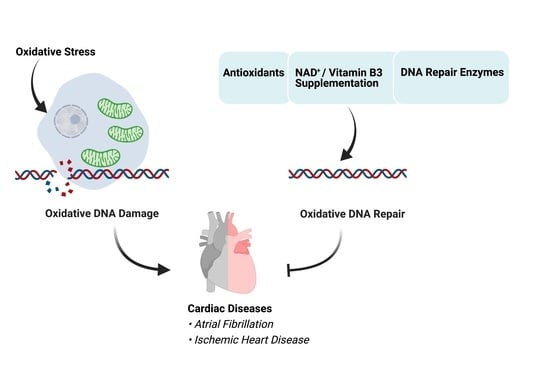
Role of Oxidative DNA Damage and Repair in Atrial Fibrillation and Ischemic Heart Disease
Abstract
:1. Introduction
2. Oxidative Stress and Oxidative DNA Damage
2.1. ROS and Oxidative Stress
2.2. Oxidative DNA Damage in Nuclei
2.3. Oxidative DNA Damage in Mitochondria
3. Oxidative DNA Repair Pathways
3.1. Base Excision Repair (BER)
3.2. Nucleotide Excision Repair (NER)
3.3. DNA Mismatch Repair (MMR)
4. Oxidative DNA Damage and Repair in AF
4.1. Oxidative Mitochondrial DNA Damage and Dysfunction in AF
4.2. The Oxidative Nuclear DNA Damage—Excessive PARP1 Activation—NAD+ Depletion Axis in AF
4.3. Novel Therapeutic Strategies for AF
4.3.1. Antioxidants: Reducing Upstream Oxidative Stress
4.3.2. PARP1 Inhibitors and NAD+ Supplements: Targeting the Pathway of Oxidative DNA Damage and Repair
5. Oxidative DNA Damage and Repair in IHD
5.1. Oxidative DNA Damage and IRI in IHD
5.2. Potential Therapies for IRI Recovery in IHD
5.2.1. Antioxidants: Attenuating the Oxidative DNA Damage in IRI
5.2.2. Novel NAD+-Based Therapeutic Approaches for IRI Recovery
5.2.3. Therapeutic Enzymes Involved in Oxidative DNA Damage and Repair Pathways
6. Summary
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chung, M.K.; Eckhardt, L.L.; Chen, L.Y.; Ahmed, H.M.; Trulock, K.M. Lifestyle and risk factor modification for reduction of atrial fibrillation: A scientific statement from the American heart association. Circulation 2020, 141, e750–e772. [Google Scholar] [CrossRef]
- Vos, T.; Allen, C.; Arora, M.; Barber, R.M.; Bhutta, Z.A.; Brown, A.; Carter, A.; Casey, D.C.; Charlson, F.J.; Chen, A.Z. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015. Lancet 2016, 388, 1545–1602. [Google Scholar] [CrossRef] [Green Version]
- Nattel, S. New ideas about atrial fibrillation 50 years on. Nature 2002, 415, 219–226. [Google Scholar] [CrossRef]
- Falk, R.H. Atrial fibrillation. N. Engl. J. Med. 2001, 344, 1067–1078. [Google Scholar] [CrossRef]
- Hannibal, G.B.; Copley, D.J.; Hill, K.M. Atrial fibrillation: A review of treatments and current guidelines. AACN Adv. Crit. Care 2016, 27, 120–128. [Google Scholar] [CrossRef]
- Gutierrez, C.; Blanchard, D.G. Atrial fibrillation: Diagnosis and treatment. Am. Fam. Physician 2011, 83, 61–68. [Google Scholar] [CrossRef] [Green Version]
- Menezes, A.R.; Lavie, C.J.; Dinicolantonio, J.J.; O’Keefe, J.; Morin, D.P.; Khatib, S.; Milani, R.V. Atrial fibrillation in the 21st century: A current understanding of risk factors and primary prevention strategies. Mayo Clin. Proc. 2013, 88, 394–409. [Google Scholar] [CrossRef] [Green Version]
- Dyg, P.O.; Peter, S.; Tonny, N.; Junge, N.S.; Steen, N.E.; Niels, F.J.; Ingolf, N.; Lars, K.; Hans, B.; Marie, S. Atrial fibrillation, ischaemic heart disease, and the risk of death in patients with heart failure. Eur. Heart J. 2006, 2866. [Google Scholar] [CrossRef] [Green Version]
- Choi, D.; Hwang, K.C.; Lee, K.Y.; Kim, Y.H. Ischemic heart diseases: Current treatments and future. J. Control Release 2009, 140, 194–202. [Google Scholar] [CrossRef]
- Dispersyn, G.D.; Ausma, J.; Thoné, F.; Flameng, W.; Vanoverschelde, J.L.; Allessie, M.A.; Ramaekers, F.C.; Borgers, M. Cardiomyocyte remodelling during myocardial hibernation and atrial fibrillation: Prelude to apoptosis. Cardiovasc. Res. 1999, 43, 947–957. [Google Scholar] [CrossRef] [Green Version]
- Everett, T.H.; Li, H.; Mangrum, J.M.; McRury, I.D.; Mitchell, M.A.; Redick, J.A.; Haines, D.E. Electrical, morphological, and ultrastructural remodeling and reverse remodeling in a canine model of chronic atrial fibrillation. Circulation 2000, 102, 1454–1460. [Google Scholar] [CrossRef] [Green Version]
- Mendis, S.; Puska, P.; Norrving, B. Global Atlas on Cardiovascular Disease Prevention and Control; Geneva World Health Organization: Geneva, Switzerland, 2011. [Google Scholar]
