRef]
- Frydman, A.; Liberman, R.; Huhman, D.V.; Carmeli-Weissberg, M.; Sapir-Mir, M.; Ophir, R.; W Sumner, L.; Eyal, Y. The molecular and enzymatic basis of bitter/non-bitter flavor of citrus fruit: Evolution of branch-forming rhamnosyltransferases under domestication. Plant J. 2013, 73, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Qi, D.; Yang, T.; Lv, H.; Guo, L.; Zhang, Y.; Zhu, Y.; Peng, Q.; Xie, D.; Tan, J.; et al. Nontargeted Analysis Using Ultraperformance Liquid Chromatography–Quadrupole Time-of-Flight Mass Spectrometry Uncovers the Effects of Harvest Season on the Metabolites and Taste Quality of Tea (Camellia sinensis L.). J. Agric. Food Chem. 2015, 63, 9869–9878. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Shi, Y.; Ma, L.; Yi, X.; Ruan, J. Metabolomic analysis using ultra-performance liquid chromatography-quadrupole-time of flight mass spectrometry (UPLC-Q-TOF MS) uncovers the effects of light intensity and temperature under shading treatments on the metabolites in tea. PLoS ONE 2014, 9, e112572. [Google Scholar] [CrossRef] [PubMed]
- Metternich, J.B.; Gilmour, R. One Photocatalyst, Activation Modes Strategy for Cascade Catalysis: Emulating Coumarin Biosynthesis with (−)-Riboflavin. J. Am. Chem. Soc. 2016, 138, 1040–1045. [Google Scholar] [CrossRef] [PubMed]
- Bourgaud, F.; Hehn, A.; Larbat, R.; Doerper, S.; Gontier, E.; Kellner, S.; Matern, U. Biosynthesis of coumarins in plants: A major pathway still to be unravelled for cytochrome P450 enzymes. Phytochem. Rev. 2006, 5, 293–308. [Google Scholar] [CrossRef]
- Arbona, V.; Iglesias, D.J.; Gómez-Cadenas, A. Non-targeted metabolite profiling of citrus juices as a tool for variety discrimination and metabolite flow analysis. BMC Plant Biol. 2015, 15, 38. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; Liu, Z.; Zhao, S.; Song, Z.; He, D.; Wang, M.; Zeng, H.; Lu, C.; Lu, A.; Liu, Y. Integrated and global pseudotargeted metabolomics strategy applied to screening for quality control markers of Citrus TCMs. Anal. Bioanal. Chem. 2017, 409, 4849–4865. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |







| No. | Retention Time (RT) | Formula | [M + H]+ m/z | [M − H]− m/z | Identification | p Value | AUC | Metabolic Pathways | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Detected | Mass Error | Detected | Mass Error | |||||||
| M1 | 4.38 | C10H13NO2 | 180.1024 | 1.9 | - | - | 3,5-Dimethylphenyl methylcarbamate | 8.77 × 10−9 | 1.0000 | Not known |
| M2 | 6.52 | C10H19O8P | 299.0892 | 0.7 | - | - | GPA(5:0/2:0)/GPA(2:0/5:0) | 0.0010 | 1.0000 | Glycerophospholipid metabolism |
| M3 | 2.91 | C11H12N2 | 173.1079 | −0.8 | 171.1077 | −1.6 | 1,2,3,4-Tetrahydro-beta-carboline | 2.63 × 10−6 | 1.0000 | Not known |
| M4 | 7.06 | C14H27O8P | 355.1523 | −0.4 | - | - | GPA(9:0/2:0)/GPA(2:0/9:0) | 1.93 × 10−9 | 1.0000 | Glycerophospholipid metabolism |
| M5 | 8.73 | C15H28O8P | 369.1681 | −0.9 | - | - | GPA(10:0/2:0)/GPA(2:0/10:0) | 3.99 × 10−7 | 1.0000 | Glycerophospholipid metabolism |
| M6 | 4.75 | C18H35O8P | 411.2168 | 4.9 | - | - | GPA(10:0/5:0)/GPA(5:0/10:0) | 1.37 × 10−8 | 1.0000 | Glycerophospholipid metabolism |
