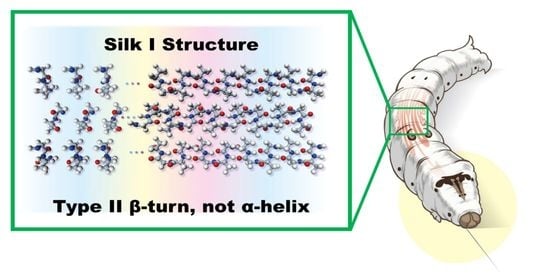
Structure of Silk I (Bombyx mori Silk Fibroin before Spinning) -Type II β-Turn, Not α-Helix-
Abstract
:1. Introduction
2. Structure of Silk Stored in the Middle Silk Gland of B. mori Silkworm
3. Determination of Silk I Structure in a Solid-State
4. Silk I Structure Determined from Solution NMR
5. Verification of Silk I Structure (Type II β-Turn)
6. Problems in Speculating Silk I Structure from the IR Spectrum
7. Conclusions and Future Aspects
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Asakura, T.; Miller, T. Biotechnology of Silk. In Biologically-Inspired Systems; Asakura, T., Miller, T., Eds.; Springer: Dordrecht, The Netherlands, 2014; Volume 5, ISBN 978-94-007-7118-5. [Google Scholar]
- Shao, Z.; Vollrath, F. Materials: Surprising strength of silkworm silk. Nature 2002, 418, 741. [Google Scholar] [CrossRef]
- Vepari, C.; Kaplan, D.L. Silk as a biomaterial. Prog. Polym. Sci. 2007, 32, 991–1007. [Google Scholar] [CrossRef]
- Koh, L.-D.; Cheng, Y.; Teng, C.-P.; Khin, Y.-W.; Loh, X.-J.; Tee, S.-Y.; Low, M.; Ye, E.; Yu, H.-D.; Zhang, Y.-W.; et al. Structures, mechanical properties and applications of silk fibroin materials. Prog. Polym. Sci. 2015, 46, 86–110. [Google Scholar] [CrossRef]
- Pereira, R.F.P.; Silva, M.M.; de Zea Bermudez, V. Bombyx mori Silk Fibers: An Outstanding Family of Materials. Macromol. Mater. Eng. 2015, 300, 1171–1198. [Google Scholar] [CrossRef]
- Thurber, A.E.; Omenetto, F.G.; Kaplan, D.L. In vivo bioresponses to silk proteins. Biomaterials 2015, 71, 145–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holland, C.; Numata, K.; Rnjak-Kovacina, J.; Seib, F.P. The Biomedical Use of Silk: Past, Present, Future. Adv. Healthc. Mater. 2018, 8, 1800465. [Google Scholar] [CrossRef] [Green Version]
- Tamara, A.B.; DeSimone, E.; Scheibel, T. Biomedical Applications of Recombinant Silk-Based Materials. Adv. Mater. 2018, 30, 1704636. [Google Scholar] [CrossRef]
- Takei, F.; Kikuchi, Y.; Kikuchi, A.; Mizuno, S.; Shimura, K. Further evidence for importance of the subunit combination of silk fibroin in its efficient secretion from the posterior silk gland cells. J. Cell Biol. 1987, 105, 175–180. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K.; Mori, K.; Mizuno, S. Immunological identification of the major disulfide-linked light component of silk fibroin. J. Biochem. 1993, 114, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Kajiyama, N.; Ishikura, K.; Waga, S.; Kikuchi, A.; Ohtomo, K.; Takagi, T.; Mizuno, S. Determination of the site of disulfide linkage between heavy and light chains of silk fibroin produced by Bombyx mori. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1999, 1432, 92–103. [Google Scholar] [CrossRef]
- Inoue, S.; Tanaka, K.; Arisaka, F.; Kimura, S.; Ohtomo, K.; Mizuno, S. Silk Fibroin of Bombyx mori is Secreted, Assembling a High Molecular Mass Elementary Unit Consisting of H-chain, L-chain, and P25, with a 6:6:1 Molar Ratio. J. Biol. Chem. 2000, 275, 40517–40528. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.-Z.; Confalonieri, F.; Medina, N.; Zivanovic, Y.; Esnault, C.; Yang, T.; Jacquet, M.; Janin, J.; Duguet, M.; Perasso, R.; et al. Fine organization of Bombyx mori fibroin heavy chain gene. Nucleic Acids Res. 2000, 28, 2413–2419. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.-Z.; Confalonieri, F.; Jacquet, M.; Perasso, R.; Li, Z.-G.; Janin, J. Silk fibroin: Structural implications of a remarkable amino acid sequence. Proteins 2001, 44, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Asakura, T.; Ohgo, K.; Ishida, T.; Taddei, P.; Monti, P.; Kishore, R. Possible Implications of Serine and Tyrosine Residues and Intermolecular Interactions on the Appearance of Silk I Structure of Bombyx mori Silk Fibroin-Derived Synthetic Peptides: High-Resolution 13C Cross-Polarization/Magic-Angle Spinni. Biomacromolecules 2005, 6, 468–474. [Google Scholar] [CrossRef]