- Jennings, R.B.; Sommers, H.M.; Smyth, G.A.; Flack, H.A.; Linn, H. Myocardial necrosis induced by temporary occlusion of a coronary artery in the dog. Arch. Pathol. 1960, 70, 68–78. [Google Scholar] [CrossRef]
- Xu, J.; Luc, J.G.Y.; Phan, K. Atrial fibrillation: Review of current treatment strategies. J. Thorac. Dis. 2016, 8, E886–E900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soares, R.O.S.; Losada, D.M.; Jordani, M.C.; Vora, P.; Castro-E-Silva, O. Ischemia/reperfusion injury revisited: An overview of the latest pharmacological strategies. Int. J. Mol. Sci. 2019, 20, 5034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, C.S.; Parise, H.; D’Agostino, R.B.; Lloyd-Jones, D.M.; Vasan, R.S.; Wang, T.J.; Levy, D.; Wolf, P.A.; Benjamin, E.J. Parental atrial fibrillation as a risk factor for atrial fibrillation in offspring. Acc. Curr. J. Rev. 2004, 291, 2851–2855. [Google Scholar] [CrossRef]
- Saffitz, J.E. Connexins, conduction, and atrial fibrillation. N. Engl. J. Med. 2006, 354, 2712–2714. [Google Scholar] [CrossRef]
- Tsang, T.S.M.; Miyasaka, Y.; Barnes, M.E.; Gersh, B.J. Epidemiological profile of atrial fibrillation: A contemporary perspective. Prog. Cardiovasc. Dis. 2005, 48, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, A.; Idelchik, M.D.P.S.; Melendez, J.A. Redox control of senescence and age-related disease. Redox Biol. 2017, 11, 91–102. [Google Scholar] [CrossRef] [Green Version]
- Young, I.; Woodside, J. Antioxidants in health and disease. J. Clin. Pathol. 2001, 54, 176–186. [Google Scholar] [CrossRef] [Green Version]
- Cadet, J.; Wagner, J.R. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb. Perspect. Biol. 2013, 5, a12559. [Google Scholar] [CrossRef]
- Lushchak, V.I. Free radicals, reactive oxygen species, oxidative stress and its classification. Chem. Biol. Interact. 2014, 224, 164–175. [Google Scholar] [CrossRef]
- Cadet, J.; Davies, K.J.A. Oxidative DNA damage & repair: An introduction. Free Radic. Biol. Med. 2017, 107, 2–12. [Google Scholar] [CrossRef]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef] [Green Version]
- Münzel, T.; Gori, T.; Bruno, R.M. Is oxidative stress a therapeutic target in cardiovascular disease? Eur. Heart J. 2010, 31, 2741–2748. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Hu, X.; Li, J.; Liu, J.; Baks-te Bulte, L.; Wiersma, M.; van Marion, D.M.; Tolouee, M.; Hoogstra-Berends, F.; Lanters, E.A. DNA damage-induced PARP1 activation confers cardiomyocyte dysfunction through NAD+ depletion in experimental atrial fibrillation. Nat. Commun. 2019, 10, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Tarry-Adkins, J.L.; Blackmore, H.L.; Martin-Gronert, M.S.; Fernandez-Twinn, D.S.; Mcconnell, J.M.; Hargreaves, I.P.; Giussani, D.A.; Ozanne, S.E. Coenzyme Q10 prevents accelerated cardiac aging in a rat model of poor maternal nutrition and accelerated postnatal growth. Mol. Metab. 2013, 2, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Lobo, V.; Patil, A.; Phatak, A.; Chandra, N. Free radicals, antioxidants and functional foods: Impact on human health. Pharmacogn. Rev. 2010, 4, 118–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilaria, L.; Gennaro, R.; Francesco, C.; Giulia, B.; Luisa, A.; David, D.M.; Gaetano, G.; Gianluca, T.; Francesco, C.; Domenico, B. Oxidative stress, aging, and diseases. Clin. Interv. Aging 2018, 13, 757–772. [Google Scholar] [CrossRef] [Green Version]
- Lin, P.H.; Lee, S.H.; Su, C.P.; Wei, Y.H. Oxidative damage to mitochondrial DNA in atrial muscle of patients with atrial fibrillation. Free Radic. Biol. Med. 2003, 35, 1310–1318. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.; DeMazumder, D.; Sidor, A.; Foster, D.B.; O’Rourke, B. Mitochondrial ROS drive sudden cardiac death and chronic proteome remodeling in heart failure. Circ. Res. 2018, 123, 356–371. [Google Scholar] [CrossRef] [PubMed]
