Lapatinib, Nilotinib and Lomitapide Inhibit Haemozoin Formation in Malaria Parasites
Abstract
:1. Introduction
2. Results and Discussion
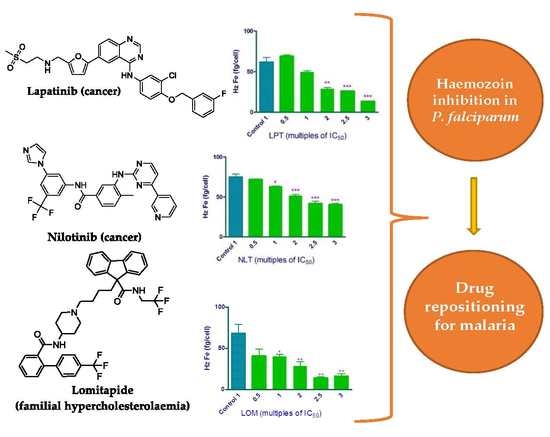
2.1. In Vitro Assays of the USFDA Approved Drugs
2.2. Structure-Based Virtual Screening Against the β-haematin Crystal
2.3. Haem Fractionation Assay
2.4. Ligand-Based Virtual Screening Using Lapatinib and Nilotinib as Templates for Shape and Eletrostatic Similarity
3. Materials and Methods
3.1. Compounds
3.2. Detergent Mediated Assay for β-haematin Inhibition
3.3. Plasmodium Lactate Dehydrogenase (pLDH) Assay
3.4. Structure-based Virtual Screening against the β-haematin Crystal
3.5. Haem Fractionation Assay
3.6. Statistical Analysis
3.7. Ligand-based Virtual Screening
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- WHO. World Malaria Report 2018; World Health Organisation: Geneva, Switzerland, 2018; Available online: https://www.who.int/malaria/publications/world-malaria-report-2018/report/en/ (accessed on 29 March 2020).
- Combrinck, J.M.; Mabotha, T.E.; Ncokazi, K.K.; Ambele, M.A.; Taylor, D.; Smith, P.J.; Hoppe, H.C.; Egan, T.J. Insights into the role of heme in the mechanism of action of antimalarials. ACS Chem. Biol. 2013, 8, 133–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buller, R.; Peterson, M.L.; Almarsson, O.; Leiserowitz, L. Quinoline binding site on malaria pigment crystal: A rational pathway for antimalaria drug design. Cryst. Growth Des. 2002, 2, 553–562. [Google Scholar] [CrossRef]
- Olafson, K.N.; Ketchum, M.A.; Rimer, J.D.; Vekilov, P.G. Mechanisms of hematin crystallization and inhibition by the antimalarial drug chloroquine. Proc. Natl. Acad. Sci. USA 2015, 112, 4946–4951. [Google Scholar] [CrossRef] [Green Version]
- Olafson, K.N.; Nguyen, T.Q.; Rimer, J.D.; Vekilov, P.G. Antimalarials inhibit hematin crystallization by unique drug-surface site interactions. Proc. Natl. Acad. Sci. USA 2017, 114, 7531–7536. [Google Scholar] [CrossRef] [Green Version]
- Fidock, D.A.; Nomura, T.; Talley, A.K.; Cooper, R.A.; Dzekunov, S.M.; Ferdig, M.T.; Ursos, L.M.; Sidhu, A.B.; Naude, B.; Deitsch, K.W.; et al. Mutations in the p. Falciparum digestive vacuole transmembrane protein pfcrt and evidence for their role in chloroquine resistance. Mol. Cell 2000, 6, 861–871. [Google Scholar] [CrossRef]
- Chaparro, M.J.; Calderon, F.; Castaneda, P.; Fernandez-Alvaro, E.; Gabarro, R.; Gamo, F.J.; Gomez-Lorenzo, M.G.; Martin, J.; Fernandez, E. Efforts aimed to reduce attrition in antimalarial drug discovery: A systematic evaluation of the current antimalarial targets portfolio. ACS Infect. Dis. 2018, 4, 568–576. [Google Scholar] [CrossRef]
- Forli, S. Charting a path to success in virtual screening. Molecules 2015, 20, 18732–18758. [Google Scholar] [CrossRef] [Green Version]
