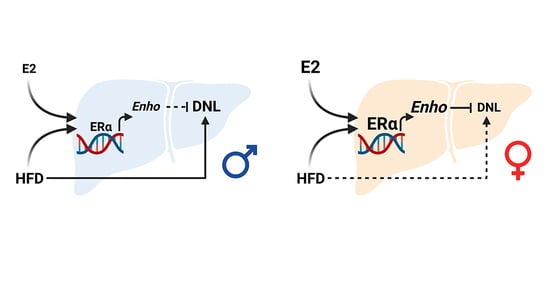
ERα-Dependent Regulation of Adropin Predicts Sex Differences in Liver Homeostasis during High-Fat Diet
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Cell Culture and Treatments
2.3. RNA-Sequencing and Transcriptomics Data Analysis
2.4. Reverse Transcription Quantitative Real-Time PCR (RT-PCR) Gene Expression Analysis
2.5. Biochemical Assays
2.6. Quantification, Statistical Analysis and Figure Preparation
3. Results
3.1. Liver mRNA Content of Hepatokines Depends on Sex and Estrogen Levels
3.2. Estrogen Treatment Regulates Hepatokine mRNA Content in Hepatic Cells
3.3. Hormone Replacement Therapy Can Restore Hepatokine Expression in a Mouse Model of Menopause
3.4. Adropin mRNA Levels Negatively Correlate with Fatty Liver in a Mouse Model of NAFLD
3.5. Sex-Specific Reprogramming of Liver Metabolism in a HFD-Induced Model of NAFLD Depends on Hepatic ERα
3.6. Adropin Induction Negatively Correlates with Lipid Synthesis in Female Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Loomba, R.; Friedman, S.L.; Shulman, G.I. Mechanisms and Disease Consequences of Nonalcoholic Fatty Liver Disease. Cell 2021, 184, 2537–2564. [Google Scholar] [CrossRef] [PubMed]
- Powell, E.E.; Wong, V.W.-S.; Rinella, M. Non-Alcoholic Fatty Liver Disease. Lancet 2021, 397, 2212–2224. [Google Scholar] [CrossRef]
- Carter, J.; Wang, S.; Friedman, S.L. Ten Thousand Points of Light: Heterogeneity Among the Stars of NASH Fibrosis. Hepatology 2021, 74, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, J.V.; Mark, H.E.; Anstee, Q.M.; Arab, J.P.; Batterham, R.L.; Castera, L.; Cortez-Pinto, H.; Crespo, J.; Cusi, K.; Dirac, M.A.; et al. Advancing the Global Public Health Agenda for NAFLD: A Consensus Statement. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 60–78. [Google Scholar] [CrossRef]
- Della Torre, S. Non-Alcoholic Fatty Liver Disease as a Canonical Example of Metabolic Inflammatory-Based Liver Disease Showing a Sex-Specific Prevalence: Relevance of Estrogen Signaling. Front. Endocrinol. 2020, 11, 572490. [Google Scholar] [CrossRef] [PubMed]
- Lonardo, A.; Nascimbeni, F.; Ballestri, S.; Fairweather, D.; Win, S.; Than, T.A.; Abdelmalek, M.F.; Suzuki, A. Sex Differences in Nonalcoholic Fatty Liver Disease: State of the Art and Identification of Research Gaps. Hepatology 2019, 70, 1457–1469. [Google Scholar] [CrossRef]
- Della Torre, S. Beyond the X Factor: Relevance of Sex Hormones in NAFLD Pathophysiology. Cells 2021, 10, 2502. [Google Scholar] [CrossRef]
- Grossmann, M.; Wierman, M.E.; Angus, P.; Handelsman, D.J. Reproductive Endocrinology of Nonalcoholic Fatty Liver Disease. Endocr. Rev. 2019, 40, 417–446. [Google Scholar] [CrossRef]
- Della Torre, S.; Mitro, N.; Meda, C.; Lolli, F.; Pedretti, S.; Barcella, M.; Ottobrini, L.; Metzger, D.; Caruso, D.; Maggi, A. Short-Term Fasting Reveals Amino Acid Metabolism as a Major Sex-Discriminating Factor in the Liver. Cell Metab. 2018, 28, 256–267.e5. [Google Scholar] [CrossRef]
- Della Torre, S.; Mitro, N.; Fontana, R.; Gomaraschi, M.; Favari, E.; Recordati, C.; Lolli, F.; Quagliarini, F.; Meda, C.; Ohlsson, C.; et al. An Essential Role for Liver ERα in Coupling Hepatic Metabolism to the Reproductive Cycle. Cell Rep. 2016, 15, 360–371. [Google Scholar] [CrossRef] [Green Version]
