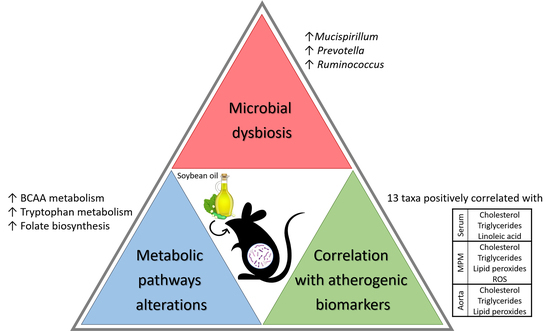
Soybean Oil Modulates the Gut Microbiota Associated with Atherogenic Biomarkers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mouse Model and Animal Feeding
2.2. Microbiota Composition Analysis
2.3. Functional Prediction of the Microbiota
2.4. Serum Analyses
2.5. Aortic Analyses
2.6. Mouse Peritoneal Macrophages (MPM) Isolation and Analyses
2.7. Statistical Analysis
3. Results
3.1. Microbial Composition and Diversity under SOE Supplementation
3.2. Key Phylotypes Shifts under SOE Supplementation
3.3. Microbial Alteration Associated with Atherosclerosis-Related Biomarkers
3.4. Microbial Metabolic Pathways Alterations
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Daniel, H.; Gholami, A.M.; Berry, D.; Desmarchelier, C.; Hahne, H.; Loh, G.; Mondot, S.; Lepage, P.; Rothballer, M.; Walker, A.; et al. High-fat diet alters gut microbiota physiology in mice. ISME J. 2014, 8, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.J.; Vangay, P.; Al-Ghalith, G.A.; Hillmann, B.M.; Ward, T.L.; Shields-Cutler, R.R.; Kim, A.D.; Shmagel, A.K.; Syed, A.N.; Walter, J.; et al. Daily Sampling Reveals Personalized Diet-Microbiome Associations in Humans. Cell Host Microbe 2019, 25, 789–802.e5. [Google Scholar] [CrossRef] [PubMed]
- Jie, Z.; Xia, H.; Zhong, S.-L.; Feng, Q.; Li, S.; Liang, S.; Zhong, H.; Liu, Z.; Gao, Y.; Zhao, H.; et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 2017, 8, 845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Prince, A.L.; Bader, D.; Hu, M.; Ganu, R.; Baquero, K.; Blundell, P.; Harris, R.A.; Frias, A.E.; Grove, K.L.; et al. High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nat. Commun. 2014, 5, 3889. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, M.; Pang, X.; Zhao, Y.; Wang, L.; Zhao, L. Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. ISME J. 2012, 6, 1848–1857. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhu, Y.; Zhao, F.; Song, S.; Li, Y.; Xu, X.; Zhou, G.; Li, C. Fish oil, lard and soybean oil differentially shape gut microbiota of middle-aged rats. Sci. Rep. 2017, 7, 826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turnbaugh, P.J.; Bäckhed, F.; Fulton, L.; Gordon, J.I. Diet-Induced Obesity Is Linked to Marked but Reversible Alterations in the Mouse Distal Gut Microbiome. Cell Host Microbe 2008, 3, 213–223. [Google Scholar] [CrossRef] [Green Version]
- Cani, P.D.; Neyrinck, A.M.; Fava, F.; Knauf, C.; Burcelin, R.G.; Tuohy, K.M.; Gibson, G.R.; Delzenne, N.M. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia 2007, 50, 2374–2383. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Xiao, G.; Yao, Y.; Guo, S.; Lu, K.; Sheng, Z. The Role of Bifidobacteria in Gut Barrier Function After Thermal Injury in Rats. J. Trauma Inj. Infect. Crit. Care 2006, 61, 650–657. [Google Scholar] [CrossRef]
- Murphy, E.F.; Cotter, P.D.; Healy, S.; Marques, T.M.; O’sullivan, O.; Fouhy, F.; Clarke, S.F.; O’toole, P.W.; Quigley, E.M.; Stanton, C.; et al. Composition and energy harvesting capacity of the gut microbiota: Relationship to diet, obesity and time in mouse models. Gut 2010, 59, 1635–1642. [Google Scholar] [CrossRef]
