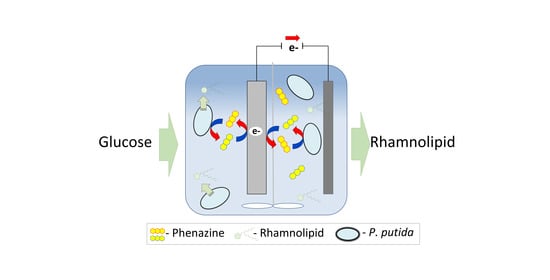
Coupling an Electroactive Pseudomonas putida KT2440 with Bioelectrochemical Rhamnolipid Production
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Strain Cultivation and Media Preparation
2.2. Combining Heterologous Rhamnolipid and Phenazine Production in P. putida
2.3. Aerobic Strain Characterization and Evaluation
2.4. Bioelectrochemical Experiments
2.5. Analysis of Sugar Metabolites
2.6. Phenazine Analysis
2.7. Rhamnolipid Analysis
3. Results
3.1. Tailoring Heterologous Mono-Rhamnolipid Synthesis with Phenazine Production in P. putida
3.2. Bioelectrochemical Production of Rhamnolipids in Oxygen-Limited BES
3.2.1. Applying Active Aeration to the Benchtop BES Reactors
3.2.2. Applying Passive Aeration to the Benchtop Single Chamber Glass BES Reactors
3.2.3. Scaling-Up and Verification of Results in 1-L Electrobioreactors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abdel-Mawgoud, A.M.; Lépine, F.; Déziel, E. Rhamnolipids: Diversity of structures, microbial origins and roles. Appl. Microbiol. Biotechnol. 2010, 86, 1323–1336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Germer, A.; Tiso, T.; Müller, C.; Behrens, B.; Vosse, C.; Scholz, K.; Froning, M.; Hayen, H.; Blank, L.M. Exploiting the natural diversity of RhlA acyltransferases for the synthesis of the rhamnolipid precursor 3-(3-hydroxyalkanoyloxy)alkanoic acid. Appl. Environ. Microbiol. 2020, 86, e02317–e02319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiso, T.; Zauter, R.; Tulke, H.; Leuchtle, B.; Li, W.-J.; Behrens, B.; Wittgens, A.; Rosenau, F.; Hayen, H.; Blank, L.M. Designer rhamnolipids by reduction of congener diversity: Production and characterization. Microb. Cell Fact. 2017, 16, 225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faivre, V.; Rosilio, V. Interest of glycolipids in drug delivery: From physicochemical properties to drug targeting. Expert Opin. Drug Deliv. 2010, 7, 1031–1048. [Google Scholar] [CrossRef] [PubMed]
- Ochsner, U.A.; Koch, A.K.; Fiechter, A.; Reiser, J. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J. Bacteriol. 1994, 176, 2044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ochsner, U.A.; Reiser, J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1995, 92, 6424–6428. [Google Scholar] [CrossRef] [Green Version]
- Pearson, J.P.; Pesci, E.C.; Iglewski, B.H. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 1997, 179, 5756. [Google Scholar] [CrossRef] [Green Version]
- Arnold, S.; Henkel, M.; Wanger, J.; Wittgens, A.; Rosenau, F.; Hausmann, R. Heterologous rhamnolipid biosynthesis by P. putida KT2440 on bio-oil derived small organic acids and fractions. AMB Express 2019, 9, 80. [Google Scholar] [CrossRef]
- Wittgens, A.; Santiago-Schuebel, B.; Henkel, M.; Tiso, T.; Blank, L.M.; Hausmann, R.; Hofmann, D.; Wilhelm, S.; Jaeger, K.E.; Rosenau, F. Heterologous production of long-chain rhamnolipids from Burkholderia glumae in Pseudomonas putida-a step forward to tailor-made rhamnolipids. Appl. Microbiol. Biotechnol. 2018, 102, 1229–1239. [Google Scholar] [CrossRef]
- Tiso, T.; Sabelhaus, P.; Behrens, B.; Wittgens, A.; Rosenau, F.; Hayen, H.; Blank, L.M. Creating metabolic demand as an engineering strategy in Pseudomonas putida—Rhamnolipid synthesis as an example. Metab. Eng. Commun. 2016, 3, 234–244. [Google Scholar] [CrossRef]