- Fransen, M.; Nordgren, M.; Wang, B.; Apanasets, O. Role of peroxisomes in ROS/RNS-metabolism: Implications for human disease. Biochim. Biophys. Acta 2012, 1822, 1363–1373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Meo, S.; Reed, T.T.; Venditti, P.; Victor, V.M. Role of ROS and RNS sources in physiological and pathological conditions. Oxid. Med. Cell. Longev. 2016, 2016, 1245049. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Fang, P.; Mai, J.; Choi, E.T. Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. J. Hematol. Oncol. 2013, 6, 19. [Google Scholar] [CrossRef] [Green Version]
- Bedard, K.; Krause, K.H. The NOX family of ROS-generating NADPH oxidases: Physiology and pathophysiology. Physiol. Rev. 2007, 87, 245–313. [Google Scholar] [CrossRef] [PubMed]
- Purushothaman, D.; Sarin, A. Cytokine-dependent regulation of NADPH oxidase activity and the consequences for activated T cell homeostasis. J. Exp. Med. 2009, 206, 1515–1523. [Google Scholar] [CrossRef] [PubMed]
- Moldogazieva, N.; Mokhosoev, I.; Mel’nikova, T.; Zavadskiy, S.; Kuz’menko, A.; Terentiev, A. Dual character of reactive oxygen, nitrogen, and halogen species: Endogenous sources, interconversions and neutralization. Biochemistry 2020, 85, 56–78. [Google Scholar] [CrossRef]
- Dikalov, S. Cross talk between mitochondria and NADPH oxidases. Free Radic. Biol. Med. 2011, 51, 1289–1301. [Google Scholar] [CrossRef] [Green Version]
- Daiber, A.; Di Lisa, F.; Oelze, M.; Kröller Schön, S.; Steven, S.; Schulz, E.; Münzel, T. Crosstalk of mitochondria with NADPH oxidase via reactive oxygen and nitrogen species signalling and its role for vascular function. Br. J. Pharmacol. 2017, 174, 1670–1689. [Google Scholar] [CrossRef] [Green Version]
- Joseph, L.C.; Barca, E.; Subramanyam, P.; Komrowski, M.; Pajvani, U.; Colecraft, H.M.; Hirano, M.; Morrow, J.P. Inhibition of NAPDH oxidase 2 (NOX2) prevents oxidative stress and mitochondrial abnormalities caused by saturated fat in cardiomyocytes. PLoS ONE 2016, 11, e0145750. [Google Scholar] [CrossRef]
- Kuroda, J.; Ago, T.; Matsushima, S.; Zhai, P.; Schneider, M.D.; Sadoshima, J. NADPH oxidase 4 (Nox4) is a major source of oxidative stress in the failing heart. Proc. Natl. Acad. Sci. USA 2010, 107, 15565–15570. [Google Scholar] [CrossRef] [Green Version]
- Schrader, M.; Fahimi, H.D. Peroxisomes and oxidative stress. Biochim. Biophys. Acta 2006, 1763, 1755–1766. [Google Scholar] [CrossRef] [Green Version]
- Marí, M.; Morales, A.; Colell, A.; García-Ruiz, C.; Fernández-Checa, J.C. Mitochondrial glutathione, a key survival antioxidant. Antioxid. Redox Signal. 2009, 11, 2685–2700. [Google Scholar] [CrossRef] [Green Version]
- Cox, A.G.; Winterbourn, C.C.; Hampton, M.B. Mitochondrial peroxiredoxin involvement in antioxidant defence and redox signalling. Biochem. J. 2009, 425, 313–325. [Google Scholar] [CrossRef] [Green Version]
- Holmgren, A. Antioxidant function of thioredoxin and glutaredoxin systems. Antioxid. Redox Signal. 2000, 2, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, P.; Nandakumar, N.; Rengarajan, T.; Palaniswami, R.; Gnanadhas, E.N.; Lakshminarasaiah, U.; Gopas, J.; Nishigaki, I. Antioxidants and human diseases. Clin. Chim. Acta 2014, 436, 332–347. [Google Scholar] [CrossRef]
- Hemnani, T.; Parihar, M.S. Reactive oxygen species and oxidative DNA damage. Indian J. Physiol. Pharmacol. 1998, 42, 440–452. [Google Scholar] [PubMed]
- Stefanis, L.; Burke, R.E.; Greene, L.A. Apoptosis in neurodegenerative disorders. Curr. Opin. Neurol. 1997, 10, 299–305. [Google Scholar] [CrossRef]