- Asakura, T.; Ogawa, T.; Naito, A.; Williamson, M.P. Chain-folded lamellar structure and dynamics of the crystalline fraction of Bombyx mori silk fibroin and of (Ala-Gly-Ser-Gly-Ala-Gly)n model peptides. Int. J. Biol. Macromol. 2020, 164, 3974–3983. [Google Scholar] [CrossRef] [PubMed]
- Lotz, B.; Cesari, F.C. The chemical structure and the crystalline structures of Bombyx mori silk fibroin. Biochimie 1979, 61, 205–214. [Google Scholar] [CrossRef]
- Marsh, R.E.; Corey, R.B.; Pauling, L. An investigation of the structure of silk fibroin. Biochim. Biophys. Acta 1955, 16, 1–34. [Google Scholar] [CrossRef]
- Asakura, T.; Aoki, A.; Komatsu, K.; Ito, C.; Suzuki, I.; Naito, A.; Kaji, H. Lamellar Structure in Alanine–Glycine Copolypeptides Studied by Solid-State NMR Spectroscopy: A Model for the Crystalline Domain of Bombyx mori Silk Fibroin in Silk II Form. Biomacromolecules 2020, 21, 3102–3111. [Google Scholar] [CrossRef]
- Fraser, B.; MacRae, T.P. Conformations of Fibrous Proteins and Related Synthetic Polypeptides; Academic Press: New York, NY, USA, 1973. [Google Scholar]
- Asakura, T.; Ashida, J.; Yamane, T.; Kameda, T.; Nakazawa, Y.; Ohgo, K.; Komatsu, K. A repeated β-turn structure in Poly(Ala-Gly) as a model for silk I of Bombyx mori silk fibroin studied with two-dimensional spin-diffusion NMR under off magic angle spinning and rotational echo double resonance. J. Mol. Biol. 2001, 306, 291–305. [Google Scholar] [CrossRef]
- Asakura, T.; Ohgo, K.; Komatsu, K.; Kanenari, M.; Okuyama, K. Refinement of Repeated β-turn Structure for Silk I Conformation of Bombyx mori Silk Fibroin Using 13C Solid-State NMR and X-ray Diffraction Methods. Macromolecules 2005, 38, 7397–7403. [Google Scholar] [CrossRef]
- Hu, X.; Kaplan, D.; Cebe, P. Dynamic Protein−Water Relationships during β-Sheet Formation. Macromolecules 2008, 41, 3939–3948. [Google Scholar] [CrossRef]
- Hu, X.; Shmelev, K.; Sun, L.; Gil, E.-S.; Park, S.-H.; Cebe, P.; Kaplan, D.L. Regulation of Silk Material Structure by Temperature-controlled Water Vapor Annealing. Biomacromolecules 2011, 12, 1686–1696. [Google Scholar] [CrossRef] [Green Version]
- Lu, Q.; Huang, Y.; Li, M.; Zuo, B.; Lu, S.; Wang, J.; Zhu, H.; Kaplan, D.L. Silk fibroin electrogelation mechanisms. Acta Biomater. 2011, 7, 2394–2400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Percot, A.; Colomban, P.; Paris, C.; Dinh, H.M.; Wojcieszak, M.; Mauchamp, B. Water dependent structural changes of silk from Bombyx mori gland to fibre as evidenced by Raman and IR spectroscopies. Vib. Spectrosc. 2014, 73, 79–89. [Google Scholar] [CrossRef]
- Dubey, P.; Murab, S.; Karmakar, S.; Chowdhury, P.K.; Ghosh, S. Modulation of Self-Assembly Process of Fibroin: An Insight for Regulating the Conformation of Silk Biomaterials. Biomacromolecules 2015, 16, 3936–3944. [Google Scholar] [CrossRef]
- Wu, X.; Wu, X.; Shao, M.; Yang, B. Structural changes of Bombyx mori fibroin from silk gland to fiber as evidenced by Terahertz spectroscopy and other methods. Int. J. Biol. Macromol. 2017, 102, 1202–1210. [Google Scholar] [CrossRef] [PubMed]
