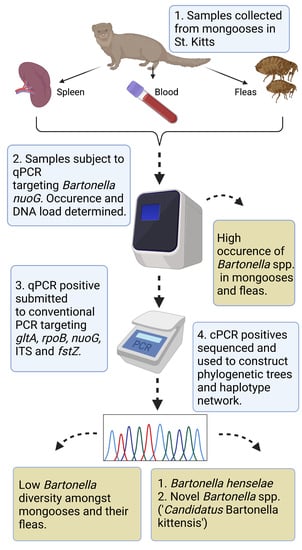
Molecular Survey and Genetic Diversity of Bartonella spp. in Small Indian Mongooses (Urva auropunctata) and Their Fleas on Saint Kitts, West Indies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics
2.2. Sampling
2.2.1. Post-Mortem Samples
2.2.2. Live Animal Samples
2.3. Flea Identification, DNA Extraction/Purification, and Quantification
2.4. DNA Integrity
2.5. Molecular Survey of Bartonella spp.
2.6. Molecular Characterization of Bartonella spp.
2.7. BLAST Analysis
2.8. Phylogenetic Analysis
2.9. Haplotype Analysis (Genetic Diversity)
3. Results
3.1. Amplifiable DNA, and Bartonella spp. Survey
3.2. Molecular Characterization and BLAST Analysis
3.3. Phylogenetic Analysis
3.4. Haplotype Analysis (Genetic Diversity)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chomel, B.B.; Boulouis, H.J.; Breitschwerdt, E.B.; Kasten, R.W.; Vayssier-Taussat, M.; Birtles, R.J.; Koehler, J.E.; Dehio, C. Ecological Fitness and Strategies of Adaptation of Bartonella Species to Their Hosts and Vectors. Vet. Res. 2009, 40, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, C.Y.; Kasten, R.W.; Paff, S.M.; Van Horn, B.A.; Vayssier-Taussat, M.; Boulouis, H.J.; Chomel, B.B. Bartonella spp. DNA Associated with Biting Flies from California. Emerg. Infect. Dis. 2004, 10, 1311–1313. [Google Scholar] [CrossRef] [PubMed]
- Breitschwerdt, E. Bartonellosis: One Health Perspectives for an Emerging Infectious Disease. ILAR J. 2014, 55, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Bergh, K.; Bevanger, L.; Hanssen, I.; Løseth, K. Low Prevalence of Bartonella henselae Infections in Norwegian Domestic and Feral Cats. APMIS 2002, 110, 309–314. [Google Scholar] [CrossRef]
- Chomel, B.B.; Carlos, E.T.; Kasten, R.W.; Yamamoto, K.; Chang, C.C.; Carlos, R.S.; Abenes, M.V.; Pajares, C.M. Bartonella henselae and Bartonella clarridgeiae Infection in Domestic Cats from the Philippines. Am. J. Trop. Med. Hyg. 1999, 60, 593–597. [Google Scholar] [CrossRef] [Green Version]
- Seubert, A.; Schulein, R.; Dehio, C. Bacterial Persistence within Erythrocytes: A Unique Pathogenic Strategy of Bartonella spp. Int. J. Med. Microbiol. 2001, 291, 555–560. [Google Scholar] [CrossRef]
- Harms, A.; Dehio, C. Intruders below the Radar: Molecular Pathogenesis of Bartonella spp. Clin. Microbiol. Rev. 2012, 25, 42–78. [Google Scholar] [CrossRef] [Green Version]
- Koehler, J.E.; Glaser, C.A.; Tappero, J.W. Rochalimaea Henselae Infection: A New Zoonosis With the Domestic Cat as Reservoir. JAMA J. Am. Med. Assoc. 1994, 271, 531–535. [Google Scholar] [CrossRef]
- Avidor, B.; Graidy, M.; Efrat, G.; Leibowitz, C.; Shapira, G.; Schattner, A.; Zimhony, O.; Giladi, M. Bartonella koehlerae, a New Cat-Associated Agent of Culture-Negative Human Endocarditis. J. Clin. Microbiol. 2004, 42, 3462–3468. [Google Scholar] [CrossRef] [Green Version]