| M7 | 9.71 | C18H36O4 | 339.2514 | 2.6 | - | - | MG(15:0/0:0/0:0) | 3.95 × 10−5 | 1.0000 | Not known |
| M8 | 9.32 | C19H35O8P | 423.2139 | −0.7 | - | - | GPA(14:1/2:0)/GPA(2:0/14:1) | 1.00 × 10−20 | 1.0000 | Glycerophospholipid metabolism |
| M9 | 4.32 | C19H36O4 | 369.2695 | 2.5 | - | - | MG(16:1/0:0/0:0) | 3.45 × 10−4 | 1.0000 | Not known |
| M10 | 6.53 | C19H36NO7P | 423.2410 | −2.1 | - | - | PE(14:1/0:0) | 7.70 × 10−9 | 1.0000 | Phosphatidylethanolamine biosynthesis |
| M11 | 9.95 | C19H38O4 | 331.2850 | 2.0 | - | - | MG(16:0/0:0/0:0) | 1.94 × 10−9 | 1.0000 | Not known |
| M12 | 2.73 | C20H37O8P | 437.2301 | 0.5 | - | - | GPA(14:1/3:0)/GPA(3:0/14:1) | 1.94 × 10−4 | 1.0000 | Glycerophospholipid metabolism |
| M13 | 10.31 | C20H38O5 | 359.2620 | 2.3 | - | - | DG(8:0/9:0/0:0)/DG(9:0/8:0/0:0) | 5.11 × 10−5 | 1.0000 | Not known |
| M14 | 11.63 | C21H34O4 | 351.2538 | 2.9 | - | - | MG(18:4/0:0/0:0) | 0.0020 | 1.0000 | Not known |
| M15 | 10.03 | C21H44NO7P | 454.2783 | 0.3 | 452.2781 | −0.3 | PE(16:0/0:0) | 1.69 × 10−5 | 1.0000 | Phosphatidylethanolamine biosynthesis |
| M16 | 11.41 | C25H44O15 | 585.2551 | −3.6 | - | - | DGDG(2:0/8:0)/DGDG(8:0/2:0) | 0.0053 | 1.0000 | DGDG synthase |
| M17 | 12.05 | C26H46O15 | 599.2709 | −3.3 | - | - | DGDG(2:0/9:0)/DGDG(9:0/2:0) | 4.64 × 10−6 | 1.0000 | DGDG synthase |
| M18 | 10.99 | C26H49O8P | 521.3257 | 0.72 | - | - | GPA(14:1/9:0)/GPA(9:0/14:1) | 3.80 × 10−7 | 1.0000 | Glycerophospholipid metabolism |
| M19 | 11.78 | C8H16O4 | 177.1126 | 2.5 | - | - | MG(5:0/0:0/0:0) | 4.90 × 10−10 | 1.0000 | Not known |
| M20 | 2.61 | C8H18OS2 | 195.0694 | 1.4 | 193.0690 | 0.4 | 3-[(2-Mercapto-1-methylpropyl)thio]-2-butanol | 2.78 × 10−9 | 1.0000 | Not known |
| M21 | 5.36 | C8H6O2 | 135.0446 | 3.9 | 133.0438 | −1.8 | Phthalide | 0.022 | 1.0000 | Not known |
| M22 | 0.87 | C8H9NO2 | 152.0708 | 1.0 | 150.0707 | 0.4 | 2-Phenylglycine | 3.94 × 10−13 | 1.0000 | Not known |
| M23 | 8.54 | C26H52O8P | - | - | 524.6784 | 0.23 | GPA(11:0/12:0)/GPA(12:0/11:0) | 5.15 × 10−4 | 1.0000 | Glycerophospholipid metabolism |
| M24 | 8.57 | C27H51O13P | - | - | 613.3005 | 1.7 | GPIns(10:0/8:0)/GPIns(8:0/10:0) | 1.01 × 10−6 | 1.0000 | Not known |
| M25 | 8.57 | C19H37O8P | - | - | 424.4562 | −3.3 | GPA(10:0/6:0)/GPA(6:0/10:0) | 7.03 × 10−12 | 1.0000 | Glycerophospholipid metabolism |
| M26 | 8.44 | C13H25O8P | - | - | 339.1223 | 2.6 | GPA(2:0/8:0)/GPA(8:0/2:0) | 2.66 × 10−8 | 1.0000 | Glycerophospholipid metabolism |
| M27 | 6.48 | C14H25O13P | - | - | 431.0977 | 4.0 | GPIns(2:0/3:0)/GPIns(3:0/2:0) | 2.14 × 10−5 | 1.0000 | Glycerophospholipid metabolism |
| M28 | 10.78 | C25H46O10 | - | - | 503.2850 | −2.4 | MGDG(14:1/2:0)/MGDG(2:0/14:1) | 0.0010 | 1.0000 | monogalactosyldiacylglycerol synthase |
| M29 | 0.92 | C5H10N2O3 | 147.0618 | 0.0 | 145.0616 | −1.7 | Glutamine | 0.0039 | 1.0000 | Inosine monophosphate biosynthesis |
| M30 | 9.16 | C21H36O4 | 353.2692 | 2.1 | - | - | MG(18:3/0:0/0:0) | 1.13 × 10−13 | 0.9941 | Not known |
| M31 | 10.96 | C21H40O4 | 357.3008 | 2.4 | - | - | MG(18:1/0:0/0:0) | 0.012 | 0.9941 | Not known |
| M32 | 5.44 | C26H30O14 | 567.1717 | 1.0 | 565.1703 | −1.4 | Mulberroside F | 0.0040 | 0.9941 | Not known |