- Kaewprasit, K.; Kobayashi, T.; Damrongsakkul, S. Thai silk fibroin gelation process enhancing by monohydric and polyhydric alcohols. Int. J. Biol. Macromol. 2018, 118, 1726–1735. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, J.; Feng, Y.; Yan, S.; Zhang, Q.; You, R. Tuning the structure and performance of silk biomaterials by combining mulberry and non-mulberry silk fibroin. Polym. Degrad. Stab. 2018, 147, 57–63. [Google Scholar] [CrossRef]
- Badillo-Sanchez, D.; Chelazzi, D.; Giorgi, R.; Cincinelli, A.; Baglioni, P. Characterization of the secondary structure of degummed Bombyx mori silk in modern and historical samples. Polym. Degrad. Stab. 2018, 157, 53–62. [Google Scholar] [CrossRef]
- Xue, Y.; Wang, F.; Torculas, M.; Lofland, S.; Hu, X. Formic Acid Regenerated Mori, Tussah, Eri, Thai, and Muga Silk Materials: Mechanism of Self-Assembly. ACS Biomater. Sci. Eng. 2019, 5, 6361–6373. [Google Scholar] [CrossRef]
- Li, X.; Wang, L.; Li, L.; Luo, Z.; Yan, S.; Zhang, Q.; You, R. Water-stable silk fibroin nerve conduits with tunable degradation prepared by a mild freezing-induced assembly. Polym. Degrad. Stab. 2019, 164, 61–68. [Google Scholar] [CrossRef]
- Yang, Y.; Greco, G.; Maniglio, D.; Mazzolai, B.; Migliaresi, C.; Pugno, N.; Motta, A. Spider (Linothele megatheloides) and silkworm (Bombyx mori) silks: Comparative physical and biological evaluation. Mater. Sci. Eng. C 2020, 107, 110197. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Jung, Y.; Li, T.-D.; Lao, J.; Tu, R.S.; Chen, X. β-Sheet Nanocrystals Dictate Water Responsiveness of Bombyx Mori Silk. Macromol. Rapid Commun. 2020, 41, 1900612. [Google Scholar] [CrossRef]
- Ling, S.; Qi, Z.; Knight, D.P.; Shao, Z.; Chen, X. Synchrotron FTIR Microspectroscopy of Single Natural Silk Fibers. Biomacromolecules 2011, 12, 3344–3349. [Google Scholar] [CrossRef]
- Zhong, J.; Liu, Y.; Ren, J.; Tang, Y.; Qi, Z.; Zhou, X.; Chen, X.; Shao, Z.; Chen, M.; Kaplan, D.L.; et al. Understanding Secondary Structures of Silk Materials via Micro- and Nano-Infrared Spectroscopies. ACS Biomater. Sci. Eng. 2019, 5, 3161–3183. [Google Scholar] [CrossRef] [PubMed]
- Vass, E.; Hollósi, M.; Besson, F.; Buchet, R. Vibrational Spectroscopic Detection of Beta- and Gamma-Turns in Synthetic and Natural Peptides and Proteins. Chem. Rev. 2003, 103, 1917–1954. [Google Scholar] [CrossRef]
- Asakura, T.; Suzuki, H.; Watanabe, Y. Conformational characterization of silk fibroin in intact Bombyx mori and Pilosamia cynthia ricini silkworms by 13C NMR spectroscopy. Macromolecules 1983, 16, 1024–1026. [Google Scholar] [CrossRef]
- Asakura, T. Nuclear Magnetic Resonance studies on the Biosynthesis of the Silk Fibroins in Silkworms. JOEL News 1987, 23A, 2–6. [Google Scholar]
- Sezutsu, H.; Yukuhiro, K. The complete nucleotide sequence of the Eri-silkworm(Samia cynthia ricini) fibroin gene. J. Insect Biotechnol. Sericol. 2014, 83, 59–70. [Google Scholar] [CrossRef]
- Asakura, T.; Murakami, T. NMR of silk fibroin. 4. Temperature- and urea-induced helix-coil transitions of the -(Ala)n- sequence in Philosamia cynthia ricini silk fibroin protein monitored by 13C NMR spectroscopy. Macromolecules 1985, 18, 2614–2619. [Google Scholar] [CrossRef]
- Asakura, T.; Kashiba, H.; Yoshimizu, H. NMR of silk fibroin. 8. 13C NMR analysis of the conformation and the conformational transition of Philosamia cynthia ricini silk fibroin protein on the basis of Bixon-Scheraga-Lifson theory. Macromolecules 1988, 21, 644–648. [Google Scholar] [CrossRef]