- Roux, V.; Eykyn, S.J.; Wyllie, S.; Raoult, D. Bartonella vinsonii subsp. Berkhoffii as an Agent of Afebrile Blood Culture-Negative Endocarditis in a Human. J. Clin. Microbiol. 2000, 38, 1698–1700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonçalves, L.R.; Favacho, A.R.D.M.; Roque, A.L.R.; Mendes, N.S.; Fidelis, O.L.; Benevenute, J.L.; Herrera, H.M.; D’Andrea, P.S.; de Lemos, E.R.S.; Machado, R.Z.; et al. Association of Bartonella Species with Wild and Synanthropic Rodents in Different Brazilian Biomes. Appl. Environ. Microbiol. 2016, 82, 7154–7164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosoy, M.; Goodrich, I. Comparative Ecology of Bartonella and Brucella Infections in Wild Carnivores. Front. Vet. Sci. 2019, 5, 322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- André, M.R.; Gutiérrez, R.; Ikeda, P.; do Amaral, R.B.; de Sousa, K.C.M.; Nachum-Biala, Y.; Lima, L.; Teixeira, M.M.G.; Machado, R.Z.; Harrus, S. Genetic Diversity of Bartonella spp. in Vampire Bats from Brazil. Transbound. Emerg. Dis. 2019, 66, 2329–2341. [Google Scholar] [CrossRef] [PubMed]
- Nellis, D.W.; Everard, C.O.R. The Biology of the Mongoose in the Caribbean. Stud. Fauna Curaçao Other Caribb. Islands 1983, 64, 1–162. [Google Scholar]
- Louppe, V.; Leroy, B.; Herrel, A.; Veron, G. The Globally Invasive Small Indian Mongoose Urva Auropunctata Is Likely to Spread with Climate Change. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Everard, C.O.R.; Everard, J.D. Mongoose Rabies. Rev. Infect. Dis. 1988, 10, S610–S614. [Google Scholar] [CrossRef]
- Shiokawa, K.; Llanes, A.; Hindoyan, A.; Cruz-Martinez, L.; Welcome, S.; Rajeev, S. Peridomestic Small Indian Mongoose: An Invasive Species Posing as Potential Zoonotic Risk for Leptospirosis in the Caribbean. Acta Trop. 2019, 190, 166–170. [Google Scholar] [CrossRef]
- Miller, S.; Zieger, U.; Ganser, C.; Satterlee, S.A.; Bankovich, B.; Amadi, V.; Hariharan, H.; Stone, D.; Wisely, S.M. Influence of Land Use and Climate on Salmonella Carrier Status in the Small Indian Mongoose (Herpestes auropunctatus) in Grenada, West Indies. J. Wildl. Dis. 2015, 51, 60–68. [Google Scholar] [CrossRef]
- Choudhary, S.; Zieger, U.; Sharma, R.N.; Chikweto, A.; Tiwari, K.P.; Ferreira, L.R.; Oliveira, S.; Barkley, L.J.; Verma, S.K.; Kwok, O.C.H.; et al. Isolation and Rflp Genotyping of Toxoplasma Gondii From the Mongoose (Herpestes auropunctatus) in Grenada, West Indies. J. Zoo Wildl. Med. 2013, 44, 1127–1130. [Google Scholar] [CrossRef]
- Rhynd, K.J.R.; Leighton, P.A.; Elcock, D.A.; Whitehall, P.J.; Rycroft, A.; Macgregor, S.K. Prevalence of Salmonella spp. and Thermophilic Campylobacter spp. in the Small Asian Mongoose (Herpestes javanicus) in Barbados, West Indies. J. Zoo Wildl. Med. 2014, 45, 911–914. [Google Scholar] [CrossRef]