- Gabriele, P.; Natasha, I.; Mariapaola, C.; Giovanni, P.; Federica, M.; Vincenzo, A.; Francesco, S.; Domenica, A.; Alessandra, B. Oxidative stress: Harms and benefits for human health. Oxidative Med. Cell. Longev. 2017, 2017, 8416763. [Google Scholar] [CrossRef]
- Whitaker, A.M.; Schaich, M.A.; Smith, M.S.; Flynn, T.S.; Freudenthal, B.D. Base excision repair of oxidative DNA damage: From mechanism to disease. Front. Biosci. 2017, 22, 1493–1522. [Google Scholar] [CrossRef] [Green Version]
- Jovanovic, S.V.; Simic, M.G. One-electron redox potentials of purines and pyrimidines. J. Phys. Chem. 1986, 90, 974–978. [Google Scholar] [CrossRef]
- Evans, M.D.; Dizdaroglu, M.; Cooke, M.S. Oxidative DNA damage and disease: Induction, repair and significance. Mutat. Res. 2004, 567, 1–61. [Google Scholar] [CrossRef]
- Malik, Q.; Herbert, K.E. Oxidative and non-oxidative DNA damage and cardiovascular disease. Free Radic. Res. 2012, 46, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Bergeron, F.; Auvré, F.; Radicella, J.P.; Ravanat, J.L. HO• radicals induce an unexpected high proportion of tandem base lesions refractory to repair by DNA glycosylases. Proc. Natl. Acad. Sci. USA 2010, 107, 5528–5533. [Google Scholar] [CrossRef] [Green Version]
- Cooke, M.S.; Evans, M.D.; Dizdaroglu, M.; Lunec, J. Oxidative DNA damage: Mechanisms, mutation, and disease. FASEB J. 2003, 17, 1195–1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjelland, S.; Seeberg, E. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat. Res. 2003, 531, 37–80. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, H. Mutagenic potentials of damaged nucleic acids produced by reactive oxygen/nitrogen species: Approaches using synthetic oligonucleotides and nucleotides: Survey and summary. Nucleic Acids Res. 2003, 31, 517–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Linthout, S.; Tschöpe, C. Inflammation–cause or consequence of heart failure or both? Curr. Heart Fail. Rep. 2017, 14, 251–265. [Google Scholar] [CrossRef] [Green Version]
- Martinet, W.; Knaapen, M.W.; De Meyer, G.R.; Herman, A.G.; Kockx, M.M. Elevated levels of oxidative DNA damage and DNA repair enzymes in human atherosclerotic plaques. Circulation 2002, 106, 927–932. [Google Scholar] [CrossRef] [Green Version]
- Douarre, C.; Sourbier, C.; Dalla Rosa, I.; Das, B.B.; Redon, C.E.; Zhang, H.; Neckers, L.; Pommier, Y. Mitochondrial topoisomerase I is critical for mitochondrial integrity and cellular energy metabolism. PLoS ONE 2012, 7, e41094. [Google Scholar] [CrossRef] [Green Version]
- Chapman, J.; Ng, Y.S.; Nicholls, T.J. The maintenance of mitochondrial DNA integrity and dynamics by mitochondrial membranes. Life 2020, 10, 164. [Google Scholar] [CrossRef]
- Yakes, F.M.; Van Houten, B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl. Acad. Sci. USA 1997, 94, 514–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albring, M.; Griffith, J.; Attardi, G. Association of a protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replication. Proc. Natl. Acad. Sci. USA 1977, 74, 1348–1352. [Google Scholar] [CrossRef] [Green Version]
- Mikhed, Y.; Daiber, A.; Steven, S. Mitochondrial oxidative stress, mitochondrial DNA damage and their role in age-related vascular dysfunction. Int. J. Mol. Sci. 2015, 16, 15918–15953. [Google Scholar] [CrossRef] [Green Version]
- Shokolenko, I.; Venediktova, N.; Bochkareva, A.; Wilson, G.L.; Alexeyev, M.F. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009, 37, 2539–2548. [Google Scholar] [CrossRef] [Green Version]
- Quan, Y.; Xin, Y.; Tian, G.; Zhou, J.; Liu, X. Mitochondrial ROS-modulated mtDNA: A potential target for cardiac aging. Oxid. Med. Cell. Longev. 2020, 2020, 9423593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gredilla, R.; Bohr, V.A.; Stevnsner, T. Mitochondrial DNA repair and association with aging—An update. Exp. Gerontol. 2010, 45, 478–488. [Google Scholar] [CrossRef] [Green Version]