- Villa, A.; Della Torre, S.; Stell, A.; Cook, J.; Brown, M.; Maggi, A. Tetradian Oscillation of Estrogen Receptor Is Necessary to Prevent Liver Lipid Deposition. Proc. Natl. Acad. Sci. USA 2012, 109, 11806–11811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Della Torre, S.; Benedusi, V.; Fontana, R.; Maggi, A. Energy Metabolism and Fertility—a Balance Preserved for Female Health. Nat. Rev. Endocrinol. 2014, 10, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Meda, C.; Barone, M.; Mitro, N.; Lolli, F.; Pedretti, S.; Caruso, D.; Maggi, A.; Della Torre, S. Hepatic ERα Accounts for Sex Differences in the Ability to Cope with an Excess of Dietary Lipids. Mol. Metab. 2020, 32, 97–108. [Google Scholar] [CrossRef]
- Erkan, G.; Yilmaz, G.; Konca Degertekin, C.; Akyol, G.; Ozenirler, S. Presence and Extent of Estrogen Receptor-Alpha Expression in Patients with Simple Steatosis and NASH. Pathol. Res. Pract. 2013, 209, 429–432. [Google Scholar] [CrossRef] [PubMed]
- Adams, L.A.; Anstee, Q.M.; Tilg, H.; Targher, G. Non-Alcoholic Fatty Liver Disease and Its Relationship with Cardiovascular Disease and Other Extrahepatic Diseases. Gut 2017, 66, 1138–1153. [Google Scholar] [CrossRef] [Green Version]
- Mikolasevic, I.; Milic, S.; Turk Wensveen, T.; Grgic, I.; Jakopcic, I.; Stimac, D.; Wensveen, F.; Orlic, L. Nonalcoholic Fatty Liver Disease—A Multisystem Disease? World J. Gastroenterol. 2016, 22, 9488. [Google Scholar] [CrossRef]
- Samuel, V.T.; Shulman, G.I. Nonalcoholic Fatty Liver Disease as a Nexus of Metabolic and Hepatic Diseases. Cell Metab. 2018, 27, 22–41. [Google Scholar] [CrossRef] [Green Version]
- Stefan, N.; Häring, H.-U.; Cusi, K. Non-Alcoholic Fatty Liver Disease: Causes, Diagnosis, Cardiometabolic Consequences, and Treatment Strategies. Lancet Diabetes Endocrinol. 2019, 7, 313–324. [Google Scholar] [CrossRef]
- Targher, G.; Corey, K.E.; Byrne, C.D.; Roden, M. The Complex Link between NAFLD and Type 2 Diabetes Mellitus—Mechanisms and Treatments. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 599–612. [Google Scholar] [CrossRef]
- Jensen-Cody, S.O.; Potthoff, M.J. Hepatokines and Metabolism: Deciphering Communication from the Liver. Mol. Metab. 2021, 44, 101138. [Google Scholar] [CrossRef]
- Kim, T.H.; Hong, D.-G.; Yang, Y.M. Hepatokines and Non-Alcoholic Fatty Liver Disease: Linking Liver Pathophysiology to Metabolism. Biomedicines 2021, 9, 1903. [Google Scholar] [CrossRef] [PubMed]
- Meex, R.C.R.; Watt, M.J. Hepatokines: Linking Nonalcoholic Fatty Liver Disease and Insulin Resistance. Nat. Rev. Endocrinol. 2017, 13, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Watt, M.J.; Miotto, P.M.; De Nardo, W.; Montgomery, M.K. The Liver as an Endocrine Organ—Linking NAFLD and Insulin Resistance. Endocr. Rev. 2019, 40, 1367–1393. [Google Scholar] [CrossRef]
- Dharmalingam, M.; Pattabhi, G. Fetuin-A as a Marker of NAFLD. J. Endocr. Soc. 2021, 5, A413–A414. [Google Scholar] [CrossRef]