- Sato, S.; Kabeya, H.; Shigematsu, Y.; Sentsui, H.; Une, Y.; Minami, M.; Murata, K.; Ogura, G.; Maruyama, S. Small Indian Mongooses and Masked Palm Civets Serve as New Reservoirs of Bartonella henselae and Potential Sources of Infection for Humans. Clin. Microbiol. Infect. 2013, 19, 1181–1187. [Google Scholar] [CrossRef] [Green Version]
- Jaffe, D.A.; Chomel, B.B.; Kasten, R.W.; Breitschwerdt, E.B.; Maggi, R.G.; McLeish, A.; Zieger, U. Bartonella henselae in Small Indian Mongooses (Herpestes auropunctatus) from Grenada, West Indies. Vet. Microbiol. 2018, 216, 119–122. [Google Scholar] [CrossRef]
- Kelly, P.J.; Moura, L.; Miller, T.; Thurk, J.; Perreault, N.; Weil, A.; Maggio, R.; Lucas, H.; Breitschwerdt, E. Feline Immunodeficiency Virus, Feline Leukemia Virus and Bartonella Species in Stray Cats on St Kitts, West Indies. J. Feline Med. Surg. 2010, 12, 447–450. [Google Scholar] [CrossRef]
- Huang, K.; Kelly, P.J.; Zhang, J.; Yang, Y.; Liu, W.; Kalalah, A.; Wang, C. Molecular Detection of Bartonella spp. In China and St. Kitts. Can. J. Infect. Dis. Med. Microbiol. 2019, 2019, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Reeves, W.K.; Beck, J.; Orlova, M.V.; Daly, J.L.; Pippin, K.; Revan, F.; Loftis, A.D. Ecology of Bats, Their Ectoparasites, and Associated Pathogens on Saint Kitts Island. J. Med. Entomol. 2016, 53, 1218–1225. [Google Scholar] [CrossRef]
- Fang, K.; Philpot, K.; Chi, X.; Ketzis, J.; Du, A.; Yao, C. Small Indian Mongooses (Herpestes auropunctatus) Serve As Reservoirs of Bartonella henselae and Rickettsia felis Vectored by Ctenocephalides felis. Vector Borne Zoonotic Dis. 2021, 21, 422–431. [Google Scholar] [CrossRef]
- Sauvé, C.; Berentsen, A.; Conan, A.; Criuz-Martinez, L.; Gilbert, A.; Leighton, P. Habitat-Specific Mongoose Density Estimates and Factors Affecting Traping Success, a Field Study in St. Kitts, West Indies. Prep 2021, 1, 1–10. [Google Scholar]
- Leary, S.; Underwood, W.; Anthony, R.; Cartner, S.; Corey, D.; Grandin, T.; Gwaltney-Brant, S.; McCrackin, M.A.; Greenacre, C.; Meyer, R.; et al. AVMA Guidelines for the Euthanasia of Animals: 2013 Edition. Sci. World 2013, 2, 1–102. [Google Scholar]
- Linardi, P.M.; Santos, J.L.C. Ctenocephalides Felis Felis vs. Ctenocephalides Canis (Siphonaptera: Pulicidae): Some Issues in Correctly Identify These Species. Rev. Bras. Parasitol. Vet. 2012, 21, 345–354. [Google Scholar] [CrossRef]
- Jansson, L.; Hedman, J. Challenging the Proposed Causes of the PCR Plateau Phase. Biomol. Detect. Quantif. 2019, 17, 100082. [Google Scholar] [CrossRef]
- Ferreira, E.C.; Gontijo, C.M.; Cruz, I.; Melo, M.N.; Silva, A.M. Alternative PCR Protocol Using a Single Primer Set for Assessing DNA Quality in Several Tissues from a Large Variety of Mammalian Species Living in Areas Endemic for Leishmaniasis. Mem. Inst. Oswaldo Cruz 2010, 105, 895–898. [Google Scholar] [CrossRef] [PubMed]