- Fradera, X.; Babaoglu, K. Overview of methods and strategies for conducting virtual small molecule screening. Curr. Protoc. Chem. Biol. 2017, 9, 196–212. [Google Scholar] [CrossRef]
- Shoichet, B.K. Virtual screening of chemical libraries. Nature 2004, 432, 862–865. [Google Scholar] [CrossRef]
- Gupta, M.; Sharma, R.; Kumar, A. Docking techniques in pharmacology: How much promising? Comput. Biol. Chem. 2018, 76, 210–217. [Google Scholar] [CrossRef]
- Slater, O.; Kontoyianni, M. The compromise of virtual screening and its impact on drug discovery. Expert Opin. Drug Discov. 2019, 14, 619–637. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, A.C.C.; Combrinck, J.M.; Maepa, K.; Egan, T.J. Virtual screening as a tool to discover new β-haematin inhibitors with activity against malaria parasites. Sci. Rep. 2020, 10, 3374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klebe, G. Virtual ligand screening: Strategies, perspectives and limitations. Drug Discov. Today 2006, 11, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Wicht, K.J.; Combrinck, J.M.; Smith, P.J.; Egan, T.J. Bayesian models trained with hts data for predicting beta-haematin inhibition and in vitro antimalarial activity. Bioorg. Med. Chem. 2015, 23, 5210–5217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandlin, R.D.; Carter, M.D.; Lee, P.J.; Auschwitz, J.M.; Leed, S.E.; Johnson, J.D.; Wright, D.W. Use of the np-40 detergent-mediated assay in discovery of inhibitors of beta-hematin crystallization. Antimicrob. Agents Chemother. 2011, 55, 3363–3369. [Google Scholar] [CrossRef] [Green Version]
- Guglielmo, S.; Bertinaria, M.; Rolando, B.; Crosetti, M.; Fruttero, R.; Yardley, V.; Croft, S.L.; Gasco, A. A new series of amodiaquine analogues modified in the basic side chain with in vitro antileishmanial and antiplasmodial activity. Eur. J. Med. Chem. 2009, 44, 5071–5079. [Google Scholar] [CrossRef]
- Carter, M.D.; Phelan, V.V.; Sandlin, R.D.; Bachmann, B.O.; Wright, D.W. Lipophilic mediated assays for beta-hematin inhibitors. Comb. Chem. High Throughput Screen. 2010, 13, 285–292. [Google Scholar] [CrossRef]
- Sandlin, R.D.; Fong, K.Y.; Wicht, K.J.; Carrell, H.M.; Egan, T.J.; Wright, D.W. Identification of beta-hematin inhibitors in a high-throughput screening effort reveals scaffolds with in vitro antimalarial activity. Int. J. Parasitol. Drugs Drug Resist. 2014, 4, 316–325. [Google Scholar] [CrossRef] [Green Version]
- Ncokazi, K.K.; Egan, T.J. A colorimetric high-throughput beta-hematin inhibition screening assay for use in the search for antimalarial compounds. Anal. Biochem. 2005, 338, 306–319. [Google Scholar] [CrossRef]
- L’Abbate, F.P.; Muller, R.; Openshaw, R.; Combrinck, J.M.; de Villiers, K.A.; Hunter, R.; Egan, T.J. Hemozoin inhibiting 2-phenylbenzimidazoles active against malaria parasites. Eur. J. Med. Chem. 2018, 159, 243–254. [Google Scholar] [CrossRef]