- Wu, H.-T.; Lu, F.-H.; Ou, H.-Y.; Su, Y.-C.; Hung, H.-C.; Wu, J.-S.; Yang, Y.-C.; Wu, C.-L.; Chang, C.-J. The Role of Hepassocin in the Development of Non-Alcoholic Fatty Liver Disease. J. Hepatol. 2013, 59, 1065–1072. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, Y.; Pan, R.; Wu, C.; Chen, S.; Li, L.; Li, Y.; Yu, C.; Meng, Z.; Xu, C. Leukocyte Cell-derived Chemotaxin 2 Promotes the Development of Nonalcoholic Fatty Liver Disease through STAT-1 Pathway in Mice. Liver Int. 2021, 41, 777–787. [Google Scholar] [CrossRef]
- Wu, C.; Borné, Y.; Gao, R.; López Rodriguez, M.; Roell, W.C.; Wilson, J.M.; Regmi, A.; Luan, C.; Aly, D.M.; Peter, A.; et al. Elevated Circulating Follistatin Associates with an Increased Risk of Type 2 Diabetes. Nat. Commun. 2021, 12, 6486. [Google Scholar] [CrossRef]
- Wang, X.; Chen, X.; Zhang, H.; Pang, J.; Lin, J.; Xu, X.; Yang, L.; Ma, J.; Ling, W.; Chen, Y. Circulating Retinol-Binding Protein 4 Is Associated with the Development and Regression of Non-Alcoholic Fatty Liver Disease. Diabetes Metab. 2020, 46, 119–128. [Google Scholar] [CrossRef]
- Chen, Y.; He, X.; Chen, X.; Li, Y.; Ke, Y. SeP Is Elevated in NAFLD and Participates in NAFLD Pathogenesis through AMPK/ACC Pathway. J. Cell Physiol. 2021, 236, 3800–3807. [Google Scholar] [CrossRef]
- Mouchiroud, M.; Camiré, É.; Aldow, M.; Caron, A.; Jubinville, É.; Turcotte, L.; Kaci, I.; Beaulieu, M.-J.; Roy, C.; Labbé, S.M.; et al. The Hepatokine Tsukushi Is Released in Response to NAFLD and Impacts Cholesterol Homeostasis. JCI Insight 2019, 4, 129492. [Google Scholar] [CrossRef] [Green Version]
- Tucker, B.; Li, H.; Long, X.; Rye, K.-A.; Ong, K.L. Fibroblast Growth Factor 21 in Non-Alcoholic Fatty Liver Disease. Metabolism 2019, 101, 153994. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Sun, X.; Shen, T.; Chen, Q.; Chen, S.; Pang, J.; Mi, J.; Tang, Y.; You, Y.; Xu, H.; et al. Lower Adropin Expression Is Associated with Oxidative Stress and Severity of Nonalcoholic Fatty Liver Disease. Free Radic. Biol. Med. 2020, 160, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Day, E.A.; Townsend, L.K.; Djordjevic, D.; Jørgensen, S.B.; Steinberg, G.R. GDF15: Emerging Biology and Therapeutic Applications for Obesity and Cardiometabolic Disease. Nat. Rev. Endocrinol. 2021, 17, 592–607. [Google Scholar] [CrossRef] [PubMed]
- Della Torre, S.; Benedusi, V.; Pepe, G.; Meda, C.; Rizzi, N.; Uhlenhaut, N.H.; Maggi, A. Dietary Essential Amino Acids Restore Liver Metabolism in Ovariectomized Mice via Hepatic Estrogen Receptor α. Nat. Commun. 2021, 12, 6883. [Google Scholar] [CrossRef]
- Della Torre, S.; Rando, G.; Meda, C.; Stell, A.; Chambon, P.; Krust, A.; Ibarra, C.; Magni, P.; Ciana, P.; Maggi, A. Amino Acid-Dependent Activation of Liver Estrogen Receptor Alpha Integrates Metabolic and Reproductive Functions via IGF-1. Cell Metab. 2011, 13, 205–214. [Google Scholar] [CrossRef] [Green Version]
- Valverde, A.M.; Burks, D.J.; Fabregat, I.; Fisher, T.L.; Carretero, J.; White, M.F.; Benito, M. Molecular Mechanisms of Insulin Resistance in IRS-2-Deficient Hepatocytes. Diabetes 2003, 52, 2239–2248. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Della Torre, S.; Maggi, A. Sex Differences: A Resultant of an Evolutionary Pressure? Cell Metab. 2017, 25, 499–505. [Google Scholar] [CrossRef] [Green Version]
- Maggi, A.; Della Torre, S. Sex, Metabolism and Health. Mol. Metab. 2018, 15, 3–7. [Google Scholar] [CrossRef]