- Asakura, T.; Yoshimizu, H.; Yoshizawa, F. NMR of Silk Fibroin. 9. Sequence and Conformation Analyses of the Silk Fibroins from Bombyx mori and Philosamia cynthia ricini by 15N NMR Spectroscopy. Macromolecules 1988, 21, 2038–2041. [Google Scholar] [CrossRef]
- Nakazawa, Y.; Asakura, T. Heterogeneous exchange behavior of Samia cynthia ricini silk fibroin during helix-coil transition studied with 13C NMR. FEBS Lett. 2002, 529, 188–192. [Google Scholar] [CrossRef] [Green Version]
- Nakazawa, Y.; Asakura, T. Structure Determination of a Peptide Model of the Repeated Helical Domain in Samia cynthia ricini Silk Fibroin before Spinning by a Combination of Advanced Solid-State NMR Methods. J. Am. Chem. Soc. 2003, 125, 7230–7237. [Google Scholar] [CrossRef]
- Suzuki, Y.; Kawanishi, S.; Yamazaki, T.; Aoki, A.; Saito, H.; Asakura, T. Structural Determination of the Tandem Repeat Motif in Samia cynthia ricini Liquid Silk by Solution NMR. Macromolecules 2015, 48, 6574–6579. [Google Scholar] [CrossRef]
- Okuyama, K.; Takanashi, K.; Nakajima, Y.; Hasegawa, Y.; Hirabayashi, K.; Nishi, N. Analysis of Silk I structure by X-ray and electron diffraction methods. J. Sericultural Sci. Jpn. 1988, 57, 23–30. [Google Scholar] [CrossRef]
- Anderson, J.P. Morphology and crystal structure of a recombinant silk-like molecule, SLP4. Biopolymers 1998, 45, 307–321. [Google Scholar] [CrossRef]
- Asakura, T.; Yamane, T.; Nakazawa, Y.; Kameda, T.; Ando, K. Structure of Bombyx mori silk fibroin before spinning in solid state studied with wide angle X-ray scattering and 13C cross-polarization/magic angle spinning NMR. Biopolymers 2001, 58, 521–525. [Google Scholar] [CrossRef]
- Fossey, S.A.; Némethy, G.; Gibson, K.D.; Scheraga, H.A. Conformational energy studies of β-sheets of model silk fibroin peptides. I. Sheets of poly(Ala-Gly) chains. Biopolymers 1991, 31, 1529–1541. [Google Scholar] [CrossRef] [PubMed]
- Monti, P.; Taddei, P.; Freddi, G.; Ohgo, K.; Asakura, T. Vibrational 13C-cross-polarization/magic angle spinning NMR spectroscopic and thermal characterization of poly(alanine-glycine) as model for silk I Bombyx mori fibroin. Biopolymers 2003, 72, 329–338. [Google Scholar] [CrossRef]
- Taddei, P.; Asakura, T.; Yao, J.; Monti, P. Raman study of poly(alanine-glycine)-based peptides containing tyrosine, valine, and serine as model for the semicrystalline domains of Bombyx mori silk fibroin. Biopolymers 2004, 75, 314–324. [Google Scholar] [CrossRef]
- Saito, H.; Tabeta, R.; Asakura, T.; Iwanaga, Y.; Shoji, A.; Ozaki, T.; Ando, I.; Saitô, H.; Tabeta, R.; Asakura, T.; et al. High-resolution 13C NMR study of silk fibroin in the solid state by the cross-polarization-magic angle spinning method. Conformational characterization of silk I and silk II type forms of Bombyx mori fibroin by the conformation-dependent. Macromolecules 1984, 17, 1405–1412. [Google Scholar] [CrossRef]
- Asakura, T.; Kuzuhara, A.; Tabeta, R.; Saito, H. Conformational characterization of Bombyx mori silk fibroin in the solid state by high-frequency 13C cross polarization-magic angle spinning NMR, X-ray diffraction, and infrared spectroscopy. Macromolecules 1985, 18, 1841–1845. [Google Scholar] [CrossRef]
- Ishida, M.; Asakura, T.; Yokoi, M.; Saito, H. Solvent- and mechanical-treatment-induced conformational transition of silk fibroins studied by high-resolution solid-state 13C NMR spectroscopy. Macromolecules 1990, 23, 88–94. [Google Scholar] [CrossRef]
- Robyr, P.; Meier, B.H.; Ernst, R.R. Radio-frequency-driven nuclear spin diffusion in solids. Chem. Phys. Lett. 1989, 162, 417–423. [Google Scholar] [CrossRef]