- Wall, R.; Ross, R.P.; Shanahan, F.; O’Mahony, L.; O’Mahony, C.; Coakley, M.; Hart, O.; Lawlor, P.; Quigley, E.M.; Kiely, B.; et al. Metabolic activity of the enteric microbiota influences the fatty acid composition of murine and porcine liver and adipose tissues. Am. J. Clin. Nutr. 2009, 89, 1393–1401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindskog Jonsson, A.; Caesar, R.; Akrami, R.; Reinhardt, C.; Fåk Hållenius, F.; Borén, J.; Bäckhed, F. Impact of Gut Microbiota and Diet on the Development of Atherosclerosis in Apoe-/- Mice. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 2318–2326. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.F.; Dwivedi, G.; O’Gara, F.; Caparros-Martin, J.; Ward, N.C. The gut microbiome and cardiovascular disease: Current knowledge and clinical potential. Am. J. Physiol. Heart Circ. Physiol. 2019, 317, H923–H938. [Google Scholar] [CrossRef] [PubMed]
- Koren, O.; Spor, A.; Felin, J.; Fåk, F.; Stombaugh, J.; Tremaroli, V.; Behre, C.J.; Knight, R.; Fagerberg, B.; Ley, R.E.; et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl. Acad. Sci. USA 2011, 108, 4592–4598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Tang, W.H.W.; Buffa, J.A.; Fu, X.; Britt, E.B.; Koeth, R.A.; Levison, B.S.; Fan, Y.; Wu, Y.; Hazen, S.L. Prognostic value of choline and betaine depends on intestinal microbiota-generated metabolite trimethylamine-N-oxide. Eur. Heart J. 2014, 35, 904–910. [Google Scholar] [CrossRef]
- Koeth, R.A.; Levison, B.S.; Culley, M.K.; Buffa, J.A.; Wang, Z.; Gregory, J.C.; Org, E.; Wu, Y.; Li, L.; Smith, J.D.; et al. γ-butyrobetaine is a proatherogenic intermediate in gut microbial metabolism of L-carnitine to TMAO. Cell Metab. 2014, 20, 799–812. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; DuGar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.-M.; et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011, 472, 57–63. [Google Scholar] [CrossRef] [Green Version]
- Koeth, R.; Wang, Z.; Levison, B.S.; Buffa, J.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [Green Version]
- Tang, W.H.; Wang, Z.; Levison, B.S.; Koeth, R.A.; Britt, E.B.; Fu, X.; Wu, Y.; Hazen, S.L. Intestinal Microbial Metabolism of Phosphatidylcholine and Cardiovascular Risk. N. Engl. J. Med. 2013, 368, 1575–1584. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.M.; Hazen, S.L. The Gut Microbial Endocrine Organ: Bacterially Derived Signals Driving Cardiometabolic Diseases. Ann. Rev. Med. 2015, 66, 343–359. [Google Scholar] [CrossRef] [Green Version]
- Macfarlane, G.T.; Macfarlane, S. Bacteria, Colonic Fermentation, and Gastrointestinal Health. J. AOAC Int. 2012, 95, 50–60. [Google Scholar] [CrossRef] [PubMed]
- den Besten, G.; Lange, K.; Havinga, R.; van Dijk, T.H.; Gerding, A.; van Eunen, K.; Müller, M.; Groen, A.K.; Hooiveld, G.J.; Bakker, B.M.; et al. Gut-derived short-chain fatty acids are vividly assimilated into host carbohydrates and lipids. Am. J. Physiol. Liver Physiol. 2013, 305, G900–G910. [Google Scholar] [CrossRef] [PubMed]
- Säemann, M.D.; Böhmig, G.A.; Osterreicher, C.H.; Burtscher, H.; Parolini, O.; Diakos, C.; Stöckl, J.; Hörl, W.H.; Zlabinger, G.J. Anti-inflammatory effects of sodium butyrate on human monocytes: Potent inhibition of IL-12 and up-regulation of IL-10 production. FASEB J. 2000, 14, 2380–2382. [Google Scholar] [CrossRef] [PubMed]
- Kimura, I.; Ozawa, K.; Inoue, D.; Imamura, T.; Kimura, K.; Maeda, T.; Terasawa, K.; Kashihara, D.; Hirano, K.; Tani, T.; et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat. Commun. 2013, 4, 1829. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.; Yin, J.; Zhang, J.; Ward, R.E.; Martin, R.J.; Lefevre, M.; Cefalu, W.T.; Ye, J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 2009, 58, 1509–1517. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, M.; Houten, S.M.; Mataki, C.; Christoffolete, M.A.; Kim, B.W.; Sato, H.; Messaddeq, N.; Harney, J.W.; Ezaki, O.; Kodama, T.; et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 2006, 439, 484–489. [Google Scholar] [CrossRef]