- Genazzani, A.R.; Monteleone, P.; Giannini, A.; Simoncini, T. Hormone Therapy in the Postmenopausal Years: Considering Benefits and Risks in Clinical Practice. Hum. Reprod. Update 2021, 27, 1115–1150. [Google Scholar] [CrossRef]
- Flores, V.A.; Pal, L.; Manson, J.E. Hormone Therapy in Menopause: Concepts, Controversies, and Approach to Treatment. Endocr. Rev. 2021, 42, 720–752. [Google Scholar] [CrossRef] [PubMed]
- Mehta, J.; Kling, J.M.; Manson, J.E. Risks, Benefits, and Treatment Modalities of Menopausal Hormone Therapy: Current Concepts. Front. Endocrinol. 2021, 12, 564781. [Google Scholar] [CrossRef] [PubMed]
- Lobo, R.A.; Gompel, A. Management of Menopause: A View towards Prevention. Lancet Diabetes Endocrinol. 2022, 10, 457–470. [Google Scholar] [CrossRef]
- Nappi, R.E.; Chedraui, P.; Lambrinoudaki, I.; Simoncini, T. Menopause: A Cardiometabolic Transition. Lancet Diabetes Endocrinol. 2022, 10, 442–456. [Google Scholar] [CrossRef]
- Gao, S.; McMillan, R.P.; Jacas, J.; Zhu, Q.; Li, X.; Kumar, G.K.; Casals, N.; Hegardt, F.G.; Robbins, P.D.; Lopaschuk, G.D.; et al. Regulation of Substrate Oxidation Preferences in Muscle by the Peptide Hormone Adropin. Diabetes 2014, 63, 3242–3252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee, S.; Ghoshal, S.; Stevens, J.R.; McCommis, K.S.; Gao, S.; Castro-Sepulveda, M.; Mizgier, M.L.; Girardet, C.; Kumar, K.G.; Galgani, J.E.; et al. Hepatocyte Expression of the Micropeptide Adropin Regulates the Liver Fasting Response and Is Enhanced by Caloric Restriction. J. Biol. Chem. 2020, 295, 13753–13768. [Google Scholar] [CrossRef]
- Jasaszwili, M.; Billert, M.; Strowski, M.Z.; Nowak, K.W.; Skrzypski, M. Adropin as A Fat-Burning Hormone with Multiple Functions—Review of a Decade of Research. Molecules 2020, 25, 549. [Google Scholar] [CrossRef] [Green Version]
- Kumar, K.G.; Trevaskis, J.L.; Lam, D.D.; Sutton, G.M.; Koza, R.A.; Chouljenko, V.N.; Kousoulas, K.G.; Rogers, P.M.; Kesterson, R.A.; Thearle, M.; et al. Identification of Adropin as a Secreted Factor Linking Dietary Macronutrient Intake with Energy Homeostasis and Lipid Metabolism. Cell Metab. 2008, 8, 468–481. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Chen, S.; Shen, T.; Yang, W.; Chen, Q.; Zhang, P.; You, Y.; Sun, X.; Xu, H.; Tang, Y.; et al. Adropin Regulates Hepatic Glucose Production via PP2A/AMPK Pathway in Insulin-Resistant Hepatocytes. FASEB J. 2020, 34, 10056–10072. [Google Scholar] [CrossRef]
- Gao, S.; Ghoshal, S.; Zhang, L.; Stevens, J.R.; McCommis, K.S.; Finck, B.N.; Lopaschuk, G.D.; Butler, A.A. The Peptide Hormone Adropin Regulates Signal Transduction Pathways Controlling Hepatic Glucose Metabolism in a Mouse Model of Diet-Induced Obesity. J. Biol. Chem. 2019, 294, 13366–13377. [Google Scholar] [CrossRef]
- Ghoshal, S.; Stevens, J.R.; Billon, C.; Girardet, C.; Sitaula, S.; Leon, A.S.; Rao, D.C.; Skinner, J.S.; Rankinen, T.; Bouchard, C.; et al. Adropin: An Endocrine Link between the Biological Clock and Cholesterol Homeostasis. Mol. Metab. 2018, 8, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Thapa, D.; Xie, B.; Manning, J.R.; Zhang, M.; Stoner, M.W.; Huckestein, B.R.; Edmunds, L.R.; Zhang, X.; Dedousis, N.L.; O’Doherty, R.M.; et al. Adropin Reduces Blood Glucose Levels in Mice by Limiting Hepatic Glucose Production. Physiol. Rep. 2019, 7, e14043. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zeng, K.; Liu, Q.; Guo, Z.; Zhang, S.; Chen, X.; Lin, J.; Wen, J.; Zhao, C.; Lin, X.; et al. Adropin Deficiency Worsens HFD-Induced Metabolic Defects. Cell Death Dis. 2017, 8, e3008. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Xue, H.; Fang, W.; Chen, K.; Chen, S.; Yang, W.; Shen, T.; Chen, X.; Zhang, P.; Ling, W. Adropin Protects against Liver Injury in Nonalcoholic Steatohepatitis via the Nrf2 Mediated Antioxidant Capacity. Redox Biol. 2019, 21, 101068. [Google Scholar] [CrossRef] [PubMed]
- Bazhan, N.; Jakovleva, T.; Feofanova, N.; Denisova, E.; Dubinina, A.; Sitnikova, N.; Makarova, E. Sex Differences in Liver, Adipose Tissue, and Muscle Transcriptional Response to Fasting and Refeeding in Mice. Cells 2019, 8, 1529. [Google Scholar] [CrossRef] [Green Version]
- Chaix, A.; Deota, S.; Bhardwaj, R.; Lin, T.; Panda, S. Sex- and Age-Dependent Outcomes of 9-Hour Time-Restricted Feeding of a Western High-Fat High-Sucrose Diet in C57BL/6J Mice. Cell Rep. 2021, 36, 109543. [Google Scholar] [CrossRef] [PubMed]
- Morris, A. Sex Differences for Fasting Levels of Glucose and Insulin: Expanding Our Understanding. Nat. Rev. Endocrinol. 2021, 17, 131. [Google Scholar] [CrossRef]
- Kane, A.E.; Sinclair, D.A.; Mitchell, J.R.; Mitchell, S.J. Sex Differences in the Response to Dietary Restriction in Rodents. Curr. Opin. Physiol. 2018, 6, 28–34. [Google Scholar] [CrossRef]
- Astafev, A.A.; Patel, S.A.; Kondratov, R.V. Calorie Restriction Effects on Circadian Rhythms in Gene Expression Are Sex Dependent. Sci. Rep. 2017, 7, 9716. [Google Scholar] [CrossRef] [Green Version]
- Larson, K.R.; Russo, K.A.; Fang, Y.; Mohajerani, N.; Goodson, M.L.; Ryan, K.K. Sex Differences in the Hormonal and Metabolic Response to Dietary Protein Dilution. Endocrinology 2017, 158, 3477–3487. [Google Scholar] [CrossRef]
- Forney, L.A.; Stone, K.P.; Gibson, A.N.; Vick, A.M.; Sims, L.C.; Fang, H.; Gettys, T.W. Sexually Dimorphic Effects of Dietary Methionine Restriction Are Dependent on Age When the Diet Is Introduced. Obesity 2020, 28, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Yang, S.E.; Miller, B.R.; Wisinski, J.A.; Sherman, D.S.; Brinkman, J.A.; Tomasiewicz, J.L.; Cummings, N.E.; Kimple, M.E.; Cryns, V.L.; et al. Short-Term Methionine Deprivation Improves Metabolic Health via Sexually Dimorphic, MTORC1-Independent Mechanisms. FASEB J. 2018, 32, 3471–3482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richardson, N.E.; Konon, E.N.; Schuster, H.S.; Mitchell, A.T.; Boyle, C.; Rodgers, A.C.; Finke, M.; Haider, L.R.; Yu, D.; Flores, V.; et al. Lifelong Restriction of Dietary Branched-Chain Amino Acids Has Sex-Specific Benefits for Frailty and Life Span in Mice. Nat. Aging 2021, 1, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Iena, F.M.; Jul, J.B.; Vegger, J.B.; Lodberg, A.; Thomsen, J.S.; Brüel, A.; Lebeck, J. Sex-Specific Effect of High-Fat Diet on Glycerol Metabolism in Murine Adipose Tissue and Liver. Front. Endocrinol. 2020, 11, 577650. [Google Scholar] [CrossRef] [PubMed]
- Glavas, M.M.; Lee, A.Y.; Miao, I.; Yang, F.; Mojibian, M.; O’Dwyer, S.M.; Kieffer, T.J. Developmental Timing of High-Fat Diet Exposure Impacts Glucose Homeostasis in Mice in a Sex-Specific Manner. Diabetes 2021, 70, 2771–2784. [Google Scholar] [CrossRef]