- Robyr, P.; Tomaselli, M.; Straka, J.; Grob-Pisano, C.; Suter, U.W.; Meier, B.H.; Ernst, R.R. RF-driven and proton-driven NMR polarization transfer for investigating local order. Mol. Phys. 1995, 84, 995–1020. [Google Scholar] [CrossRef]
- Kümmerlen, J.; van Beek, J.D.; Vollrath, F.; Meier, B.H. Local Structure in Spider Dragline Silk Investigated by Two-Dimensional Spin-Diffusion Nuclear Magnetic Resonance. Macromolecules 1996, 29, 2920–2928. [Google Scholar] [CrossRef] [Green Version]
- Levitt, M.H.; Raleigh, D.P.; Creuzet, F.; Griffin, R.G. Theory and simulations of homonuclear spin pair systems in rotating solids. J. Chem. Phys. 1990, 92, 6347–6364. [Google Scholar] [CrossRef]
- Meier, B.H.; van Beek, J.D.; Beaulieu, L.; Schäfer, H.; Demura, M.; Asakura, T. Solid-state NMR determination of the secondary structure of Samia cynthia ricini silk. Nature 2000, 405, 1077–1079. [Google Scholar] [CrossRef]
- Ashida, J.; Ohgo, K.; Asakura, T. Determination of the Torsion Angles of Alanine and Glycine Residues of Bombyx Mori Silk Fibroin and the Model Peptides in the Silk I and Silk II Forms Using 2D Spin Diffusion Solid-State NMR under Off Magic Angle Spinning. J. Phys. Chem. B 2002, 106, 9434–9439. [Google Scholar] [CrossRef]
- Asakura, T.; Ashida, J.; Yamane, T. Structure of Bombyx mori Silk Fibroin before Spinning in Silkworm. In NMR Spectroscopy of Polymers in Solution and in the Solid State; American Chemical Society: Washington, DC, USA, 2003; pp. 71–82. ISBN 9780841238084. [Google Scholar]
- Asakura, T.; Suzuki, Y.; Nakazawa, Y.; Yazawa, K.; Holland, G.P.; Yarger, J.L. Silk structure studied with nuclear magnetic resonance. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 69, 23–68. [Google Scholar] [CrossRef] [PubMed]
- Asakura, T.; Suzuki, Y.; Nakazawa, Y.; Holland, G.P.; Yarger, J.L. Elucidating silk structure using solid-state NMR. Soft Matter 2013, 9, 11440–11450. [Google Scholar] [CrossRef]
- Asakura, T.; Okushita, K.; Williamson, M.P. Analysis of the structure of Bombyx mori silk fibroin by NMR. Macromolecules 2015, 48, 2345–2357. [Google Scholar] [CrossRef]
- Gullion, T.; Schaefer, J. Detection of Weak Heteronuclear Dipolar Coupling by Rotational-Echo Double-Resonance Nuclear Magnetic Resonance. In Advances in Magnetic and Optical Resonance; Warren, W.S., Ed.; Academic Press: Cambridge, MA, USA, 1989; Volume 13, pp. 57–83. [Google Scholar]
- Gullion, T.; Baker, D.B.; Conradi, M.S. New, compensated Carr-Purcell sequences. J. Magn. Reson. 1990, 89, 479–484. [Google Scholar] [CrossRef]
- Gullion, T.; Schaefer, J. Elimination of resonance offset effects in rotational-echo, double-resonance NMR. J. Magn. Reson. 1991, 92, 439–442. [Google Scholar] [CrossRef]
- Asakura, T.; Aoki, A.; Demura, M.; Joers, J.M.; Rosanske, R.C.; Gullion, T.; James, M.J.; Richard, C.R.; Terry, G. Structure of Bombyx mori Silk Fibroin Studied by REDOR NMR Spectroscopy. Polym. J. 1994, 26, 1405–1408. [Google Scholar] [CrossRef] [Green Version]
- Terry, G.; Raghuvansh, K.; Asakura, T.; Gullion, T.; Kishore, R.; Asakura, T. Determining dihedral angles and local structure in silk peptide by 13C 2H REDOR. J. Am. Chem. Soc. 2003, 125, 7510–7511. [Google Scholar] [CrossRef]
- Gullion, T.; Yamauchi, K.; Okonogi, M.; Asakura, T.; Terry, G.; Yamauchi, K.; Okonogi, M.; Asakura, T. 13C-17O REAPDOR NMR as a tool for determining secondary structure in polyamides. Macromolecules 2007, 40, 1363–1365. [Google Scholar] [CrossRef]
- Lotz, B.; Keith, H.D. Crystal structure of poly(l-Ala-Gly)II. J. Mol. Biol. 1971, 61, 201–215. [Google Scholar] [CrossRef]