- Pols, T.W.H.; Nomura, M.; Harach, T.; Lo Sasso, G.; Oosterveer, M.H.; Thomas, C.; Rizzo, G.; Gioiello, A.; Adorini, L.; Pellicciari, R.; et al. TGR5 Activation Inhibits Atherosclerosis by Reducing Macrophage Inflammation and Lipid Loading. Cell Metab. 2011, 14, 747–757. [Google Scholar] [CrossRef] [Green Version]
- Thomas, C.; Gioiello, A.; Noriega, L.; Strehle, A.; Oury, J.; Rizzo, G.; Macchiarulo, A.; Yamamoto, H.; Mataki, C.; Pruzanski, M.; et al. TGR5-Mediated Bile Acid Sensing Controls Glucose Homeostasis. Cell Metab. 2009, 10, 167–177. [Google Scholar] [CrossRef] [Green Version]
- Ma, K.; Saha, P.K.; Chan, L.; Moore, D.D. Farnesoid X receptor is essential for normal glucose homeostasis. J. Clin. Investig. 2006, 116, 1102–1109. [Google Scholar] [CrossRef] [Green Version]
- Downes, M.; Verdecia, M.A.; Roecker, A.J.; Hughes, R.; Hogenesch, J.B.; Kast-Woelbern, H.R.; Bowman, M.E.; Ferrer, J.-L.; Anisfeld, A.M.; Edwards, P.A.; et al. A Chemical, Genetic, and Structural Analysis of the Nuclear Bile Acid Receptor FXR. Mol. Cell 2003, 11, 1079–1092. [Google Scholar] [CrossRef]
- Sayin, S.I.; Wahlström, A.; Felin, J.; Jäntti, S.; Marschall, H.-U.; Bamberg, K.; Angelin, B.; Hyötyläinen, T.; Orešič, M.; Bäckhed, F. Gut Microbiota Regulates Bile Acid Metabolism by Reducing the Levels of Tauro-beta-muricholic Acid, a Naturally Occurring FXR Antagonist. Cell Metab. 2013, 17, 225–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An Immunomodulatory Molecule of Symbiotic Bacteria Directs Maturation of the Host Immune System. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsiao, E.Y.; McBride, S.W.; Hsien, S.; Sharon, G.; Hyde, E.R.; McCue, T.; Codelli, J.A.; Chow, J.; Reisman, S.E.; Petrosino, J.F.; et al. Microbiota Modulate Behavioral and Physiological Abnormalities Associated with Neurodevelopmental Disorders. Cell 2013, 155, 1451–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asano, Y.; Hiramoto, T.; Nishino, R.; Aiba, Y.; Kimura, T.; Yoshihara, K.; Koga, Y.; Sudo, N. Critical role of gut microbiota in the production of biologically active, free catecholamines in the gut lumen of mice. Am. J. Physiol. Liver Physiol. 2012, 303, G1288–G1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blasbalg, T.L.; Hibbeln, J.R.; Ramsden, C.E.; Majchrzak, S.F.; Rawlings, R.R. Changes in consumption of omega-3 and omega-6 fatty acids in the United States during the 20th century. Am. J. Clin. Nutr. 2011, 93, 950–962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clemente, T.E.; Cahoon, E.B. Update on Soybean Oil Soybean Oil: Genetic Approaches for Modification of Functionality and Total Content. Plant Physiol. 2009, 151, 1030–1040. [Google Scholar] [CrossRef] [Green Version]
- Michas, G.; Micha, R.; Zampelas, A. Dietary fats and cardiovascular disease: Putting together the pieces of a complicated puzzle. Atherosclerosis 2014, 234, 320–328. [Google Scholar] [CrossRef]
- Harris, W.S.; Shearer, G.C. Omega-6 fatty acids and cardiovascular disease: Friend, not foe? Circulation 2014, 130, 1562–1564. [Google Scholar] [CrossRef] [Green Version]