- Simon, R.; Meller, R.; Yang, T.; Pearson, A.; Wilson, G. Enhancing base excision repair of mitochondrial DNA to reduce ischemic injury following reperfusion. Transl. Stroke Res. 2019, 10, 664–671. [Google Scholar] [CrossRef] [Green Version]
- Cadenas, E.; Davies, K.J. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic. Biol. Med. 2000, 29, 222–230. [Google Scholar] [CrossRef]
- Peng, Y.; Jing, S.; Liu, L.; Ma, F.; Zhang, Y.; Wang, C.; Duan, H.; Zhou, K.; Hua, Y.; Wu, G.; et al. Association between mitochondrial DNA copy number and cardiovascular disease: Current evidence based on a systematic review and meta-analysis. PLoS ONE 2018, 13, e0206003. [Google Scholar] [CrossRef]
- Hollensworth, S.B.; Shen, C.-C.; Sim, J.E.; Spitz, D.R.; Wilson, G.L.; LeDoux, S.P. Glial cell type-specific responses to menadione-induced oxidative stress. Free Radic. Biol. Med. 2000, 28, 1161–1174. [Google Scholar] [CrossRef]
- Bohr, V.A. Repair of oxidative DNA damage in nuclear and mitochondrial DNA, and some changes with aging in mammalian cells. Free Radic. Biol. Med. 2002, 32, 804–812. [Google Scholar] [CrossRef]
- Saki, M.; Prakash, A. DNA damage related crosstalk between the nucleus and mitochondria. Free Radic. Biol. Med. 2017, 107, 216–227. [Google Scholar] [CrossRef]
- Jazwinski, S.M. The retrograde response: A conserved compensatory reaction to damage from within and from without. Prog. Mol. Biol. Transl. Sci. 2014, 127, 133–154. [Google Scholar] [CrossRef] [Green Version]
- Schrepper, A.; Schwarzer, M.; Schöpe, M.; Amorim, P.A.; Doenst, T. Biphasic response of skeletal muscle mitochondria to chronic cardiac pressure overload—Role of respiratory chain complex activity. J. Mol. Cell. Cardiol. 2012, 52, 125–135. [Google Scholar] [CrossRef]
- Corral-Debrinski, M.; Stepien, G.; Shoffner, J.M.; Lott, M.T.; Kanter, K.; Wallace, D.C. Hypoxemia is associated with mitochondrial DNA damage and gene induction: Implications for cardiac disease. JAMA 1991, 266, 1812–1816. [Google Scholar] [CrossRef]
- Cicchillitti, L.; Corrado, G.; De Angeli, M.; Mancini, E.; Baiocco, E.; Patrizi, L.; Zampa, A.; Merola, R.; Martayan, A.; Conti, L. Circulating cell-free DNA content as blood based biomarker in endometrial cancer. Oncotarget 2017, 8, 115230–115243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiersma, M.; van Marion, D.; Bouman, E.J.; Li, J.; Zhang, D.; Ramos, K.S.; Lanters, E.A.; de Groot, N.; Brundel, B.J. Cell-free circulating mitochondrial DNA: A potential blood-based marker for atrial fibrillation. Cells 2020, 9, 1159. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; He, H.; Kelley, M.R.; Georgiadis, M.M. Redox regulation of DNA repair: Implications for human health and cancer therapeutic development. Antioxid. Redox Signal. 2010, 12, 1247–1269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, A.B.; Klungland, A.; Rognes, T.; Leiros, I. DNA repair in mammalian cells: Base excision repair: The long and short of it. Cell. Mol. Life Sci. 2009, 66, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Slupphaug, G.; Kavli, B.; Krokan, H.E. The interacting pathways for prevention and repair of oxidative DNA damage. Mutat. Res. 2003, 531, 231–251. [Google Scholar] [CrossRef]
- Khodyreva, S.; Prasad, R.; Ilina, E.; Sukhanova, M.; Kutuzov, M.; Liu, Y.; Hou, E.; Wilson, S.; Lavrik, O. Apurinic/apyrimidinic (AP) site recognition by the 5′-dRP/AP lyase in poly(ADP-ribose) polymerase-1 (PARP-1). Proc. Natl. Acad. Sci. USA 2010, 107, 22090–22095. [Google Scholar] [CrossRef] [Green Version]