- Woodland, J.G.; Hunter, R.; Smith, P.J.; Egan, T.J. Shining new light on ancient drugs: Preparation and subcellular localisation of novel fluorescent analogues of cinchona alkaloids in intraerythrocytic plasmodium falciparum. Org. Biomol. Chem. 2017, 15, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Woodland, J.G.; Hunter, R.; Smith, P.J.; Egan, T.J. Chemical proteomics and super-resolution imaging reveal that chloroquine interacts with plasmodium falciparum multidrug resistance-associated protein and lipids. ACS Chem. Biol. 2018, 13, 2939–2948. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, S.; Waters, N.C.; Chavchich, M. Leveraging cell cycle analysis in anticancer drug discovery to identify novel plasmodial drug targets. Infect. Disord. Drug Targets 2010, 10, 165–190. [Google Scholar] [CrossRef]
- Bhullar, K.S.; Lagaron, N.O.; McGowan, E.M.; Parmar, I.; Jha, A.; Hubbard, B.P.; Rupasinghe, H.P.V. Kinase-targeted cancer therapies: Progress, challenges and future directions. Mol. Cancer 2018, 17, 48. [Google Scholar] [CrossRef]
- Cabrera, D.G.; Horatscheck, A.; Wilson, C.R.; Basarab, G.; Eyermann, C.J.; Chibale, K. Plasmodial kinase inhibitors: License to cure? J. Med. Chem. 2018, 61, 8061–8077. [Google Scholar] [CrossRef]
- Pathak, V.; Colah, R.; Ghosh, K. Tyrosine kinase inhibitors: New class of antimalarials on the horizon? Blood Cells Mol. Dis. 2015, 55, 119–126. [Google Scholar] [CrossRef]
- Kesely, K.R.; Pantaleo, A.; Turrini, F.M.; Olupot-Olupot, P.; Low, P.S. Inhibition of an erythrocyte tyrosine kinase with imatinib prevents plasmodium falciparum egress and terminates parasitemia. PLoS ONE 2016, 11, e0164895. [Google Scholar] [CrossRef]
- Lotharius, J.; Gamo-Benito, F.J.; Angulo-Barturen, I.; Clark, J.; Connelly, M.; Ferrer-Bazaga, S.; Parkinson, T.; Viswanath, P.; Bandodkar, B.; Rautela, N.; et al. Repositioning: The fast track to new anti-malarial medicines? Malar. J. 2014, 13, 143. [Google Scholar] [CrossRef] [Green Version]
- Ishiyama, A.; Iwatsuki, M.; Hokari, R.; Sawa, M.; Omura, S.; Otoguro, K. Antimalarial activity of kinase inhibitor, nilotinib, in vitro and in vivo. J. Antibiot. (Tokyo) 2015, 68, 469–472. [Google Scholar] [CrossRef]
- Irwin, J.J.; Shoichet, B.K. Zinc—A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef] [Green Version]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with pyrx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar] [PubMed]
- Derbyshire, E.R.; Prudencio, M.; Mota, M.M.; Clardy, J. Liver-stage malaria parasites vulnerable to diverse chemical scaffolds. Proc. Natl. Acad. Sci. USA 2012, 109, 8511–8516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goulooze, S.C.; Cohen, A.F.; Rissmann, R. Lomitapide. Br. J. Clin. Pharm. 2015, 80, 179–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Njoroge, M.; Njuguna, N.M.; Mutai, P.; Ongarora, D.S.; Smith, P.W.; Chibale, K. Recent approaches to chemical discovery and development against malaria and the neglected tropical diseases human african trypanosomiasis and schistosomiasis. Chem. Rev. 2014, 114, 11138–11163. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef]