- Reif, K.E.; Stout, R.W.; Henry, G.C.; Foil, L.D.; Macaluso, K.R. Prevalence and Infection Load Dynamics of Rickettsia Felis in Actively Feeding Cat Fleas. PLoS ONE 2008, 3, e2805. [Google Scholar] [CrossRef]
- André, M.R.; Dumler, J.S.; Herrera, H.M.; Gonçalves, L.R.; de Sousa, K.C.M.; Scorpio, D.G.; de Santis, A.C.G.A.; Domingos, I.H.; de Macedo, G.C.; Machado, R.Z. Assessment of a Quantitative 5’ Nuclease Real-Time Polymerase Chain Reaction Using the Nicotinamide Adenine Dinucleotide Dehydrogenase Gamma Subunit (NuoG) for Bartonella Species in Domiciled and Stray Cats in Brazil. J. Feline Med. Surg. 2015, 18, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Müller, A.; Rodríguez, E.; Walker, R.; Bittencourt, P.; Pérez-Macchi, S.; Gonçalves, L.R.; Machado, R.Z.; André, M.R. Occurrence and Genetic Diversity of Bartonella spp. (Rhizobiales: Bartonellaceae) and Rickettsia spp. (Rickettsiales: Rickettsiaceae) in Cat Fleas (Siphonaptera: Pulicidae) From Chile. J. Med. Entomol. 2018, 55, 1627–1632. [Google Scholar] [CrossRef] [PubMed]
- Billeter, S.A.; Gundi, V.A.K.B.; Rood, M.P.; Kosoy, M.Y. Molecular Detection and Identification of Bartonella Species in Xenopsylla Cheopis Fleas (Siphonaptera: Pulicidae) Collected from Rattus Norvegicus Rats in Los Angeles, California. Appl. Environ. Microbiol. 2011, 77, 7850–7852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paziewska, A.; Harris, P.D.; Zwolińska, L.; Bajer, A.; Siński, E. Recombination Within and Between Species of the Alpha Proteobacterium Bartonella Infecting Rodents. Microb. Ecol. 2011, 61, 134–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colborn, J.M.; Kosoy, M.Y.; Motin, V.L.; Telepnev, M.V.; Valbuena, G.; Myint, K.S.; Fofanov, Y.; Putonti, C.; Feng, C.; Peruski, L. Improved Detection of Bartonella DNA in Mammalian Hosts and Arthropod Vectors by Real-Time PCR Using the NADH Dehydrogenase Gamma Subunit (NuoG). J. Clin. Microbiol. 2010, 48, 4630–4633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dillon, B.; Iredell, J.; Breitschwerdt, E.B.; Maggi, R.G. Potential Limitations of the 16S-23S RRNA Intergenic Region for Molecular Detection of Bartonella Species [5] (Multiple Letters). J. Clin. Microbiol. 2005, 43, 4921–4922. [Google Scholar] [CrossRef] [Green Version]
- Ewing, B.; Green, P. Base-Calling of Automated Sequencer Traces Using Phred. II. Error Probabilities. Genome Res. 1998, 8, 186–194. [Google Scholar] [CrossRef] [Green Version]
- Ewing, B.; Hillier, L.D.; Wendl, M.C.; Green, P. Base-Calling of Automated Sequencer Traces Using Phred. I. Accuracy Assessment. Genome Res. 1998, 8, 175–185. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. High-Performance Computing Selection of Models of DNA Substitution for Multicore Clusters. Int. J. High Perform. Comput. Appl. 2014, 28, 112–125. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the 2010 Gateway Computing Environments Workshop, GCE 2010, New Orleans, LA, USA, 14 November 2010. [Google Scholar]