- Gredilla, R. DNA damage and base excision repair in mitochondria and their role in aging. J. Aging Res. 2010, 2011, 257093. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, A.L.; Schär, P. DNA glycosylases: In DNA repair and beyond. Chromosoma 2012, 121, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Ko, H.L.; Ren, E.C. Functional aspects of PARP1 in DNA repair and transcription. Biomolecules 2012, 2, 524–548. [Google Scholar] [CrossRef] [Green Version]
- Bazzani, V.; Barchiesi, A.; Radecka, D.; Pravisani, R.; Guadagno, A.; Di Loreto, C.; Baccarani, U.; Vascotto, C. Mitochondrial apurinic/apyrimidinic endonuclease 1 enhances mtDNA repair contributing to cell proliferation and mitochondrial integrity in early stages of hepatocellular carcinoma. BMC Cancer 2020, 20, 969. [Google Scholar] [CrossRef] [PubMed]
- Nemec, A.A.; Wallace, S.S.; Sweasy, J.B. Variant base excision repair proteins: Contributors to genomic instability. Semin. Cancer Biol. 2010, 20, 320–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, S.S.; Copeland, W.C. DNA polymerase gamma and mitochondrial disease: Understanding the consequence of POLG mutations. Biochim. Biophys. Acta 2009, 1787, 312–319. [Google Scholar] [CrossRef] [Green Version]
- Kelley, M.; Fishel, M. Overview of DNA repair pathways, current targets, and clinical trials bench to clinic. In DNA Repair in Cancer Therapy; Elsevier: Amsterdam, The Netherlands, 2016; pp. 1–54. [Google Scholar] [CrossRef]
- Xu, G.; Herzig, M.; Rotrekl, V.; Walter, C.A. Base excision repair, aging and health span. Mech. Ageing Dev. 2008, 129, 366–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fortini, P.; Pascucci, B.; Parlanti, E.; D’errico, M.; Simonelli, V.; Dogliotti, E. The base excision repair: Mechanisms and its relevance for cancer susceptibility. Biochimie 2003, 85, 1053–1071. [Google Scholar] [CrossRef] [PubMed]
- Yndestad, A.; Neurauter, C.G.; Øie, E.; Forstrøm, R.J.; Vinge, L.E.; Eide, L.; Luna, L.; Aukrust, P.; Bjørås, M. Up-regulation of myocardial DNA base excision repair activities in experimental heart failure. Mutat. Res. 2009, 666, 32–38. [Google Scholar] [CrossRef]
- Shah, A.; Gray, K.; Figg, N.; Finigan, A.; Starks, L.; Bennett, M. Defective base excision repair of oxidative DNA damage in vascular smooth muscle cells promotes atherosclerosis. Circulation 2018, 138, 1446–1462. [Google Scholar] [CrossRef]
- Herrington, W.; Lacey, B.; Sherliker, P.; Armitage, J.; Lewington, S. Epidemiology of atherosclerosis and the potential to reduce the global burden of atherothrombotic disease. Circ. Res. 2016, 118, 535–546. [Google Scholar] [CrossRef]
- Melis, J.P.; van Steeg, H.; Luijten, M. Oxidative DNA damage and nucleotide excision repair. Antioxid. Redox Signal. 2013, 18, 2409–2419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, N.B.; Rasmussen, M.; Rasmussen, L.J. Nuclear and mitochondrial DNA repair: Similar pathways? Mitochondrion 2005, 5, 89–108. [Google Scholar] [CrossRef] [PubMed]
- Schärer, O.D. Nucleotide excision repair in eukaryotes. Cold Spring Harb. Perspect. Biol. 2013, 5, a012609. [Google Scholar] [CrossRef] [PubMed]
- Durik, M.; Kavousi, M.; van der Pluijm, I.; Isaacs, A.; Cheng, C.; Verdonk, K.; Loot, A.E.; Oeseburg, H.; Bhaggoe, U.M.; Leijten, F. Nucleotide excision DNA repair is associated with age-related vascular dysfunction. Circulation 2012, 126, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Curtin, N.J. DNA repair dysregulation from cancer driver to therapeutic target. Nat. Rev. Cancer 2012, 12, 801–817. [Google Scholar] [CrossRef]
- Martin, S.A.; Lord, C.J.; Ashworth, A. Therapeutic targeting of the DNA mismatch repair pathway. Clin. Cancer Res. 2010, 16, 5107–5113. [Google Scholar] [CrossRef] [Green Version]
- Brierley, D.J.; Martin, S.A. Oxidative stress and the DNA mismatch repair pathway. Antioxid. Redox Signal. 2013, 18, 2420–2428. [Google Scholar] [CrossRef]