- Wittgens, A.; Tiso, T.; Arndt, T.T.; Wenk, P.; Hemmerich, J.; Müller, C.; Wichmann, R.; Kupper, B.; Zwick, M.; Wilhelm, S.; et al. Growth independent rhamnolipid production from glucose using the non-pathogenic Pseudomonas putida KT2440. Microb. Cell Fact. 2011, 10, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Küpper, B.; Mause, A.; Halka, L.; Imhoff, A.; Nowacki, C.; Wichmann, R. Fermentative Produktion von Monorhamnolipiden im Pilotmaßstab—Herausforderungen der Maßstabsvergrößerung. Chem. Ing. Tech. 2013, 85, 834–840. [Google Scholar] [CrossRef]
- Beuker, J.; Steier, A.; Wittgens, A.; Rosenau, F.; Henkel, M.; Hausmann, R. Integrated foam fractionation for heterologous rhamnolipid production with recombinant Pseudomonas putida in a bioreactor. AMB Express 2016, 6, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blesken, C.C.; Bator, I.; Eberlein, C.; Heipieper, H.J.; Tiso, T.; Blank, L.M. Genetic cell-surface modification for optimized foam fractionation. Front. Bioeng. Biotechnol. 2020, 8. [Google Scholar] [CrossRef]
- Bator, I.; Karmainski, T.; Tiso, T.; Blank, L.M. Killing two birds with one stone—Strain engineering facilitates the development of a unique rhamnolipid production process. Front. Bioeng. Biotechnol. 2020, 8. [Google Scholar] [CrossRef]
- Tiso, T.; Ihling, N.; Kubicki, S.; Biselli, A.; Schonhoff, A.; Bator, I.; Thies, S.; Karmainski, T.; Kruth, S.; Willenbrink, A.-L.; et al. Integration of genetic and process engineering for optimized rhamnolipid production using Pseudomonas putida. Front. Bioeng. Biotechnol. 2020, 8. [Google Scholar] [CrossRef]
- Schröder, U.; Harnisch, F.; Angenent, L.T. Microbial electrochemistry and technology: Terminology and classification. Energy Environ. Sci. 2015, 8, 513–519. [Google Scholar] [CrossRef]
- Schmitz, S.; Nies, S.; Wierckx, N.; Blank, L.M.; Rosenbaum, M.A. Engineering mediator-based electroactivity in the obligate aerobic bacterium Pseudomonas putida KT2440. Front. Microbiol. 2015, 6, 284. [Google Scholar] [CrossRef] [Green Version]
- Askitosari, T.D.; Boto, S.T.; Blank, L.M.; Rosenbaum, M.A. Boosting heterologous phenazine production in Pseudomonas putida KT2440 through the exploration of the natural sequence space. Front. Microbiol. 2019, 10, 1990. [Google Scholar] [CrossRef]
- Rosa, L.F.M.; Hunger, S.; Gimkiewicz, C.; Zehnsdorf, A.; Harnisch, F. Paving the way for bioelectrotechnology: Integrating electrochemistry into bioreactors. Eng. Life Sci. 2017, 17, 77–85. [Google Scholar] [CrossRef]
- Hartmans, S.; Smits, J.P.; van der Werf, M.J.; Volkering, F.; de Bont, J.A. Metabolism of Styrene Oxide and 2-Phenylethanol in the Styrene-Degrading Xanthobacter Strain 124X. Appl. Environ. Microbiol. 1989, 55, 2850–2855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.-H.; Kumar, A.; Schweizer, H.P. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: Application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 2006, 64, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Rosa, L.F.M.; Hunger, S.; Zschernitz, T.; Strehlitz, B.; Harnisch, F. Integrating electrochemistry into bioreactors: Effect of the upgrade kit on mass transfer, mixing time and sterilizability. Front. Energy Res. 2019, 7, 98. [Google Scholar] [CrossRef]