- Posada, D.; Buckley, T.R. Model Selection and Model Averaging in Phylogenetics: Advantages of Akaike Information Criterion and Bayesian Approaches over Likelihood Ratio Tests. Syst. Biol. 2004, 53, 793–808. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian Phylogenetic Inference under Mixed Models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stöver, B.C.; Müller, K.F. TreeGraph 2: Combining and Visualizing Evidence from Different Phylogenetic Analyses. BMC Bioinform. 2010, 11, 7. [Google Scholar] [CrossRef] [Green Version]
- Librado, P.; Rozas, J. DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [Green Version]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A Computer Program to Estimate Gene Genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef] [Green Version]
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 2005, 23, 254–267. [Google Scholar] [CrossRef]
- Chomel, B.B.; Kasten, R.W.; Floyd-Hawkins, K.; Chi, B.; Yamamoto, K.; Roberts-Wilson, J.; Gurfield, A.N.; Abbott, R.C.; Pedersen, N.C.; Koehler, J.E. Experimental Transmission of Bartonella henselae by the Cat Flea. J. Clin. Microbiol. 1996, 34, 1952–1956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, T.; Halper, B.; Siebert, J.; Cruz-Martinez, L.; Chapwanya, A.; Kelly, P.; Ketzis, J.K.; Vessell, J.; Köster, L.; Yao, C. Parasites of Small Indian Mongoose, Herpestes Auropunctatus, on St. Kitts, West Indies. Parasitol. Res. 2018, 117, 989–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omasits, U.; Varadarajan, A.R.; Schmid, M.; Goetze, S.; Melidis, D.; Bourqui, M.; Nikolayeva, O.; Québatte, M.; Patrignani, A.; Dehio, C.; et al. An Integrative Strategy to Identify the Entire Protein Coding Potential of Prokaryotic Genomes by Proteogenomics. Genome Res. 2017, 27, 2083–2095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedrassani, D.; Biolchi, J.; Gonçalves, L.R.; Mendes, N.S.; Zanatto, D.C.D.S.; Calchi, A.C.; Machado, R.Z.; André, M.R. Molecular Detection of Vector-Borne Agents in Cats in Southern Brazil. Rev. Bras. Parasitol. Vet. 2019, 28, 632–643. [Google Scholar] [CrossRef]
- Boulouis, H.J.; Chang, C.C.; Henn, J.B.; Kasten, R.W.; Chomel, B.B. Factors Associated with the Rapid Emergence of Zoonotic Bartonella Infections. Vet. Res. 2005, 36, 383–410. [Google Scholar] [CrossRef] [Green Version]
- Kosoy, M.; McKee, C.; Albayrak, L.; Fofanov, Y. Genotyping of Bartonella Bacteria and Their Animal Hosts: Current Status and Perspectives. Parasitology 2018, 145, 543–562. [Google Scholar] [CrossRef] [Green Version]
- La Scola, B.; Zeaiter, Z.; Khamis, A.; Raoult, D. Gene-Sequence-Based Criteria for Species Definition in Bacteriology: The Bartonella Paradigm. Trends Microbiol. 2003, 11, 318–321. [Google Scholar] [CrossRef]