- Gu, Y.; Parker, A.; Wilson, T.M.; Bai, H.; Chang, D.-Y.; Lu, A.-L. Human MutY homolog, a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins human MutS homolog 2/human MutS homolog 6. J. Biol. Chem. 2002, 277, 11135–11142. [Google Scholar] [CrossRef] [Green Version]
- Lin, R.; Gao, D.; Gu, Y.; Bonde, P.; Fitton, T.P.; Hare, J.M.; Conte, J.V.; Williams, G.M.; Wei, C. Oxidative DNA damage and DNA mismatch repair pathway play an important role in failing human myocardium. J. Cardiothorac. Ren. Res. 2006, 1, 41–49. [Google Scholar] [CrossRef]
- Pascucci, B.; D’errico, M.; Parlanti, E.; Giovannini, S.; Dogliotti, E. Role of nucleotide excision repair proteins in oxidative DNA damage repair: An updating. Biochemistry 2011, 76, 4–15. [Google Scholar] [CrossRef]
- Allessie, M.A.; de Groot, N.M.; Houben, R.P.; Schotten, U.; Boersma, E.; Smeets, J.L.; Crijns, H.J. Electropathological substrate of long-standing persistent atrial fibrillation in patients with structural heart disease: Longitudinal dissociation. Circ. Arrhythm. Electrophysiol. 2010, 3, 606–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meijering, R.A.M.; Zhang, D.; Hoogstra-Berends, F.; Henning, R.; Brundel, B. Loss of proteostatic control as a substrate for atrial fibrillation: A novel target for upstream therapy by heat shock proteins. Front. Physiol. 2012, 3, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.J.; Parise, H.; Levy, D.; D’Agostino, R.B.; Wolf, P.A.; Vasan, R.S.; Benjamin, E.J. Obesity and the risk of new-onset atrial fibrillation. JAMA 2004, 292, 2471–2477. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, H.; Tanabe, N.; Watanabe, T.; Darbar, D.; Roden, D.M.; Sasaki, S.; Aizawa, Y. Metabolic syndrome and risk of development of atrial fibrillation: The Niigata preventive medicine study. Circulation 2008, 117, 1255–1260. [Google Scholar] [CrossRef] [Green Version]
- Feinberg, W.M.; Blackshear, J.L.; Laupacis, A.; Kronmal, R.; Hart, R.G. Prevalence, age distribution, and gender of patients with atrial fibrillation: Analysis and implications. Arch. Intern. Med. 1995, 155, 469–473. [Google Scholar] [CrossRef]
- Kotecha, D.; Piccini, J.P. Atrial fibrillation in heart failure: What should we do? Eur. Heart J. 2015, 36, 3250–3257. [Google Scholar] [CrossRef] [Green Version]
- Wijffels, M.C.; Kirchhof, C.J.; Dorland, R.; Allessie, M.A. Atrial fibrillation begets atrial fibrillation: A study in awake chronically instrumented goats. Circulation 1995, 92, 1954–1968. [Google Scholar] [CrossRef]
- Nattel, S.; Burstein, B.; Dobrev, D. Atrial remodeling and atrial fibrillation: Mechanisms and implications. Circ. Arrhythm. Electrophysiol. 2008, 1, 62–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liaquat, M.T.; Makaryus, A.N. Cardiac Electrical and Structural Remodeling; StatPearls Publishing LLC: Tampa, FL, USA, 2020. [Google Scholar]
- Prabhu, S.; Voskoboinik, A.; Kaye, D.M.; Kistler, P.M. Atrial Fibrillation and heart failure—Cause or effect? Heart Lung Circ. 2017, 26, 967–974. [Google Scholar] [CrossRef]
- Soliman, E.Z.; Safford, M.M.; Muntner, P.; Khodneva, Y.; Dawood, F.Z.; Zakai, N.A.; Thacker, E.L.; Judd, S.; Howard, V.J.; Howard, G. Atrial fibrillation and the risk of myocardial infarction. JAMA Intern. Med. 2014, 174, 107–114. [Google Scholar] [CrossRef]
- Yang, X.; An, N.; Zhong, C.; Guan, M.; Jiang, Y.; Li, X.; Zhang, H.; Wang, L.; Ruan, Y.; Gao, Y. Enhanced cardiomyocyte reactive oxygen species signaling promotes ibrutinib-induced atrial fibrillation. Redox Boil. 2020, 30, 101432. [Google Scholar] [CrossRef]