- Farvid, M.S.; Ding, M.; Pan, A.; Sun, Q.; Chiuve, S.E.; Steffen, L.M.; Willett, W.C.; Hu, F.B. Dietary Linoleic Acid and Risk of Coronary Heart Disease: A Systematic Review and Meta-Analysis of Prospective Cohort Studies. Circulation 2017, 130, 1568–1578. [Google Scholar] [CrossRef]
- Ramsden, C.E.; Zamora, D.; Leelarthaepin, B.; Majchrzak-Hong, S.F.; Faurot, K.R.; Suchindran, C.M.; Ringel, A.; Davis, J.M.; Hibbeln, J.R. Use of dietary linoleic acid for secondary prevention of coronary heart disease and death: Evaluation of recovered data from the Sydney Diet Heart Study and updated meta-analysis. BMJ 2013, 346, e8707. [Google Scholar] [CrossRef] [Green Version]
- da Silva Afonso, M.; Castilho, G.; Lavrador, M.S.; Passarelli, M.; Nakandakare, E.R.; Lottenberg, S.A.; Lottenberg, A.M. The impact of dietary fatty acids on macrophage cholesterol homeostasis. J. Nutr. Biochem. 2014, 25, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Rom, O.; Jeries, H.; Hayek, T.; Aviram, M. Supplementation with linoleic acid-rich soybean oil stimulates macrophage foam cell formation via increased oxidative stress and diacylglycerol acyltransferase1-mediated triglyceride biosynthesis. BioFactors 2017, 43, 100–116. [Google Scholar] [CrossRef] [PubMed]
- Paigen, B.; Holmes, P.A.; Mitchell, D.; Albee, D. Comparison of atherosclerotic lesions and HDL-lipid levels in male, female, and testosterone-treated female mice from strains C57BL/6, BALB/c, and C3H. Atherosclerosis 1987, 64, 215–221. [Google Scholar] [CrossRef]
- Whitman, S.C. A Practical Approach to Using Mice in Atherosclerosis Research. Clin. Biochem. Rev. 2004, 25, 81–93. [Google Scholar] [PubMed]
- Lewis, S.R.; Dym, C.; Chai, C.; Singh, A.; Kest, B.; Bodnar, R.J. Genetic variance contributes to ingestive processes: A survey of eleven inbred mouse strains for fat (Intralipid) intake. Physiol. Behav. 2007, 90, 82–94. [Google Scholar] [CrossRef]
- Le Roux, C.W.; Bueter, M.; Theis, N.; Werling, M.; Ashrafian, H.; Löwenstein, C.; Athanasiou, T.; Bloom, S.R.; Spector, A.C.; Olbers, T.; et al. Gastric bypass reduces fat intake and preference. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 301, 1057–1066. [Google Scholar] [CrossRef] [Green Version]
- Laukens, D.; Brinkman, B.M.; Raes, J.; De Vos, M.; Vandenabeele, P. Heterogeneity of the gut microbiome in mice: Guidelines for optimizing experimental design. FEMS Microbiol. Rev. 2016, 036, 117–132. [Google Scholar] [CrossRef] [Green Version]
- Korach-Rechtman, H.; Freilich, S.; Gerassy-Vainberg, S.; Buhnik-Rosenblau, K.; Danin-Poleg, Y.; Bar, H.; Kashi, Y. Murine Genetic Background has a Stronger Impact on the Composition of the Gut Microbiota than Maternal Inoculation or Exposure to Unlike Exogenous Microbiota. Appl. Environ. Microbiol. 2019, 85, e00826-19. [Google Scholar] [CrossRef] [Green Version]
- Green, S.J.; Venkatramanan, R.; Naqib, A. Deconstructing the polymerase chain reaction: Understanding and correcting bias associated with primer degeneracies and primer-template mismatches. PLoS ONE 2015, 10, e0128122. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- I Langille, M.G.; Zaneveld, J.; Gregory Caporaso, J.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Rom, O.; Korach-Rechtman, H.; Hayek, T.; Danin-Poleg, Y.; Bar, H.; Kashi, Y.; Aviram, M. Acrolein increases macrophage atherogenicity in association with gut microbiota remodeling in atherosclerotic mice: Protective role for the polyphenol-rich pomegranate juice. Arch. Toxicol. 2016, 91, 1709–1725. [Google Scholar] [CrossRef] [PubMed]