- Egan, T.J.; Warner, D.F. Editorial overview: Tuberculosis, malaria and schistosomiasis; understanding resistance and development of new drugs. Curr. Opin. Pharm. 2018, 42, iv–vi. [Google Scholar] [CrossRef]
- Chua, M.J.; Arnold, M.S.; Xu, W.; Lancelot, J.; Lamotte, S.; Spath, G.F.; Prina, E.; Pierce, R.J.; Fairlie, D.P.; Skinner-Adams, T.S.; et al. Effect of clinically approved hdac inhibitors on plasmodium, leishmania and schistosoma parasite growth. Int. J. Parasitol. Drugs Drug Resist. 2017, 7, 42–50. [Google Scholar] [CrossRef]
- Kurosawa, Y.; Dorn, A.; Kitsuji-Shirane, M.; Shimada, H.; Satoh, T.; Matile, H.; Hofheinz, W.; Masciadri, R.; Kansy, M.; Ridley, R.G. Hematin polymerization assay as a high-throughput screen for identification of new antimalarial pharmacophores. Antimicrob. Agents Chemother. 2000, 44, 2638–2644. [Google Scholar] [CrossRef] [Green Version]
- Rush, M.A.; Baniecki, M.L.; Mazitschek, R.; Cortese, J.F.; Wiegand, R.; Clardy, J.; Wirth, D.F. Colorimetric high-throughput screen for detection of heme crystallization inhibitors. Antimicrob. Agents Chemother. 2009, 53, 2564–2568. [Google Scholar] [CrossRef] [Green Version]
- Fong, K.Y.; Sandlin, R.D.; Wright, D.W. Identification of beta-hematin inhibitors in the mmv malaria box. Int. J. Parasitol. Drugs Drug Resist. 2015, 5, 84–91. [Google Scholar] [CrossRef] [Green Version]
- Huy, N.T.; Chi, P.L.; Nagai, J.; Dang, T.N.; Mbanefo, E.C.; Ahmed, A.M.; Long, N.P.; Thoa, L.T.; Hung, L.P.; Titouna, A.; et al. High-throughput screening and prediction model building for novel hemozoin inhibitors using physicochemical properties. Antimicrob. Agents Chemother. 2017, 61, e01607-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Combrinck, J.M.; Fong, K.Y.; Gibhard, L.; Smith, P.J.; Wright, D.W.; Egan, T.J. Optimization of a multi-well colorimetric assay to determine haem species in plasmodium falciparum in the presence of anti-malarials. Malar. J. 2015, 14, 253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sander, T.; Freyss, J.; von Korff, M.; Rufener, C. Datawarrior: An open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Nie, A.; An, J.; Huang, Z. Structure-based virtual screening of chemical libraries for drug discovery. Curr. Opin. Chem. Biol. 2006, 10, 194–202. [Google Scholar] [CrossRef]
- Naylor, E.; Arredouani, A.; Vasudevan, S.R.; Lewis, A.M.; Parkesh, R.; Mizote, A.; Rosen, D.; Thomas, J.M.; Izumi, M.; Ganesan, A.; et al. Identification of a chemical probe for naadp by virtual screening. Nat. Chem. Biol. 2009, 5, 220–226. [Google Scholar] [CrossRef] [Green Version]