- Gutiérrez, R.; Vayssier-Taussat, M.; Buffet, J.P.; Harrus, S. Guidelines for the Isolation, Molecular Detection, and Characterization of Bartonella Species. Vector Borne Zoonotic Dis. 2017, 17, 42–50. [Google Scholar] [CrossRef] [Green Version]
- Lin, E.Y.; Tsigrelis, C.; Baddour, L.M.; Lepidi, H.; Rolain, J.M.; Patel, R.; Raoult, D. Candidatus Bartonella Mayotimonensis and Endocarditis. Emerg. Infect. Dis. 2010, 16, 500–503. [Google Scholar] [CrossRef]
- Chomel, B.B.; McMillan-Cole, A.C.; Kasten, R.W.; Stuckey, M.J.; Sato, S.; Maruyama, S.; Diniz, P.P.V.P.; Breitschwerdt, E.B. Candidatus Bartonella Merieuxii, a Potential New Zoonotic Bartonella Species in Canids from Iraq. PLoS Negl. Trop. Dis. 2012, 6, e1843. [Google Scholar] [CrossRef]
- Gutiérrez, R.; Morick, D.; Cohen, C.; Hawlena, H.; Harrus, S. The Effect of Ecological and Temporal Factors on the Composition of Bartonella Infection in Rodents and Their Fleas. ISME J. 2014, 8, 1598–1608. [Google Scholar] [CrossRef] [Green Version]
- Pérez, C.; Maggi, R.G.; Diniz, P.P.V.P.; Breitschwerdt, E.B. Molecular and Serological Diagnosis of Bartonella Infection in 61 Dogs from the United States. J. Vet. Intern. Med. 2011, 25, 805–810. [Google Scholar] [CrossRef]
- Gutiérrez, R.; Morick, D.; Gross, I.; Winkler, R.; Abdeen, Z.; Harrus, S. Bartonellae in Domestic and Stray Cats from Israel: Comparison of Bacterial Cultures and High-Resolution Melt Real-Time PCR as Diagnostic Methods. Vector Borne Zoonotic Dis. 2013, 13, 857–864. [Google Scholar] [CrossRef]
- Gurfield, A.N.; Boulouis, H.J.; Chomel, B.B.; Heller, R.; Kasten, R.W.; Yamamoto, K.; Piemont, Y. Coinfection with Bartonella clarridgeiae and Bartonella henselae and with Different Bartonella henselae Strains in Domestic Cats. J. Clin. Microbiol. 1997, 35, 2120–2123. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Gilbert, A.; Fox, K.; Osikowicz, L.; Kosoy, M. Bartonella rochalimae and B. Vinsonii Subsp. Berkhoffii in Wild Carnivores from Colorado, USA. J. Wildl. Dis. 2016, 52, 844–849. [Google Scholar] [CrossRef]
- Gerrikagoitia, X.; Gil, H.; García-Esteban, C.; Anda, P.; Juste, R.A.; Barral, M. Presence of Bartonella Species in Wild Carnivores of Northern Spain. Appl. Environ. Microbiol. 2012, 78, 885–888. [Google Scholar] [CrossRef] [Green Version]
- López-Pérez, A.M.; Osikowicz, L.; Bai, Y.; Montenieri, J.; Rubio, A.; Moreno, K.; Gage, K.; Suzán, G.; Kosoy, M. Prevalence and Phylogenetic Analysis of Bartonella Species of Wild Carnivores and Their Fleas in Northwestern Mexico. Ecohealth 2017, 14, 116–129. [Google Scholar] [CrossRef]
- Agan, B.K.; Dolan, M.J. Laboratory Diagnosis of Bartonella Infections. Clin. Lab. Med. 2002, 22, 937–962. [Google Scholar] [CrossRef]