- Rom, O.; Grajeda-Iglesias, C.; Najjar, M.; Abu-Saleh, N.; Volkova, N.; Dar, D.E.; Hayek, T.; Aviram, M. Atherogenicity of amino acids in the lipid-laden macrophage model system in vitro and in atherosclerotic mice: A key role for triglyceride metabolism. J. Nutr. Biochem. 2017, 45, 24–38. [Google Scholar] [CrossRef]
- El-Saadani, M.; Esterbauer, H.; El-Sayed, M.; Goher, M.; Nassar, A.Y.; Jurgens, G. A spectrophotometric assay for lipid peroxides in serum lipoproteins using a commercially available reagent. J. Lipid Res. 1989, 30, 627–630. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef] [Green Version]
- Lozupone, C.; Knight, R. UniFrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [Green Version]
- Ley, R.; Turnbaugh, P.; Klein, S.; Gordon, J. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Biagi, E.; Nylund, L.; Candela, M.; Ostan, R.; Bucci, L.; Pini, E.; Nikkïla, J.; Monti, D.; Satokari, R.; Franceschi, C.; et al. Through Ageing, and Beyond: Gut Microbiota and Inflammatory Status in Seniors and Centenarians. PLoS ONE 2010, 5, e10667. [Google Scholar] [CrossRef]
- Mariat, D.; Firmesse, O.; Levenez, F.; Guimarăes, V.; Sokol, H.; Doré, J.; Corthier, G.; Furet, J.-P. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009, 9, 123. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Yang, D.; Tao, X.; Yu, J.; Xiong, H.; Wei, H. Enterobacter aerogenes ZDY01 attenuates choline-induced Trimethylamine N-oxide levels via remodeling gut microbiota in mice. J. Microbiol. Biotechnol. 2017, 27, 1491–1499. [Google Scholar] [PubMed]
- Liu, S.; Bennett, D.C.; Tun, H.M.; Kim, J.-E.; Cheng, K.M.; Zhang, H.; Leung, F.C. The effect of diet and host genotype on ceca microbiota of Japanese quail fed a cholesterol enriched diet. Front. Microbiol. 2015, 6, 1092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.-J.; Tang, X.-D.; Yu, J.; Zhang, H.-Y.; Li, X.-R. Gut microbiota alterations from different Lactobacillus probiotic-fermented yoghurt treatments in slow-transit constipation. J. Funct. Foods. 2017, 38, 110–118. [Google Scholar] [CrossRef]
- Romano, K.A.; Vivas, E.I.; Amador-noguez, D.; Rey, F.E. Intestinal Microbiota Composition Modulates Choline Bioavailability. MBio 2015, 6, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Jonsson, A.L.; Bäckhed, F. Role of gut microbiota in atherosclerosis. Nat. Rev. Cardiol. 2016, 14, 79–87. [Google Scholar] [CrossRef]
- Emoto, T.; Yamashita, T.; Sasaki, N.; Hirota, Y.; Hayashi, T.; So, A.; Kasahara, K.; Yodoi, K.; Matsumoto, T.; Mizoguchi, T.; et al. Analysis of Gut Microbiota in Coronary Artery Disease Patients: A Possible Link between Gut Microbiota and Coronary Artery Disease. J. Atheroscler. Thromb. 2016, 23, 908–921. [Google Scholar] [CrossRef] [Green Version]
- Libby, P.; Ridker, P.M.; Hansson, G.K. Progress and challenges in translating the biology of atherosclerosis. Nature 2011, 473, 317–325. [Google Scholar] [CrossRef]
- Moreira, A.P.B.; Texeira, T.F.S.; Ferreira, A.B.; do Carmo Gouveia Peluzio, M.; de Cássia Gonçalves Alfenas, R. Influence of a high-fat diet on gut microbiota, intestinal permeability and metabolic endotoxaemia. Br. J. Nutr. 2012, 108, 801–809. [Google Scholar] [CrossRef]