- Rush, T.S., 3rd; Grant, J.A.; Mosyak, L.; Nicholls, A. A shape-based 3-d scaffold hopping method and its application to a bacterial protein-protein interaction. J. Med. Chem. 2005, 48, 1489–1495. [Google Scholar] [CrossRef]
- Da Costa, G.V.; Ferreira, E.F.B.; Ramos, R.S.; da Silva, L.B.; de Sa, E.M.F.; da Silva, A.K.P.; Lobato, C.M.; Souto, R.N.P.; da Silva, C.H.T.P.; Federico, L.B.; et al. Hierarchical virtual screening of potential insectides inhibitors of acetylcholinesterase and juvenile hormone from temephos. Pharmaceuticals (Basel) 2019, 12, 61. [Google Scholar] [CrossRef] [Green Version]
- Trager, W.; Jensen, J.B. Human malaria parasites in continuous culture. Science 1976, 193, 673–675. [Google Scholar] [CrossRef]
- Makler, M.T.; Ries, J.M.; Williams, J.A.; Bancroft, J.E.; Piper, R.C.; Gibbins, B.L.; Hinrichs, D.J. Parasite lactate dehydrogenase as an assay for plasmodium falciparum drug sensitivity. Am. J. Trop. Med. Hyg. 1993, 48, 739–741. [Google Scholar] [CrossRef]
- Pagola, S.; Stephens, P.W.; Bohle, D.S.; Kosar, A.D.; Madsen, S.K. The structure of malaria pigment (β-haematin). Nature 2000, 404, 307–310. [Google Scholar] [CrossRef]
- Schrodinger, LLC. The Pymol Molecular Graphics System, version 2.2.0; Schrödinger: New York, NY, USA, 2015. [Google Scholar]
Sample Availability: All materials were obtained from specified commercial sources and are available for purchase. |



| Bayesian Rank | Drug | β-haematin IC50 (μM) | PfNF54 IC50 (µM) | PfK1 IC50 (µM) | RI 2 |
|---|---|---|---|---|---|
| 1 | Lapatinib | 5.43 ± 0.03 | 0.26 ± 0.07 | 0.85 ± 0.02 | 3.3 |
| 2 | Amodiaquine | 21 4 | NT 3 | 0.0086 5 | - |
| 3 | Imatinib | 99 ± 5 | 3.9 ± 0.1 | NT 3 | - |
| 4 | Nafarelin | 48 ± 5 | NA 1 | NT 3 | - |
| 5 | Nilotinib | 7 ± 1 | 0.28 ± 0.02 | 0.38 ± 0.06 | 1.4 |
| 6 | Antrafenine | NT 3 | NT 3 | NT 3 | - |
| 7 | Vapreotide | 48 ± 7 | NA 1 | NT 3 | - |
| 8 | Gefitinib | 20.4 ± 0.8 | 11.5 ± 0.4 | NT 3 | - |
| 13 | Thiabendazole | > 500 | NA 1 | NT 3 | - |
| 14 | Erlotinib | 17.6 ± 0.2 | 13.3 ± 0.4 | NT 3 | - |
| 19 | Lomitapide | 68.2 ± 0.9 | 0.56 ± 0.10 | 0.69 ± 0.14 | 1.2 |
| 21 | Chloroquine | 26 ± 0.3 | 0.015 ± 0.001 | 0.272 ± 0.005 | 18 |
| 1505 | Dantrolene | >500 | NT 3 | NT 3 | - |
| 1506 | Nitrendipine | >500 | NT 3 | NT 3 | - |
| 1508 | Nimodipine | >500 | NT 3 | NT 3 | - |
| 1510 | Acenocumarol | >500 | NT 3 | NT 3 | - |
| Zinc Code | Binding Affinity | Drug | β-haematin IC50 (μM) | PfNF54 IC50 (µM) |
|---|---|---|---|---|
| zinc000001550499 | −13.4 | Cinacalcet | NA 1 | - |
| zinc000052716421 | −12.9 | Flibanserin | NA 1 | - |
| zinc000000601229 | −12.3 | Azelastine | NA 1 | - |
| zinc000012503187 | −12.3 | Conivaptan | NA 1 | - |
| zinc000022010649 | −12.2 | Panobinostat | - 2 | - |
| zinc000003816514 | −12 | Rolapitant | - 2 | - |