| M33 | 4.12 | C10H10O2 | 163.0758 | 2.2 | 161.0749 | −3.4 | 4,5-Dihydro-1-benzoxepin-3(2H)-one | 5.98 × 10−8 | 0.9912 | Not known |
| M34 | 0.80 | C9H13N2O9P | 321.0289 | 0.7 | 323.0291 | 1.3 | UMP | 9.79 × 10−15 | 0.9912 | Uridine monophosphate biosynthesis |
| M35 | 3.38 | C10H20NO8P | 314.1928 | 0.3 | 312.1916 | −2.4 | GPEtn(2:0/3:0)/GPEtn(3:0/2:0) | 1.63 × 10−8 | 0.9883 | Glycerophospholipid metabolism |
| M36 | 2.73 | C16H25N5O6 | 384.1865 | −3.3 | - | - | Dihydrozeatin-O-glucoside | 5.77 × 10−5 | 0.9854 | Not known |
| M37 | 3.44 | C10H19O10P | - | - | 330.1346 | 1.9 | GPGro(2:0/2:0) | 8.11 × 10−8 | 0.9854 | Glycerophospholipid metabolism |
| M38 | 3.26 | C11H22NO8P | - | - | 326.2290 | −2.2 | GPEtn(2:0/4:0)/GPEtn(4:0/2:0) | 2.28 × 10−9 | 0.9795 | Glycerophospholipid metabolism |
| M39 | 2.92 | C8H18NO7P | - | - | 270.1774 | −1.2 | PE(3:0/0:0) | 4.11 × 10−4 | 0.9795 | Phosphatidylethanolamine biosynthesis |
| M40 | 6.28 | C20H20O9 | 405.1185 | 1.3 | 405.1174 | −1.3 | cis-resveratrol 4′-O-glucuronide | 1.26 × 10−7 | 0.9737 | Not known |
| M41 | 0.87 | C7H14N2O3 | 175.0920 | −4.5 | 173.0929 | −1.4 | Theanine | 1.21 × 10−8 | 0.9737 | Theanine synthetase |
| M42 | 0.67 | C4H8N2O3 | 133.0461 | −1.3 | 131.0458 | −3.0 | Asparagine | 1.58 × 10−13 | 0.9503 | Asparagine synthase |
| M43 | 9.15 | C18H34O4 | 315.2357 | 1.5 | - | - | MG(15:1/0:0/0:0) | 0.016 | 0.9386 | Not known |
| M44 | 9.67 | C26H50NO7P | 520.3330 | 1.4 | - | - | PC(18:2/0:0) | 5.44 × 10−5 | 0.9211 | Phosphatidylcholine biosynthesis |
| M45 | 3.37 | C20H23NO9 | 422.1449 | 0.2 | - | - | 5-O-cis-Coumaroylnigrumin | 7.01 × 10−4 | 0.9152 | Not known |
| M46 | 10.50 | C26H52NO7P | 522.3487 | 1.1 | - | - | PC(18:1/0:0)/PE(P-16:0/5:0)/PE(P-18:0/3:0) | 2.20 × 10−5 | 0.9006 | Phosphatidylcholine biosynthesis |
| M47 | 2.86 | C7H10O5 | 175.0454 | −0.4 | 173.0456 | 0.4 | Shikimic acid | 1.73 × 10−5 | 0.8918 | TCA cycle |
| M48 | 0.81 | C4H7NO4 | 133.0300 | −1.4 | 133.0303 | 0.7 | Aspartic acid | 1.44 × 10−4 | 0.8801 | TCA cycle |
| No. | RT (min) | Formula | [M + H]+ m/z | [M − H]− m/z | Mw (Da) | Identification | VIP (p-Value) | AUC | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Detected | Mass Erorr | Detected | Mass Erorr | |||||||
| Flavones and flavones glycoside | ||||||||||
| 1 | 3.37 | C15H10O8 | 319.0446 | −0.6 | 317.0447 | −0.3 | 318.0448 | Myricetin | 1.22 (2.08 × 10−5) a, 1.17 (2.99 × 10−5) c | 0.9778 a |
| 2 | 3.83 | C21H20O10 | 433.0988 | 1.2 | 431.0978 | −1.1 | 432.0983 | Vitexin | 1.78 (1.46 × 10−4) c | 0.9181 c |
| 3 * | 3.86 | C22H22O12 | 479.1185 | 0.2 | 477.1175 | −1.9 | 478.1184 | Isorhamnetin-3-O-glucoside | 1.24 (6.37 × 10−4) a, 2.11 (0.019) c | 0.9000 a |
| 4 * | 3.93 | C22H22O11 | 463.1156 | −1.1 | 461.1183 | 4.7 | 462.1161 | Diosmetin-7-O-glucoside | 2.23(9.87 × 10−9) b | 1.0000 b |
| 5 | 4.09 | C27H30O14 | 579.1630 | −1.0 | 577.1652 | 2.8 | 578.1636 | Isorhoifolin | 1.48 (0.005) a, 1.40 (0.031) c | 0.9111 a |
| 6 | 4.13 | C21H20O11 | 449.1003 | −0.4 | 447.0989 | −3.6 | 448.1005 | Orientin | 2.68 (1.52 × 10−9) a, 1.25 (0.017) c | 1.0000 a |
| 7 | 4.18 | C26H28O14 | 565.1475 | −0.7 | 563.1483 | 0.7 | 564.1479 | Apiin | 13.68 (9.24 × 10−15) c | 1.0000 c |