- Okuyama, K.; Somashekar, R.; Noguchi, K.; Ichimura, S. Refined molecular and crystal structure of silk I based on Ala–Gly and (Ala–Gly)2—Ser–Gly peptide sequence. Biopolymers 2001, 59, 310–319. [Google Scholar] [CrossRef]
- Asakura, T. NMR of Silk Fibroin, 6. Structure of Bombyx mori silk fibroin in aqueous solution. Makromol. Chem. Rapid Commun. 1986, 7, 755–759. [Google Scholar] [CrossRef]
- Suzuki, Y.Y.; Yamazaki, T.; Aoki, A.; Shindo, H.; Asakura, T. NMR Study of the Structures of Repeated Sequences, GAGXGA (X = S, Y, V), in Bombyx mori Liquid Silk. Biomacromolecules 2014, 15, 104–112. [Google Scholar] [CrossRef]
- Shen, Y.; Bax, A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 2013, 56, 227–241. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Bax, A. Identification of helix capping and β-turn motifs from NMR chemical shifts. J. Biomol. NMR 2012, 52, 211–232. [Google Scholar] [CrossRef]
- Asakura, T.; Watanabe, Y.; Uchida, A.; Minagawa, H. NMR of silk fibroin. 2. 13C NMR study of the chain dynamics and solution structure of Bombyx mori silk fibroin. Macromolecules 1984, 17, 1075–1081. [Google Scholar] [CrossRef]
- Asakura, T.; Watanabe, Y.; Itoh, T. NMR of Silk Fibroin. 3. Assignment of Carbonyl Carbon Resonances and Their Dependence on Sequence and Conformation in Bombyx mori Silk Fibroin Using Selective Isotopic Labeling. Macromolecules 1984, 17, 2421–2426. [Google Scholar] [CrossRef]
- Asakura, T.; Demura, M.; Date, T.; Miyashita, N.; Ogawa, K.; Williamson, M.P. NMR study of silk I structure of Bombyx mori silk fibroin with 15N- and 13C-NMR chemical shift contour plots. Biopolymers 1997, 41, 193–203. [Google Scholar] [CrossRef]
- Yamane, T.; Umemura, K.; Asakura, T. The structural characteristics of Bombyx mori silk fibroin before spinning as studied with molecular dynamics simulation. Macromolecules 2002, 35, 8831–8838. [Google Scholar] [CrossRef]
- Laity, P.R.; Holland, C. The Rheology behind Stress-Induced Solidification in Native Silk Feedstocks. Int. J. Mol. Sci. 2016, 17, 1812. [Google Scholar] [CrossRef] [Green Version]
- Schaefer, C.; Laity, P.R.; Holland, C.; McLeish, T.C.B. Silk Protein Solution: A Natural Example of Sticky Reptation. Macromolecules 2020, 53, 2669–2676. [Google Scholar] [CrossRef] [Green Version]
- Schaefer, C.; Laity, P.R.; Holland, C.; McLeish, T.C.B. Stretching of Bombyx mori Silk Protein in Flow. Molecules 2021, 26, 1663. [Google Scholar] [CrossRef]
- Asakura, T.; Suzuki, Y.; Yazawa, K.; Aoki, A.; Nishiyama, Y.; Nishimura, K.; Suzuki, F.; Kaji, H. Determination of Accurate 1H Positions of (Ala-Gly)n as a Sequential Peptide Model of Bombyx mori Silk Fibroin before Spinning (Silk I). Macromolecules 2013, 46, 8046–8050. [Google Scholar] [CrossRef]
- Yamauchi, K.; Yamasaki, S.; Takahashi, R.; Asakura, T. Microscopic structural analysis of fractured silk fibers from Bombyx mori and Samia cynthia ricini using 13C CP/MAS NMR with a 1 mm microcoil MAS NMR probehead. Solid State Nucl. Magn. Reson. 2010, 38, 27–30. [Google Scholar] [CrossRef]
- Yates, J.R.; Pickard, C.J.; Mauri, F. Calculation of NMR chemical shifts for extended systems using ultrasoft pseudopotentials. Phys. Rev. B 2007, 76, 024401. [Google Scholar] [CrossRef]
- Brown, S.P. Probing proton–proton proximities in the solid state. Prog. Nucl. Magn. Reson. Spectrosc. 2007, 50, 199–251. [Google Scholar] [CrossRef]
- Brinkmann, A.; Litvinov, V.M.; Kentgens, A.P.M. Environmentally friendly flame retardants. A detailed solid-state NMR study of melamine orthophosphate. Magn. Reson. Chem. 2007, 45, S231–S246. [Google Scholar] [CrossRef]