| zinc000003784182 | −12 | Adapalene | NA 1 | - |
| zinc000003978005 | −12 | Dihydroergotamine | NA 1 | - |
| zinc000019632618 | −12 | Imatinib | 99.0 ± 4.76 | 3.9 ± 0.14 |
| zinc000000968263 | −11.9 | Cyclobenzaprine | NA 1 | - |
| zinc000003816287 | −11.9 | Axitinib | NA 1 | - |
| zinc000027990463 | −11.8 | Lomitapide | 68.2 ± 0.85 | 0.56 ± 0.10 |
| zinc000006716957 | −11.8 | Nilotinib | 7.3 ± 1.2 | 0.28 ± 0.02 |
| zinc000001530977 | −11.8 | Naftifine | NA 1 | - |
| zinc000011681563 | −11.8 | Netupitant | - 2 | - |
| zinc000000025958 | −11.7 | Rucaparib | NA 1 | - |
| zinc000000538312 | −11.6 | Risperidone | NA 1 | - |
| zinc000068202099 | −11.6 | Sonidegib | NA 1 | - |
| zinc000003776633 | −11.5 | Acrivastine | NA 1 | - |
| zinc000003798734 | −11.5 | Acitretin | NA 1 | - |
| zinc000000968264 | −11.5 | Cyproheptadine | NA 1 | - |
| zinc000000001261 | −11.4 | Desloratadine | NA 1 | - |
| zinc000001550477 | −11.4 | Lapatinib | 5.43 ± 0.03 | 0.26 ± 0.07 |
| zinc000003939013 | −11.3 | Fosaprepitant | NA 1 | - |
| zinc000070466416 | −11.2 | Cabozantinib | - 2 | - |
| zinc000001530886 | −11.2 | Telmisartan | 51.07 ± 0.67 | 4.99 ± 1.7 |
| zinc000019144231 | −7.4 | (S)-Chloroquine 3 | - 2 | - |
| zinc000019144226 | −8 | (R)-Chloroquine 3 | - 2 | - |
| - | - | (±)-Chloroquine 3 | 26 ± 0.26 | 0.015 ± 0.001 |
| Rank EON | Chemdiv Code | Structure | β-haematin IC50 (μM) | PfNF54 IC50 (µM) |
|---|---|---|---|---|
| Query molecule | Lapatinib |  | 5.43 ± 0.03 | 0.26 ± 0.07 |
| 17 | F424-1065 |  | 353.9 ± 59.96 | 4.91 ± 0.44 |
| 24 | C514-0466 |  | 143.55 ± 32.88 | NA 1 |
| 27 | C514-0233 |  | 254.8 ± 59.26 | NA 1 |
| 48 | G637-3522 |  | 290.05 ± 42.07 | NA 1 |
| 54 | F731-0128 |  | 207.15 ± 19.02 | NA 1 |
| 62 | C514-0129 |  | 183.5 ± 4.67 | NA 1 |
| Query molecule | Nilotinib |  | 7.3 ± 1.2 | 0.28 ± 0.02 |
| 2 | F871-0652 |  | 156.15 ± 1.63 | 6.42 ± 0.57 |
| 16 | F871-0649 |  | 373.4 ± 98.0 | 8.09 ± 0.12 |
| 42 | F725-0689 |  | 85.73 ± 40.41 | 3.9 ± 0.14 |
| 79 | G416-5459 |  | 100.95 ± 22.85 | NA 1 |
| 80 | D444-0908 |  | 68.25 ± 6.41 | 3.84 ± 0.33 |
| 113 | F545-1038 |  | 243.9 ± 11.46 | NA 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Sousa, A.C.C.; Maepa, K.; Combrinck, J.M.; Egan, T.J. Lapatinib, Nilotinib and Lomitapide Inhibit Haemozoin Formation in Malaria Parasites. Molecules 2020, 25, 1571. https://doi.org/10.3390/molecules25071571
de Sousa ACC, Maepa K, Combrinck JM, Egan TJ. Lapatinib, Nilotinib and Lomitapide Inhibit Haemozoin Formation in Malaria Parasites. Molecules. 2020; 25(7):1571. https://doi.org/10.3390/molecules25071571
Chicago/Turabian Stylede Sousa, Ana Carolina C., Keletso Maepa, Jill M. Combrinck, and Timothy J. Egan. 2020. "Lapatinib, Nilotinib and Lomitapide Inhibit Haemozoin Formation in Malaria Parasites" Molecules 25, no. 7: 1571. https://doi.org/10.3390/molecules25071571