- Müller, A.; Walker, R.; Bittencourt, P.; MacHado, R.Z.; Benevenute, J.L.; Do Amaral, R.B.; Gonçalves, L.R.; André, M.R. Prevalence, Hematological Findings and Genetic Diversity of Bartonella spp. in Domestic Cats from Valdivia, Southern Chile. Parasitology 2017, 144, 773–782. [Google Scholar] [CrossRef] [Green Version]
- Deng, H.K.; Le Rhun, D.; Lecuelle, B.; Le Naour, E.; Vayssier-Taussat, M. Role of the Spleen in Bartonella spp. Infection. FEMS Immunol. Med. Microbiol. 2012, 64, 143–145. [Google Scholar] [CrossRef] [Green Version]
- Schülein, R.; Seubert, A.; Gille, C.; Lanz, C.; Hansmann, Y.; Piémont, Y.; Dehio, C. Invasion and Persistent Intracellular Colonization of Erythrocytes: A Unique Parasitic Strategy of the Emerging Pathogen Bartonella. J. Exp. Med. 2001, 193, 1077–1086. [Google Scholar] [CrossRef] [Green Version]
- Guy, L.; Nystedt, B.; Toft, C.; Zaremba-Niedzwiedzka, K.; Berglund, E.C.; Granberg, F.; Näslund, K.; Eriksson, A.S.; Andersson, S.G.E. A Gene Transfer Agent and a Dynamic Repertoire of Secretion Systems Hold the Keys to the Explosive Radiation of the Emerging Pathogen Bartonella. PLoS Genet. 2013, 9, e1003393. [Google Scholar] [CrossRef]










| Target Locus | Primers (5′-3′) | Amplification Conditions | Size (bp) | Reference | ||
|---|---|---|---|---|---|---|
| irbp Mammalian species | F:TCCAACACCACCACTGAGATCTGGAC R:GTGAGGAAGAAATCGGACTGGCC | 95 °C for 4 m 94 °C for 30 s 57 °C for 30 s 72 °C for 1 m 72 °C for 5 m |  | 35 cycles | 227 | [31] |
| 18S rRNA Ctenocephalides felis | F:TGCTCACCGTTTGACTTGG R:GTTTCTCAGGCTCCCTCTCC | 95 °C for 3 m 94 °C for 30 s 60 °C for 45 s 72 °C for 1 m 72 °C for 7 m |  | 35 cycles | 179 | [32] |
| gltA Bartonella spp. | F:GCTATGTCTGCATTCTATCA R:GATCYTCAATCATTTCTTTCCA | 95 °C for 2 m 94 °C for 30 s 52 °C for 30 s 72 °C for 1 m 72 °C for 5 m |  | 40 cycles | 767 | [36] |
| rpoB Bartonella spp. | F:CGATTYGCATCATCATTTTCC R:CGCATTATGGTCGTATTTGTCC | 95 °C for 5 m 94 °C for 45 s 52 °C for 45 s 72 °C for 45 s 72 °C for 7 m |  | 40 cycles | 333 | [37] |
| ftsZ Bartonella spp. | F:CATATGGTTTTCATTACTGCYGGTATGG R:TTCTTCGCGAATACGATTAGCAGCTTC | 95 °C for 5 m 94 °C for 45 s 61 °C for 45 s 72 °C for 45 s 72 °C for 7 m |  | 40 cycles | 515 | [37] |
| nuoG Bartonella spp. | F:GGCGTGATTGTTCTCGTTA R:CACGACCACGGCTATCAAT | 94 °C for 5 m 94 °C for 30 s 53 °C for 30 s 72 °C for 30 s 72 °C for 7 m |  | 35 cycles | 400 | [38] |
| 16-23S rRNA ITS Bartonella spp. | F:CTTCAGATGATGATCCCAAGCCTTYTGGCG R:GAACCGACGACCCCTGCTTGCAAAGCA | 95 °C for 5 m 94 °C for 15 s 66 °C for 15 s 72 °C for 15 s 72 °C for 5 m |  | 55 cycles | 453–717 | [39] |
| Locus | Occurrence | Identity by BLASTn | % Blast Identity | Accession Numbers |
|---|---|---|---|---|
| gltA (n = 42) | 11/42 | B. henselae from cats in Chile | 97.80–100.00% | KY913625, KY913626, KY913627 |
| 16/42 | B. henselae from cat in Brazil | 99.64–100.00% | MN107415 | |
| 2/42 | B. henselae strain Houston-1 from human in Germany | 99.87–100.00% | CP020742 | |