- Harnisch, F.; Hunger, S.; Zehnsdorf, A.; Beyer, D.; Rosa, L.F.M. Expansion Kit for Bioreactors Used for Performing Microbial Bio-Electrosynthesis. U.S. Patent 10,767,150 B2, 8 September 2020. [Google Scholar]
- Kern, S.E.; Newman, D.K. Measurement of Phenazines in Bacterial Cultures. In Pseudomonas Methods and Protocols; Filloux, A., Ramos, J.-L., Eds.; Springer: New York, NY, USA, 2014; pp. 303–310. [Google Scholar]
- Duetz, W.A.; Rüedi, L.; Hermann, R.; Connor, K.; Büchs, J.; Witholt, B. Methods for intense aeration, growth, storage, and replication of bacterial strains in microtiter plates. Appl. Environ. Microbiol. 2000, 66, 2641. [Google Scholar] [CrossRef] [Green Version]
- Nikel, P.I.; Chavarría, M.; Fuhrer, T.; Sauer, U.; de Lorenzo, V. Pseudomonas putida KT2440 strain metabolizes glucose through a cycle formed by enzymes of the Entner-Doudoroff, Embden-Meyerhof-Parnas, and pentose phosphate pathways. J. Biol. Chem. 2015, 290, 25920–25932. [Google Scholar] [CrossRef] [Green Version]
- Martin, C.H.; Wu, D.; Prather, K.L. Integrated bioprocessing for the pH-dependent production of 4-valerolactone from levulinate in Pseudomonas putida KT2440. Appl. Environ. Microbiol. 2010, 76, 417–424. [Google Scholar] [CrossRef] [Green Version]
- Costa, K.C.; Bergkessel, M.; Saunders, S.; Korlach, J.; Newman, D.K. Enzymatic degradation of phenazines can generate energy and protect sensitive organisms from toxicity. mBio 2015, 6, e01520. [Google Scholar] [CrossRef] [Green Version]
- Meirelles, L.A.; Newman, D.K. Both toxic and beneficial effects of pyocyanin contribute to the lifecycle of Pseudomonas aeruginosa. Mol. Microbiol. 2018, 110, 995–1010. [Google Scholar] [CrossRef] [Green Version]
- Mentel, M.; Ahuja, E.G.; Mavrodi, D.V.; Breinbauer, R.; Thomashow, L.S.; Blankenfeldt, W. Of two make one: The biosynthesis of phenazines. ChemBioChem 2009, 10, 2295–2304. [Google Scholar] [CrossRef]
- Behrens, B.; Engelen, J.; Tiso, T.; Blank, L.M.; Hayen, H. Characterization of rhamnolipids by liquid chromatography/mass spectrometry after solid-phase extraction. Anal. Bioanal. Chem. 2016, 408, 2505–2514. [Google Scholar] [CrossRef] [PubMed]
- Kampers, L.F.C.; van Heck, R.G.A.; Donati, S.; Saccenti, E.; Volkers, R.J.M.; Schaap, P.J.; Suarez-Diez, M.; Nikel, P.I.; Martins dos Santos, V.A.P. In silico-guided engineering of Pseudomonas putida towards growth under micro-oxic conditions. Microb. Cell Fact. 2019, 18. [Google Scholar] [CrossRef] [PubMed]
- Bosire, E.M.; Blank, L.M.; Rosenbaum, M.A. Strain- and substrate-dependent redox mediator and electricity production by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2016, 82, 5026–5038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glasser, N.R.; Kern, S.E.; Newman, D.K. Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Mol. Microbiol. 2014, 92, 399–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Kern, S.E.; Newman, D.K. Endogenous phenazine antibiotics promote anaerobic survival of Pseudomonas aeruginosa via extracellular electron transfer. J. Bacteriol. 2010, 192, 365–369. [Google Scholar] [CrossRef] [Green Version]
- Beuker, J.; Barth, T.; Steier, A.; Wittgens, A.; Rosenau, F.; Henkel, M.; Hausmann, R. High titer heterologous rhamnolipid production. AMB Express 2016, 6. [Google Scholar] [CrossRef] [Green Version]