| 8 * | 4.2 | C27H30O14 | 579.1626 | −1.7 | 577.1655 | 3.3 | 578.1636 | Rhoifolin | 9.35 (3.86 × 10−11) c | 1.0000 c |
| 9 | 4.28 | C28H32O15 | 609.1745 | 0.7 | 607.1720 | −3.5 | 608.1741 | Diosmin | 1.32 (0.013) a, 2.52 (0.0021) b, 4.39 (8.97 × 10−10) c | 0.9012 b, 0.9737 c |
| 10 | 4.37 | C28H32O15 | 609.1745 | 0.7 | 607.1720 | −3.5 | 608.1741 | Neodiosmin | 4.52 (1.80 × 10−5) b, 5.89 (3.21 × 10−10) c | 1.0000 b, 0.9971 c |
| 11 | 4.42 | C27H30O16 | 611.1526 | −1.3 | 609.1530 | −0.7 | 610.1534 | Luteolin-3′,7-di-O-glucoside | 7.28 (0.0033) b, 10.19 (7.85 × 10−12) c | 0.8642 b, 0.9883 c |
| 12 | 4.47 | C21H20O11 | 449.1009 | 0.9 | 447.0989 | −3.6 | 448.1005 | Cynaroside | 4.52 (0.0035) b, 8.87 (1.83 × 10−13) c | 0.9912 c |
| 13 | 4.47 | C28H32O14 | 593.1786 | −1.0 | 591.1785 | −1.2 | 592.1792 | Linarin | 1.20 (0.049) b, 2.0 (1.19 × 10−12) c | 0.9883 c |
| 14 | 5.21 | C16H12O6 | 301.0629 | −1.7 | 299.0621 | −4.3 | 300.0634 | Chrysoeriol | 7.02 (7.09 × 10−4) b, 12.07 (9.40 × 10−13) c | 0.9136 b, 1.0000 c |
| 15 | 5.21 | C30H18O10 | 539.0896 | −0.7 | 537.0911 | 2.0 | 538.0900 | Amentoflavone | 1.93 (6.36 × 10−5) b, 2.21 (2.30 × 10−7) c | 0.9630 b, 1.0000 c |
| 16 * | 5.51 | C21H20O10 | 433.1040 | −3.9 | 431.1037 | −4.6 | 432.1057 | Apigetrin | 1.52 (8.6 × 10−4) b, 4.66 (1.83 × 10−17) c | 1.0000 c |
| 17 | 5.51 | C21H20O11 | 449.1015 | 2.2 | 447.0989 | −3.6 | 448.1005 | Luteolin-4′-O-glucoside | 1.68 (0.0019) b, 3.96 (1.17 × 10-15) c | 0.8889 b, 1.0000 c |
| 18 | 5.51 | C27H30O15 | 595.1589 | 0.8 | 593.1577 | −1.2 | 594.1584 | Saponarin | 2.44 (0.0012) b, 5.54 (1.04 × 10−15) c | 0.9136 b, 1.0000 c |
| 19 * | 5.58 | C15H10O6 | 287.0476 | −0.3 | 285.0477 | 0.0 | 286.0477 | Luteolin | 1.39 (0.0016) a, 2.46 (0.035) b, 8.67 (3.48 × 10−17) c | 1.0000 c |
| 20 | 6.3 | C15H10O5 | 271.0526 | −0.7 | 269.0524 | −1.5 | 270.0528 | Apigenin | 3.43 (0.032) a, 2.27 (7.13 × 10−4) c | 0.9667 a, 0.8889 c |
| 21 * | 6.58 | C16H12O6 | 301.0637 | 1.0 | 299.0630 | −1.3 | 300.0634 | Diosmetin | 3.42 (6.16 × 10−4) b, 2.37 (0.0020) c | 0.8778 a, 0.9753 b, 0.8597 c |
| 22 * | 6.95 | C16H12O5 | 285.0678 | −2.5 | 283.0690 | 1.8 | 284.0685 | Acacetin | 1.24 (0.0030) b, 1.49 (3.95 × 10−7) c | 0.8667 a, 0.8642 b, 1.0000 c |
| 23 * | 7.75 | C20H20O7 | 373.1212 | 0.8 | 371.1208 | −0.3 | 372.1209 | Sinensetin | 8.98 (0.0083) a, 4.79 (0.016) b | - |
| 24 * | 8.35 | C21H22O8 | 403.1320 | 1.2 | - | - | 402.1315 | Nobiletin | 16.79 (0.025) a, 14.60 (0.0087) b | - |
| 25 | 8.38 | C19H18O6 | 343.1108 | 1.5 | - | - | 342.1103 | Tetramethyl-isoscutellarein | 9.15 (0.0059) a, 9.34 (0.035) b, 4.48 (0.046) c | - |
| 26 * | 8.88 | C20H20O7 | 373.1201 | −2.1 | - | - | 372.1209 | Tangeretin | 17.13 (0.015) a, 16.34 (0.0094) b | 0.8642 b |
| 27 * | 9.31 | C20H20O8 | 389.1162 | 1.0 | - | - | 388.1158 | 5-demethylnobiletin | 11.83 (0.0045) a, 8.46 (1.63 × 10−6) b, 3.89 (0.050) c | 0.8667 a, 0.9260 b |
| Flavanones and flavanone glycoside | ||||||||||
| 28 * | 3.71 | C27H32O15 | 597.1745 | 0.7 | 595.1745 | 0.7 | 596.1741 | Eriocitrin | 2.71 (2.70 × 10−8) c | 0.9357 c |
| 29 | 3.73 | C21H22O11 | 451.1156 | −1.3 | 449.1164 | 0.4 | 450.1162 | Pyracanthoside | 2.85 (9.22 × 10−16) c | 0.9971 c |