- Grajeda-Iglesias, C.; Rom, O.; Hamoud, S.; Volkova, N.; Hayek, T.; Abu-Saleh, N.; Aviram, M. Leucine supplementation attenuates macrophage foam-cell formation: Studies in humans, mice, and cultured macrophages. BioFactors 2018, 44, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Grajeda-Iglesias, C.; Rom, O.; Aviram, M. Branched-chain amino acids and atherosclerosis: Friends or foes? Curr. Opin. Lipidol. 2018, 29, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Rom, O.; Villacorta, L.; Zhang, J.; Chen, Y.E.; Aviram, M. Emerging therapeutic potential of glycine in cardiometabolic diseases: Dual benefits in lipid and glucose metabolism. Curr. Opin. Lipidol. 2018, 29, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Rom, O.; Aviram, M. It is not just lipids: Proatherogenic vs. antiatherogenic roles for amino acids in macrophage foam cell formation. Curr. Opin. Lipidol. 2017, 28, 85–87. [Google Scholar] [PubMed]
- Yang, R.Y.; Wang, S.M.; Sun, L.; Liu, J.M.; Li, H.X.; Sui, X.F.; Wang, M.; Xiu, H.L.; Wang, S.; He, Q.; et al. Association of branched-chain amino acids with coronary artery disease: A matched-pair case-control study. Nutr. Metab. Cardiovasc. Dis. 2015, 25, 937–942. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Granger, C.B.; Craig, D.; Haynes, C.; Bain, J.; Stevens, R.D.; Hauser, E.R.; Newgard, C.B.; Kraus, W.E.; Newby, L.K.; et al. Validation of the association between a branched chain amino acid metabolite profile and extremes of coronary artery disease in patients referred for cardiac catheterization. Atherosclerosis 2014, 232, 191–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, R.; Dong, J.; Zhao, H.; Li, H.; Guo, H.; Wang, S.; Zhang, C.; Wang, S.; Wang, M.; Yu, S.; et al. Association of branched-chain amino acids with carotid intima-media thickness and coronary artery disease risk factors. PLoS ONE 2014, 9, e99598. [Google Scholar] [CrossRef] [Green Version]
- Pedersen, H.K.; Gudmundsdottir, V.; Nielsen, H.B.; Hyotylainen, T.; Nielsen, T.; Jensen, B.A.H.; Forslund, K.; Hildebrand, F.; Prifti, E.; Falony, G.; et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016, 535, 376–381. [Google Scholar] [CrossRef]
- Bogiatzi, C.; Gloor, G.; Allen-Vercoe, E.; Reid, G.; Wong, R.G.; Urquhart, B.L.; Dinculescu, V.; Ruetz, K.N.; Velenosi, T.J.; Pignanelli, M.; et al. Metabolic products of the intestinal microbiome and extremes of atherosclerosis. Atherosclerosis 2018, 273, 91–97. [Google Scholar] [CrossRef]
- Loria, C.M.; Ingram, D.D.; Feldman, J.J.; Wright, J.D.; Madans, J.H. Serum Folate and Cardiovascular Disease Mortality among US Men and Women. Arch. Intern. Med. 2000, 160, 3258. [Google Scholar] [CrossRef] [Green Version]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Korach-Rechtman, H.; Rom, O.; Mazouz, L.; Freilich, S.; Jeries, H.; Hayek, T.; Aviram, M.; Kashi, Y. Soybean Oil Modulates the Gut Microbiota Associated with Atherogenic Biomarkers. Microorganisms 2020, 8, 486. https://doi.org/10.3390/microorganisms8040486
Korach-Rechtman H, Rom O, Mazouz L, Freilich S, Jeries H, Hayek T, Aviram M, Kashi Y. Soybean Oil Modulates the Gut Microbiota Associated with Atherogenic Biomarkers. Microorganisms. 2020; 8(4):486. https://doi.org/10.3390/microorganisms8040486
Chicago/Turabian StyleKorach-Rechtman, Hila, Oren Rom, Lirane Mazouz, Shay Freilich, Helana Jeries, Tony Hayek, Michael Aviram, and Yechezkel Kashi. 2020. "Soybean Oil Modulates the Gut Microbiota Associated with Atherogenic Biomarkers" Microorganisms 8, no. 4: 486. https://doi.org/10.3390/microorganisms8040486