| Strains/Plasmids | Characteristics | Source |
|---|---|---|
| Strains | ||
| P. aeruginosa PAO1 | Wildtype | DSMZ |
| P. putida 14phz2 | PCA producer | [19] |
| P. putida RL | RL producer | this study |
| P. putida RL-PCA | PCA and RL producer | this study |
| P. putida RL-MS | PhzMS and RL producer | this study |
| P. putida RL-PYO | PCA, PYO, and RL producer | this study |
| Plasmids | ||
| pJNN.rhlAB | oriRO1600 for Pseudomonas and oriCoIE1 for E. coli; gentamycin resistance-cassette, salicylate-inducible nagR/pNagAa promoter, rhlA and rhlB genes (P. aeruginosa PAO1) | this study |
| pJNN.rhlAB.phzMS | oriRO1600 for Pseudomonas and oriCoIE1 for E. coli; gentamycin resistance-cassette, salicylate-inducible nagR/pNagAa promoter, phzM, phzS, rhlA and rhlB genes (P. aeruginosa PAO1) | this study |
| pBNT.14phz2 | ORI: ori/IHF for replication in E. coli and Pseudomonas; kanamycin resistance-cassette, salicylate-inducible nagR/pNagAa promoter, phzA2-G2 genes (P. aeruginosa PA14) | [19] |
| pSK02 | oriR6K-pir dependent origin of replication; oriI -origin of transfer; riboJ gene; kanamycin resistance-cassette, rhlA and rhlB genes (P. aeruginosa PAO1) | [16] |
| Organism | Medium | Substrate (conc. in g/L) | Duration | Maximal RL Titer (g/L) | Growth Condition | Ref. |
|---|---|---|---|---|---|---|
| P. putida KT42C1 pVLT31_rhlAB | LB | Glucose (10) | 42 h | 1.5 | Flask, aerobic | [11] |
| P. putida KT2440 pSynPro8 | LB | Glucose (10) | 22 h | 3.22 | Flask, aerobic | [10] |
| P. putida KT2440 * pSynPro8oT | ModR | Glucose (253) | 72 h | 14.9 | Benchtop bioreactor, aerobic | [38] |
| P. putida KT2440 Δflag SK4 | Delft | Glucose (11) | 10 h | 1.48 | Benchtop bioreactor, aerobic | [16] |
| P. putida RL KT2440 pJNN.rhlAB | LB | Glucose (10) | 30 h | 0.73 | Microscale, aerobic | this study |
| P. putida RL KT2440 pJNN.rhlAB | Delft | Glucose (10) | 30 h 72 h | 0.36 0.11 | Microscale, aerobic Flask, aerobic | this study |
| P. putida RL-PCA KT2440 pJNN.rhlAB_pBNT.14phz2 | Delft | Glucose (10) | 30 h 72 h | 0.11 0.15 | Microscale, aerobic Flask, aerobic | this study |
| P. putida RL-PCA KT2440 pJNN.rhlAB_pBNT.14phz2 | Delft | Glucose (5) | 10 d | 0.02 | Benchtop BES, PA ** | this study |
| P. putida RL-PCA KT2440 pJNN.rhlAB_ pBNT.14phz2 | Delft | Glucose (5) | 25 d | 0.03 | Electrobioreactor BES, PA ** | this study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Askitosari, T.D.; Berger, C.; Tiso, T.; Harnisch, F.; Blank, L.M.; Rosenbaum, M.A. Coupling an Electroactive Pseudomonas putida KT2440 with Bioelectrochemical Rhamnolipid Production. Microorganisms 2020, 8, 1959. https://doi.org/10.3390/microorganisms8121959
Askitosari TD, Berger C, Tiso T, Harnisch F, Blank LM, Rosenbaum MA. Coupling an Electroactive Pseudomonas putida KT2440 with Bioelectrochemical Rhamnolipid Production. Microorganisms. 2020; 8(12):1959. https://doi.org/10.3390/microorganisms8121959
Chicago/Turabian StyleAskitosari, Theresia D., Carola Berger, Till Tiso, Falk Harnisch, Lars M. Blank, and Miriam A. Rosenbaum. 2020. "Coupling an Electroactive Pseudomonas putida KT2440 with Bioelectrochemical Rhamnolipid Production" Microorganisms 8, no. 12: 1959. https://doi.org/10.3390/microorganisms8121959