| 30 | 3.74 | C28H34O14 | 595.1940 | −1.5 | 593.1945 | −0.7 | 594.1949 | Neoponcirin | 2.74 (0.0012) a, 1.58 (0.0047) c | 0.8778 a |
| 31 | 3.82 | C27H32O15 | 597.1746 | 0.8 | 595.1735 | −1.0 | 596.1741 | Neoeriocitrin | 1.73 (0.050) b, 3.88 (5.98 × 10−11) c | 1.0000 c |
| 32 * | 3.84 | C28H34O14 | 595.1935 | −2.3 | 593.1955 | 1.0 | 594.1949 | Poncirin | 1.38 (3.97× 10−11) a, 1.24 (5.44 × 10−8) c | 1.0000 a, 0.9620 c |
| 33 * | 4.1 | C27H32O14 | 581.1786 | −1.0 | 579.1780 | −2.1 | 580.1792 | Narirutin | 2.06 (0.010) a, 1.07 (0.044) b, 5.38 (5.12 × 10−20) c | - |
| 34 * | 4.26 | C27H32O14 | 581.1802 | 1.7 | 579.1797 | 0.9 | 580.1792 | Naringin | 1.43 (0.013) a, 2.52 (0.032) b, 13.64 (5.97 × 10−19) c | 1.0000 c |
| 35 * | 4.34 | C28H34O15 | 611.1882 | −2.5 | 609.1896 | −0.2 | 610.1897 | Hesperidin | 1.39 (0.031) a, 2.00 (0.0092) b, 2.89 (2.81 × 10−5) c | 0.9912 c |
| 36 * | 4.49 | C28H34O15 | 611.1889 | −1.3 | 609.1905 | 1.3 | 610.1897 | Neohesperidin | 1.91 (0.041) a, 7.92 (0.0082) b, 16.60 (5.1 × 10−5) c | - |
| 37 | 4.69 | C16H14O6 | 303.0792 | 0.7 | 301.0785 | −1.7 | 302.0790 | Homoeriodictyol | 4.85 (5.43 × 10−4) b, 8.93 (3.55 × 10−12) c | 0.8778 a, 0.9506 b, 0.9912 c |
| 38 * | 4.82 | C15H12O6 | 289.0628 | −2.1 | 287.0628 | −2.1 | 288.0634 | Eriodictyol | 5.99 (8.49 × 10−14) c | 1.0000 c |
| 39 | 5.51 | C16H14O5 | 287.0835 | −2.1 | 285.0854 | 4.5 | 286.0841 | Sakuranetin | 1.46 (0.022) a, 2.47 (0.035) b, 9.26 (3.48 × 10−17) c | 1.0000 c |
| 40 * | 6.3 | C15H12O5 | 273.0673 | −4.4 | 271.0691 | 2.2 | 272.0685 | Naringenin | 4.64 (0.028) a, 8.95 (9.7 × 10−4) b, 9.37 (2.30 × 10−5) c | 0.9240 c |
| 41 * | 6.62 | C16H14O6 | 303.0795 | 1.7 | 301.0785 | −1.7 | 302.0790 | Hesperetin | 2.31 (0.0011) a, 8.57 (0.049) b, 7.07 (3.52 × 10−13) c | - |
| Flavonols | ||||||||||
| 42 * | 3.39 | C27H30O14 | 579.1630 | −1.0 | 577.1629 | −1.2 | 578.1636 | Kaempferitrin | 1.66 (2.16 × 10−7) a, 1.74 (0.025) b | 1.0000 a, 0.8518 b |
| 43 * | 3.67 | C27H30O16 | 611.1543 | 1.5 | 609.1541 | 1.1 | 610.1534 | Rutin | 2.59 (6.42 × 10−9) a, 2.08 (1.22 × 10−4) c | 1.0000 a, 0.8742 c |
| 44 * | 5.60 | C15H10O7 | 303.0425 | −0.7 | 301.0425 | −0.7 | 302.0427 | Quercetin | 2.56 (2.77 × 10−4) c | 0.9506 b, 0.8684 c |
| Cyanidin | ||||||||||
| 45 | 2.98 | C21H21ClO11 | 485.0782 | 2.1 | 483.0770 | −0.4 | 484.0772 | Cyanidin-3-O-glucoside | 2.42 (0.0061) a | 1.0000 a |
| Coumarins | ||||||||||
| 46 | 3.46 | C11H10O4 | 207.0577 | −1.0 | 205.0585 | 2.9 | 206.0579 | Limettin | 3.06 (1.10 × 10−8) a, 2.08 (6.4 × 10−4) c | 1.0000 a |
| 47 * | 4.35 | C9H6O3 | 163.0312 | −3.1 | 161.0310 | −4.3 | 162.0317 | Umbelliferone | 1.96 (7.53 × 10−5) b, 1.89 (2.86 × 10−5) c | 1.0000 b, 0.9270 c |
| 48 * | 5.34 | C11H10O4 | 207.0570 | −4.4 | - | - | 206.0579 | Scoparone | 1.09 (0.0077) b | - |
| 49 | 6.15 | C20H24O4 | 329.1681 | 2.1 | - | - | 328.1674 | 5-Geranyloxy-7-Methoxycoumarin | 2.17 (0.0089) a, 2.04 (0.0212) b | 0.9012 b |
| 50 | 7.85 | C19H22O4 | 315.1507 | −3.5 | - | - | 314.1518 | Epoxyaurapten | 1.16 (0.0075) a, 4.62 (1.73 × 10−7) b, 4.12 (6.73 × 10−8) c | 0.8667 a, 1.0000 b, 0.9737 c |
| 51 | 9.60 | C15H16O3 | 245.1102 | 1.2 | - | - | 244.1099 | Osthol | 3.6 (0.016) b, 4.12 (2.12 × 10−8) c | 1.0000 c |