- Low, W.; Cornfield, T.; Charlton, C.; Tomlinson, J.; Hodson, L. Sex Differences in Hepatic De Novo Lipogenesis with Acute Fructose Feeding. Nutrients 2018, 10, 1263. [Google Scholar] [CrossRef] [Green Version]
- Elstgeest, L.E.M.; Schaap, L.A.; Heymans, M.W.; Hengeveld, L.M.; Naumann, E.; Houston, D.K.; Kritchevsky, S.B.; Simonsick, E.M.; Newman, A.B.; Farsijani, S.; et al. Sex-and Race-Specific Associations of Protein Intake with Change in Muscle Mass and Physical Function in Older Adults: The Health, Aging, and Body Composition (Health ABC) Study. Am. J. Clin. Nutr. 2020, 112, 84–95. [Google Scholar] [CrossRef]
- Sadagurski, M.; Debarba, L.K.; Werneck-de-Castro, J.P.; Ali Awada, A.; Baker, T.A.; Bernal-Mizrachi, E. Sexual Dimorphism in Hypothalamic Inflammation in the Offspring of Dams Exposed to a Diet Rich in High Fat and Branched-Chain Amino Acids. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E526–E534. [Google Scholar] [CrossRef]
- Bideyan, L.; Nagari, R.; Tontonoz, P. Hepatic Transcriptional Responses to Fasting and Feeding. Genes Dev. 2021, 35, 635–657. [Google Scholar] [CrossRef]
- Geisler, C.E.; Hepler, C.; Higgins, M.R.; Renquist, B.J. Hepatic Adaptations to Maintain Metabolic Homeostasis in Response to Fasting and Refeeding in Mice. Nutr. Metab. 2016, 13, 62. [Google Scholar] [CrossRef] [Green Version]
- Smith, R.L.; Soeters, M.R.; Wüst, R.C.I.; Houtkooper, R.H. Metabolic Flexibility as an Adaptation to Energy Resources and Requirements in Health and Disease. Endocr. Rev. 2018, 39, 489–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allard, C.; Bonnet, F.; Xu, B.; Coons, L.; Albarado, D.; Hill, C.; Fagherazzi, G.; Korach, K.S.; Levin, E.R.; Lefante, J.; et al. Activation of Hepatic Estrogen Receptor-α Increases Energy Expenditure by Stimulating the Production of Fibroblast Growth Factor 21 in Female Mice. Mol. Metab. 2019, 22, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Sumida, Y.; Yoneda, M. Current and Future Pharmacological Therapies for NAFLD/NASH. J. Gastroenterol. 2018, 53, 362–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talukdar, S.; Kharitonenkov, A. FGF19 and FGF21: In NASH We Trust. Mol. Metab. 2021, 46, 101152. [Google Scholar] [CrossRef]
- Tillman, E.J.; Rolph, T. FGF21: An Emerging Therapeutic Target for Non-Alcoholic Steatohepatitis and Related Metabolic Diseases. Front. Endocrinol. 2020, 11, 601290. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meda, C.; Dolce, A.; Vegeto, E.; Maggi, A.; Della Torre, S. ERα-Dependent Regulation of Adropin Predicts Sex Differences in Liver Homeostasis during High-Fat Diet. Nutrients 2022, 14, 3262. https://doi.org/10.3390/nu14163262
Meda C, Dolce A, Vegeto E, Maggi A, Della Torre S. ERα-Dependent Regulation of Adropin Predicts Sex Differences in Liver Homeostasis during High-Fat Diet. Nutrients. 2022; 14(16):3262. https://doi.org/10.3390/nu14163262
Chicago/Turabian StyleMeda, Clara, Arianna Dolce, Elisabetta Vegeto, Adriana Maggi, and Sara Della Torre. 2022. "ERα-Dependent Regulation of Adropin Predicts Sex Differences in Liver Homeostasis during High-Fat Diet" Nutrients 14, no. 16: 3262. https://doi.org/10.3390/nu14163262