- Bradley, J.P.; Tripon, C.; Filip, C.; Brown, S.P. Determining relative proton-proton proximities from the build-up of two-dimensional correlation peaks in 1H double-quantum MAS NMR: Insight from multi-spin density-matrix simulations. Phys. Chem. Chem. Phys. 2009, 11, 6941–6952. [Google Scholar] [CrossRef]
- Harris, R.K.; Hodgkinson, P.; Zorin, V.; Dumez, J.-N.; Elena-Herrmann, B.; Emsley, L.; Salager, E.; Stein, R.S. Computation and NMR crystallography of terbutaline sulfate. Magn. Reson. Chem. 2010, 48, S103–S112. [Google Scholar] [CrossRef]
- Webber, A.L.; Elena, B.; Griffin, J.M.; Yates, J.R.; Pham, T.N.; Mauri, F.; Pickard, C.J.; Gil, A.M.; Stein, R.; Lesage, A.; et al. Complete 1H resonance assignment of β-maltose from 1H–1H DQ-SQ CRAMPS and 1H (DQ-DUMBO)–13CSQ refocused INEPT 2D solid-state NMR spectra and first principles GIPAW calculations. Phys. Chem. Chem. Phys. 2010, 12, 6970–6983. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.P. Applications of high-resolution 1H solid-state NMR. Solid State Nucl. Magn. Reson. 2012, 41, 1–27. [Google Scholar] [CrossRef]
- Yazawa, K.; Suzuki, F.; Nishiyama, Y.; Ohata, T.; Aoki, A.; Nishimura, K.; Kaji, H.; Shimizu, T.; Asakura, T. Determination of accurate 1H positions of an alanine tripeptide with anti-parallel and parallel β-sheet structures by high resolution 1H solid state NMR and GIPAW chemical shift calculation. Chem. Commun. 2012, 48, 11199–11201. [Google Scholar] [CrossRef]
- Asakura, T.; Yazawa, K.; Horiguchi, K.; Suzuki, F.; Nishiyama, Y.; Nishimura, K.; Kaji, H. Difference in the structures of alanine tri- and tetra-peptides with antiparallel β-sheet assessed by X-ray diffraction, solid-state NMR and chemical shift calculations by GIPAW. Biopolymers 2014, 101, 13–20. [Google Scholar] [CrossRef]
- Pickard, C.J.; Mauri, F. All-electron magnetic response with pseudopotentials: NMR chemical shifts. Phys. Rev. B 2001, 63, 245101. [Google Scholar] [CrossRef] [Green Version]
- Sezutsu, H.; Yukuhiro, K. Dynamic Rearrangement Within the Antheraea pernyi Silk Fibroin Gene Is Associated with Four Types of Repetitive Units. J. Mol. Evol. 2000, 51, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, Y.; Asakura, T. High-Resolution 13C CP/MAS NMR Study on Structure and Structural Transition of Antheraea pernyi Silk Fibroin Containing Poly(L-alanine) and Gly-Rich Regions. Macromolecules 2002, 35, 2393–2400. [Google Scholar] [CrossRef]
- Lu, Q.; Hu, X.; Wang, X.; Kluge, J.A.; Lu, S.; Cebe, P.; Kaplan, D.L. Water-insoluble silk films with silk I structure. Acta Biomater. 2010, 6, 1380–1387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monti, P.; Taddei, P.; Freddi, G.; Asakura, T.; Tsukada, M. Raman spectroscopic characterization of Bombyx mori silk fibroin: Raman spectrum of Silk I. J. Raman Spectrosc. 2001, 32, 103–107. [Google Scholar] [CrossRef]
- Rousseau, M.-E.; Lefèvre, T.; Beaulieu, L.; Asakura, T.; Pézolet, M. Study of protein conformation and orientation in silkworm and spider silk fibers using Raman microspectroscopy. Biomacromolecules 2004, 5, 2247–2257. [Google Scholar] [CrossRef]
- Chen, X.; Shao, Z.; Marinkovic, N.S.; Miller, L.M.; Zhou, P.; Chance, M.R. Conformation transition kinetics of regenerated Bombyx mori silk fibroin membrane monitored by time-resolved FTIR spectroscopy. Biophys. Chem. 2001, 89, 25–34. [Google Scholar] [CrossRef]
- Taddei, P.; Monti, P. Vibrational infrared conformational studies of model peptides representing the semicrystalline domains of Bombyx mori silk fibroin. Biopolymers 2005, 78, 249–258. [Google Scholar] [CrossRef]