| 52 | 10.67 | C19H22O3 | 299.1571 | 0.7 | - | - | 298.1569 | Aurapten | 4.76 (2.16 × 10−5) b, 5.17 (4.83 × 10−7) c | 0.9753 b |
| 53 * | 11.00 | C19H22O3 | 299.1565 | −1.3 | 297.1568 | −0.3 | 298.1569 | Auraptene | 10.50 (1.39 × 10−4) b, 12.57 (6.83 × 10−8) c | 0.8556 a, 0.9506 b, 1.0000 c |
| Furanocoumarins | ||||||||||
| 54 | 4.56 | C17H18O7 | 335.1053 | 0.3 | - | - | 334.1052 | Byakangelicin | 1.23 (3.19 × 10−4) b, 1.33 (0.0016) c | 0.8889 a, 1.0000 b |
| 55 | 5.31 | C16H16O6 | 305.0941 | −2.0 | - | - | 304.0947 | Heraclenol | 2.47 (2.79 × 10−6) b, 1.01 (0.0015) c | 1.0000 b, 0.9591 c |
| 56 | 5.51 | C16H14O5 | 287.0845 | 1.4 | - | - | 286.0841 | Heraclenin | 1.05 (0.022) a, 5.37 (1.41 × 10−17) c | - |
| 57 | 5.72 | C16H16O6 | 305.0940 | −2.3 | - | - | 304.0947 | Oxypeucedanin hydrate | 1.01 (0.0016) c | 0.9532 c |
| 58 | 6.75 | C11H6O3 | 187.0318 | 0.5 | - | - | 186.0317 | Psoralen | 1.27 (0.0078) b, 1.72 (5.99 × 10−9) c | 0.8642 b |
| 59 | 7.06 | C12H8O4 | 217.0420 | −0.9 | 215.0418 | −1.9 | 216.0422 | Xanthotoxin | 1.01 (2.09× 10−7) b,1.02 (3.07 × 10−7) c | 0.9971 c |
| 60 | 7.06 | C21H22O5 | 355.1472 | 1.4 | - | - | 354.1467 | Epoxybergamottin | 1.00 (0.0305) a, 6.77 (7.82 × 10−7) b, 8.57 (2.94 × 10−9) c | 0.8667 a, 1.0000 b, 1.0000 c |
| 61 * | 7.43 | C12H8O4 | 217.0421 | −0.5 | - | - | 216.0422 | Bergapten | 3.07 (0.019) b, 2.82 (0.0063) c | 0.9673 c |
| 62 | 7.48 | C13H10O5 | 247.0531 | 1.2 | - | - | 246.0528 | Isopimpinellin | 1.03 (2.10 × 10−4) b | 0.9591 c |
| 63 * | 7.89 | C11H6O4 | 203.0274 | 4.0 | 201.0270 | 2.0 | 202.0266 | Xanthotoxol | 6.44 (0.027) a, 2.73 (2.82× 10−7) b, 6.43 (1.6× 10−4) c | - |
| 64 | 7.89 | C17H16O5 | 301.1007 | 3.0 | - | - | 300.0998 | Cnidilin | 1.01 (6.16 × 10−4) b, 1.09 (0.0020) c | 0.8778 a, 0.9753 b, 0.8597 c |
| 65 | 8.22 | C21H24O6 | 373.1581 | 2.1 | - | - | 372.1573 | 6′-7′-dihydroxybergamottin | 1.11 (4.49 × 10−5) b, 1.02 (5.61 × 10−6) c | 0.8778 a, 0.9753 b, 0.8655 c |
| 66 | 8.24 | C16H14O5 | 287.0832 | −3.1 | - | - | 286.0841 | Oxypeucedanin | 1.46 (0.016) a, 2.04 (0.0083) b, 1.94 (0.0047) c | 0.8778 a |
| 67 | 8.45 | C17H16O6 | 317.0956 | 2.8 | - | - | 316.0947 | Byakangelicol | 1.05 (0.0063) b | 0.9383 b |
| 68 * | 9.27 | C16H14O4 | 271.0895 | 1.1 | - | - | 270.0892 | Imperatorin | 1.62 (0.0060) b, 1.52 (7.13 × 10−4) c | 0.8889 c |
| 69 | 9.54 | C17H16O5 | 301.0997 | −0.3 | - | - | 300.0998 | Phellopterin | 1.14 (7.09 × 10−4) b | 0.9136 b |
| 70 | 9.76 | C16H14O4 | 271.0892 | 0.0 | - | - | 270.0892 | Isoimperatorin | 1.84 (0.032) b, 1.67 (0.023) c | - |
| 71 | 10.37 | C21H22O5 | 355.1465 | −0.6 | 353.1460 | −2.0 | 354.1467 | Cnidicin | 1.39 (0.017) b, 2.61 (3.33E−7) c | 1.0000 c |
| 72 | 10.87 | C21H22O4 | 339.1513 | −1.5 | - | - | 338.1518 | 8-geranyloxypsoralen | 1.07 (0.0078) b | 0.8642 b, 0.9357 c |
| 73 | 11.26 | C11H6O4 | 203.0267 | 0.5 | 201.0271 | 2.5 | 202.0266 | Bergaptol | 3.70 (0.046) a, 3.14 (1.16 × 10−4) b, 3.70 (5.51 × 10−7) c | 0.9506 b |
| 74 * | 11.56 | C21H22O4 | 339.1522 | 1.2 | 337.1510 | 0.5 | 338.1518 | Bergamottin | 1.47 (6.11 × 10−5) b, 1.42 (2.14 × 10−5) c | 0.9630 b, 1.0000 c |
| Limonin | ||||||||||