| 13/42 | Bartonella sp. from Meles Anakuma in Japan | 98.37–99.01% | CP019788 | |
| rpoB (n = 50) | 16/50 | B. henselae from a cat in Brazil | 98.16–100.00% | MN107418 |
| 6/50 | B. henselae from a lion in South Africa | 99.71–99.72% | KX499338 | |
| 28/50 | Bartonella sp. from Meles Anakuma in Japan | 99.71–99.72% | CP019788 | |
| nuoG (n = 21) | 17/21 | B. henselae strain Houston-1 from human in Germany | 99.43–100.00% | CP020742 |
| 4/21 | Bartonella sp. from Meles Anakuma in Japan | 96.74–96.78% | CP019788 | |
| ITS (n = 21) | 29/31 | B. henselae in a flea from Iran (KT314216), cheetah from Zimbabwe (KX499346) and cat from Brazil (MT095053) | 99.58–100.00% | KT314216, KX499346, MT095053 |
| 2/31 | Bartonella sp. in a flea from St. Kitts | 100.00% | MT048286 | |
| fstZ (n = 21) | 21/21 | B. henselae strain Houston-1 from human in Germany | 99.81–100.00% | CP020742 |
| Sample | B. henselae | Unidentified Bartonella sp. | Co-Positive |
|---|---|---|---|
| Spleen (n = 16) | 5 (31.2%) | 9 (56.3%) | 2 (12.5%) |
| Blood (n = 34) | 21 (61.8%) | 4 (11.8%) | 9 (26.4%) |
| Fleas (n = 15) | 7 (46.7%) | 4 (26.7%) | 4 (26.7%) |
| Overall (n = 65) | 33 (50.8%) | 17(26.2%) | 15 (23.1%) |
| Gene/Region | bp | N | VS | GC% | h | hd (Mean ± SD) | π (Mean ± SD) | K |
|---|---|---|---|---|---|---|---|---|
| gltA | 568 | 50 | 70 | 36.8 | 6 | 0.547 ± 0.069 | 0.04864 ± 0.00670 | 27.62776 |
| rpoB | 318 | 77 | 40 | 38.0 | 4 | 0.595 ± 0.032 | 0.05454 ± 0.00325 | 17.34245 |
| fstZ | 491 | 31 | 5 | 44.9 | 5 | 0.544 ± 0.087 | 0.00151 ± 0.00040 | 0.73978 |
| nuoG | 336 | 26 | 51 | 40.2 | 5 | 0.655 ± 0.077 | 0.04744 ± 0.01376 | 15.93846 |
| ITS | 547 | 33 | 183 | 37.6 | 4 | 0.619 ± 0.049 | 0.08856 ± 0.03474 | 37.28409 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mau, A.; Calchi, A.C.; Bittencourt, P.; Navarrete-Talloni, M.J.; Sauvé, C.; Conan, A.; André, M.R.; Kelly, P.; Müller, A. Molecular Survey and Genetic Diversity of Bartonella spp. in Small Indian Mongooses (Urva auropunctata) and Their Fleas on Saint Kitts, West Indies. Microorganisms 2021, 9, 1350. https://doi.org/10.3390/microorganisms9071350
Mau A, Calchi AC, Bittencourt P, Navarrete-Talloni MJ, Sauvé C, Conan A, André MR, Kelly P, Müller A. Molecular Survey and Genetic Diversity of Bartonella spp. in Small Indian Mongooses (Urva auropunctata) and Their Fleas on Saint Kitts, West Indies. Microorganisms. 2021; 9(7):1350. https://doi.org/10.3390/microorganisms9071350
Chicago/Turabian StyleMau, Alex, Ana Cláudia Calchi, Pedro Bittencourt, Maria Jose Navarrete-Talloni, Caroline Sauvé, Anne Conan, Marcos Rogério André, Patrick Kelly, and Ananda Müller. 2021. "Molecular Survey and Genetic Diversity of Bartonella spp. in Small Indian Mongooses (Urva auropunctata) and Their Fleas on Saint Kitts, West Indies" Microorganisms 9, no. 7: 1350. https://doi.org/10.3390/microorganisms9071350