- Ming, J.; Pan, F.; Zuo, B. Influence factors analysis on the formation of silk I structure. Int. J. Biol. Macromol. 2015, 75, 398–401. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Han, H.; Huang, X.; Xu, W.; Kaplan, D.L.; Zhu, H.; Lu, Q. Silk scaffolds with tunable mechanical capability for cell differentiation. Acta Biomater. 2015, 20, 22–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua, J.; You, H.; Li, X.; You, R.; Ma, L. Cu(II) ion loading in silk fibroin scaffolds with silk I structure. Int. J. Biol. Macromol. 2020, 158, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Carissimi, G.; Baronio, C.M.; Montalbán, M.G.; Víllora, G.; Barth, A. On the Secondary Structure of Silk Fibroin Nanoparticles Obtained Using Ionic Liquids: An Infrared Spectroscopy Study. Polymers 2020, 12, 1294. [Google Scholar] [CrossRef]













| Data No. | (hkl) | Iobs | Icald | Data No. | (hkl) | Iobs | Icald | Data No. | (hkl) | Iobs | Icald |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | (011) | 75 | 89 | 11 | (122) | 76 | 75 | 17 | (043) | 121 | 100 |
| (020) | (201) | ||||||||||
| 12 | (013) | 49 | 20 | (211) | |||||||
| 2 | (021) | 42 | 45 | (140) | (004) | ||||||
| (220) | |||||||||||
| 3 | (002) | 151 | 164 | 13 | (042) | 115 | 108 | (133) | |||
| (110) | (023) | (014) | |||||||||
| (051) | |||||||||||
| 4 | (012) | 93 | 81 | (141) | 18 | (221) | 68 | 83 | |||
| (031) | (132) | (152) | |||||||||
| (101) | (160) | ||||||||||
| 14 | (033) | 58 | 46 | ||||||||
| 5 | (111) | 32 | 79 | (103) | 19 | (230) | 70 | 47 | |||
| (120) | (113) | (062) | |||||||||
| (024) | |||||||||||
| 6 | (022) | 37 | 42 | 15 | (150) | 142 | 144 | ||||
| (052) | 20 | (161) | 106 | 116 | |||||||
| 7 | (121) | 156 | 128 | (142) | (231) | ||||||
| (040) | (060) | (202) | |||||||||
| (151) | (212) | ||||||||||
| 8 | (130) | 42 | 35 | (123) | (053) | ||||||
| (041) | (200) | (043) | |||||||||
| 9 | (032) | 43 | 49 | 16 | (210) | 63 | 36 | 21 | (034) | 74 | 63 |
| (102) | (061) | (104) | |||||||||
| (071) | |||||||||||
| 10 | (112) | 123 | 129 | ||||||||
| (131) |
| DQ Correlation | Intra-Molecular | Inter-Molecular | ||||
|---|---|---|---|---|---|---|
| ① | G-Hɴ | A-Hɴ | 2.83 | (5 3) | 3.43 | (4 3) |
| ② | G-Hɴ | A-Hα | 2.12 | (5 2) | 3.46 | (4 3) |
| ③ | G-Hɴ | G-Hα1 | 2.30 | (5 3) | ||
| ④ | G-Hɴ | G-Hα2 | 2.88 | (5 3) | 2.76, 3.42 | (2 5),(4 3) |
| ⑤ | A-Hɴ | A-Hα | 2.86, 3.52 | (5 3),(5 2) | 2.96 | (4 3) |
| ⑥ | A-Hɴ | G-Hα1 | 3.28, 3.99 | (5 3),(5 2) | ||
| ⑦ | A-Hɴ | G-Hα2 | 3.47 | (5 3) | ||
| ⑧ | G-Hɴ | A-Hβ | 3.89 | (5 2) | ||
| ⑨ | A-Hɴ | A-Hβ | 2.87 | (5 3) | ||
| ⑩ | A-Hα | G-Hα1 | 3.23 | (6 3) | ||
| ⑪ | A-Hα | G-Hα2 | 3.66 | (9 4) | ||
| ⑫ | A-Hα | A-Hβ | 2.70 | (5 3) | ||
| ⑬ | G-Hα1 | A-Hβ | 3.65 | (5 4) | 3.56 | (2 4) |
| ⑭ | G-Hα2 | A-Hβ | 3.60 | (3 4) | ||
| ⑮ | G-Hα1 | G-Hα2 | 1.77 | (5 3) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asakura, T. Structure of Silk I (Bombyx mori Silk Fibroin before Spinning) -Type II β-Turn, Not α-Helix-. Molecules 2021, 26, 3706. https://doi.org/10.3390/molecules26123706
Asakura T. Structure of Silk I (Bombyx mori Silk Fibroin before Spinning) -Type II β-Turn, Not α-Helix-. Molecules. 2021; 26(12):3706. https://doi.org/10.3390/molecules26123706
Chicago/Turabian StyleAsakura, Tetsuo. 2021. "Structure of Silk I (Bombyx mori Silk Fibroin before Spinning) -Type II β-Turn, Not α-Helix-" Molecules 26, no. 12: 3706. https://doi.org/10.3390/molecules26123706