| 75 | 3.93 | C32H42O14 | 651.2430 | −1.2 | 649.2441 | 0.5 | 650.2438 | Limonin glycoside | 4.45 (3.45 × 10−5) a, 3.01 (0.0055) b | - |
| 76 * | 7.82 | C26H30O8 | 471.2033 | 0.4 | 469.2035 | 0.9 | 470.2031 | Limonin | 3.01 (0.0023) a, 5.66 (1,8 × 10−5) b, 9.63 (3.04 × 10−5) c | 0.8510 c |
| 77 | 7.86 | C34H46O15 | 695.2499 | 0.6 | 693.2490 | −0.7 | 694.2495 | Nomilin glycoside | 2.10 (0.012) a, 1.42 (0.0010) b, 1.44 (0.0090) c | - |
| 78 * | 8.38 | C28H34O9 | 515.2447 | −0.2 | 513.2444 | −0.8 | 514.2448 | Nomilin | 6.40 (0.0011) b, 4.05 (0.0063) c | - |
| Abscisic acid | ||||||||||
| 79 * | 4.14 | C15H20O4 | 265.1380 | 2.3 | 263.1373 | −0.4 | 264.1374 | Abscisic acid | 1.62 (3.95 × 10−5)a, 5.74 (0.035) b, 7.54 (1.72 × 10−5) c | - |
| No. | Original Plant | Local Name | Location and Collection Time | Growing Environment |
|---|---|---|---|---|
| Pericarps of the ripe fruit of Citrus reticulate Blanco | ||||
| 1 | Citrus reticulate Blanco | Chenpi | Fengjie, Sichuan; August 2015 | Field margins (29° N 106° E; Alt.200–230 m) |
| 2 | Citrus reticulate Blanco | Chenpi | Qinglong, Guizhou; September 2015 | Hillsides (25° N 105° E; Alt.1200–1301 m) |
| 3 | Citrus reticulate Blanco | Chenpi | Jiangjin, Sichuan; September 2014 | Hillsides (29° N 106° E; Alt.231 m) |
| 4 | Citrus reticulate Blanco | Chenpi | Jiangjin, Sichuan; October 2014 | Field margins (29° N 106° E; Alt.200–230 m) |
| 5 | Citrus reticulate Blanco | Chenpi | Ganzhou, Jiangxi; September 2015 | Plain (26° N 115° E; Alt.500 m) |
| 6 | Citrus reticulate Blanco | Chenpi | Ganzhou, Jiangxi; September 2015 | Plain (25° N 115° E; Alt.520 m) |
| 7 | Citrus reticulate Blanco | Chenpi | Qinglong, Guizhou; October 2015 | Hillsides (25° N 105° E; Alt.1200–1300 m) |
| 8 | Citrus reticulate Blanco | Chenpi | Huangyan, Zhejiang; September 2013 | Field margins (28° N 121° E; Alt.45m) |
| 9 | Citrus reticulate Blanco | Chenpi | Jiangjin, Sichuan; August 2015 | Hillsides (29° N 106° E; Alt.238 m) |
| Pericarps of the young or immature fruit of Citrus reticulate Blanco | ||||
| 10 | Citrus reticulate Blanco | Qingpi | Xingan, Jiangxi; May 2015 | Hillsides (27° N 115° E; Alt.50–60 m) |
| 11 | Citrus reticulate Blanco | Qingpi | Jiangjin, Sichuan; May 2015 | Hillsides (29° N 106° E; Alt.220 m) |
| 12 | Citrus reticulate Blanco | Qingpi | Xingan, Jiangxi; June 2015 | Hillsides (27° N 115° E; Alt.50–60 m) |
| 13 | Citrus reticulate Blanco | Qingpi | Xingan, Jiangxi; May 2014 | Hillsides (27° N 115° E; Alt.50–60 m) |
| 14 | Citrus reticulate Blanco | Qingpi | Jiangjin, Sichuan; May 2014 | Hillsides (29° N 106° E; Alt.220 m) |
| 15 | Citrus reticulate Blanco | Qingpi | Xingan, Jiangxi; June 2014 | Hillsides (27° N 115° E; Alt.50–60 m) |
| 16 | Citrus reticulate Blanco | Qingpi | Xingan, Jiangxi; May 2013 | Hillsides (27° N 115° E; Alt.50–60 m) |
| 17 | Citrus reticulate Blanco | Qingpi | Jiangjin, Sichuan; May 2013 | Hillsides (29° N 106° E; Alt.220 m) |
| 18 | Citrus reticulate Blanco | Qingpi | Xingan, Jiangxi; June 2013 | Hillsides (27° N 115° E; Alt.50–60 m) |
| 19 | Citrus reticulate Blanco | Qingpi | Ganzhou, Jiangxi; June 2015 | Hillsides (27° N 115° E; Alt.50–60 m) |
| Immature fruit of Citrus aurantium L. | ||||
| 20 | Citrus aurantium cv Xiucheng | Xiucheng | Xingan, Jiangxi; July 2014 | Plain (27° N 115° E; Alt.20–30 m) |
| 21 | Citrus aurantium L. | Sour orange | Ezhou, Hubei; July 2013 | Hillsides (21° N 110° E; Alt.300 m) |
| 22 | Citrus aurantium cv Xiucheng | Xiucheng | Xingan, Jiangxi; July 2015 | Plain (27° N 115° E; Alt.20–30 m) |
| 23 | Citrus aurantium L. | Jiangjin sour orange | Jiangjin, Sichuan; July 2014 | Hillsides (29° N 106° E; Alt.231 m) |
| 24 | Citrus aurantium cv Jizicheng | Jizhicheng | Zhangshu, Jiangxi; July 2015 | Plain (27° N 115° E; Alt.20–30 m) |
| 25 | Citrus aurantium L. | Sour orange | Jian, Jiangxi; July 2015 | Plain (27° N 115° E; Alt.20–30 m) |
| 26 | Citrus aurantium L. | Sour orange | Zhangshu, Jiangxi; July 2014 | Plain (27° N 115° E; Alt.20–30 m) |
| 27 | Citrus aurantium L. | Sour orange | Jian, Jiangxi; July 2014 | Plain (27°N115°E; Alt.20–30 m) |
| 28 | Citrus aurantium × P. trifoliata | Citrange | Yuanjiang, Hunan; July 2014 | Hillsides (29° N 112° E; Alt.310 m) |
| Young fruit of Citrus aurantium L. | ||||
| 29 | Citrus aurantium cv Xiucheng | Xiucheng | Xingan, Jiangxi; May 2014 | Plain (27° N 115° E; Alt.20–30 m) |
| 30 | Citrus aurantium L. | Sour orange | Jiangjin, Sichuan; June 2014 | Field margins (29° N 106° E; Alt.200–230 m) |
| 31 | Citrus aurantium cv Xiucheng | Xiucheng | Xingan, Jiangxi; May 2015 | Plain (27° N 115° E; Alt.20–30 m) |
| 32 | Citrus aurantium L. | Jiangjin sour orange | Jiangjin, Sichuan; June 2014 | Hillsides (29° N 106° E; Alt.231 m) |
| 33 | Citrus aurantium cv Daidai | Daidai | Jiangjin, Sichuan; May 2014 | Field margins (29° N 106° E; Alt.200–230 m) |
| 34 | Citrus aurantium cv Xiucheng | Xiucheng | Zhangshu, Jiangxi; June 2014 | Plain (27° N 115° E; Alt.20–30 m) |
| 35 | Citrus aurantium cv Morocco sour orange | Morocco sour orange | Huangyan, Zhejiang; May 2013 | Field margins (28° N 121° E; Alt.45 m) |
| 36 | Citrus aurantium L. | Xiucheng | Qingjiang, Jiangxi; June 2012 | Plain (27° N 114° E; Alt.100–200 m) |
| 37 | Citrus aurantium cv Xiucheng | Xiucheng | Xingan, Jiangxi; June 2015 | Plain (27° N 115° E; Alt.20–30 m) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, S.-Y.; Liu, Z.-L.; Shu, Y.-S.; Wang, M.-L.; He, D.; Song, Z.-Q.; Zeng, H.-L.; Ning, Z.-C.; Lu, C.; Lu, A.-P.; et al. Chemotaxonomic Classification Applied to the Identification of Two Closely-Related Citrus TCMs Using UPLC-Q-TOF-MS-Based Metabolomics. Molecules 2017, 22, 1721. https://doi.org/10.3390/molecules22101721
Zhao S-Y, Liu Z-L, Shu Y-S, Wang M-L, He D, Song Z-Q, Zeng H-L, Ning Z-C, Lu C, Lu A-P, et al. Chemotaxonomic Classification Applied to the Identification of Two Closely-Related Citrus TCMs Using UPLC-Q-TOF-MS-Based Metabolomics. Molecules. 2017; 22(10):1721. https://doi.org/10.3390/molecules22101721
Chicago/Turabian StyleZhao, Si-Yu, Zhen-Li Liu, Yi-Song Shu, Meng-Lei Wang, Dan He, Zhi-Qian Song, Hong-Lian Zeng, Zhang-Chi Ning, Cheng Lu, Ai-Ping Lu, and et al. 2017. "Chemotaxonomic Classification Applied to the Identification of Two Closely-Related Citrus TCMs Using UPLC-Q-TOF-MS-Based Metabolomics" Molecules 22, no. 10: 1721. https://doi.org/10.3390/molecules22101721