- Friedrichs, K.; Baldus, S.; Klinke, A. Fibrosis in atrial fibrillation–role of reactive species and MPO. Front. Physiol. 2012, 3, 214. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Zhang, D.; Ramos, K.S.; Baks, L.; Wiersma, M.; Lanters, E.A.; Bogers, A.J.; De Groot, N.M.; Brundel, B.J. Blood-based 8-hydroxy-2′-deoxyguanosine level: A potential diagnostic biomarker for atrial fibrillation. Heart Rhythm. 2021, 18, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Laredo, M.; Waldmann, V.; Khairy, P.; Nattel, S. Age as a critical determinant of atrial fibrillation: A two-sided relationship. Can. J. Cardiol. 2018, 34, 1396–1406. [Google Scholar] [CrossRef] [PubMed]
- Baris, O.R.; Ederer, S.; Neuhaus, J.F.; von Kleist-Retzow, J.C.; Wunderlich, C.M.; Pal, M.; Wunderlich, F.T.; Peeva, V.; Zsurka, G.; Kunz, W.S. Mosaic deficiency in mitochondrial oxidative metabolism promotes cardiac arrhythmia during aging. Cell. Metab. 2015, 21, 667–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuboi, M.; Hisatome, I.; Morisaki, T.; Tanaka, M.; Tomikura, Y.; Takeda, S.; Shimoyama, M.; Ohtahara, A.; Ogino, K.; Igawa, O. Mitochondrial DNA deletion associated with the reduction of adenine nucleotides in human atrium and atrial fibrillation. Eur. J. Clin. Investig. 2001, 31, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Lai, L.P.; Tsai, C.C.; Su, M.-J.; Lin, J.L.; Chen, Y.S.; Tseng, Y.Z.; Huang, S.K.S. Atrial fibrillation is associated with accumulation of aging-related common type mitochondrial DNA deletion mutation in human atrial tissue. Chest 2003, 123, 539–544. [Google Scholar] [CrossRef]
- Montaigne, D.; Marechal, X.; Lefebvre, P.; Modine, T.; Fayad, G.; Dehondt, H.; Hurt, C.; Coisne, A.; Koussa, M.; Remy-Jouet, I. Mitochondrial dysfunction as an arrhythmogenic substrate: A translational proof-of-concept study in patients with metabolic syndrome in whom post-operative atrial fibrillation develops. J. Am. Coll. Cardiol. 2013, 62, 1466–1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, W.; Santulli, G.; Reiken, S.R.; Yuan, Q.; Osborne, B.W.; Chen, B.X.; Marks, A.R. Mitochondrial oxidative stress promotes atrial fibrillation. Sci. Rep. 2015, 5, 11427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeganathan, J.; Saraf, R.; Mahmood, F.; Pal, A.; Bhasin, M.K.; Huang, T.; Mittel, A.; Knio, Z.; Simons, R.; Khabbaz, K. Mitochondrial dysfunction in atrial tissue of patients developing postoperative atrial fibrillation. Ann. Thorac. Surg. 2017, 104, 1547–1555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiersma, M.; van Marion, D.; Wüst, R.C.; Houtkooper, R.H.; Zhang, D.; de Groot, N.; Henning, R.H.; Brundel, B.J. Mitochondrial dysfunction underlies cardiomyocyte remodeling in experimental and clinical atrial fibrillation. Cells 2019, 8, 1202. [Google Scholar] [CrossRef] [Green Version]
- Caldecott, K.W. Single-strand break repair and genetic disease. Nat. Rev. Genet. 2008, 9, 619–631. [Google Scholar] [CrossRef]
- Tao, Z.; Gao, P.; Liu, H. Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: Analysis and implications. J. Am. Chem. Soc. 2009, 131, 14258–14260. [Google Scholar] [CrossRef] [PubMed]
- Skidmore, C.J.; Davies, M.I.; Goodwin, P.M.; Halldorsson, H.; Lewis, P.J.; Shall, S.; Zia’ee, A.A. The involvement of poly(ADP-ribose) polymerase in the degradation of NAD caused by gamma-radiation and N-methyl-N-nitrosourea. Eur. J. Biochem. 1979, 101, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Dantzer, F.; Amé, J.C.; Schreiber, V.; Nakamura, J.; Ménissier de Murcia, J.; de Murcia, G. Poly (ADP-ribose) polymerase-1 activation during DNA damage and repair. Methods Enzymol. 2006, 409, 493–510. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Koh, D.W. Roles of poly(ADP-ribose)glycohydrolase in DNA damage and apoptosis. Int. Rev. Cell Mol. Biol. 2013, 304, 227–281. [Google Scholar] [CrossRef]
- Bürkle, A.; Virág, L. Poly(adp-ribose): Paradigms and paradoxes. Mol. Aspects Med. 2013, 34, 1046–1065. [Google Scholar] [CrossRef] [Green Version]
- Horváth, E.M.; Szabó, C. Poly (ADP-ribose) polymerase as a drug target for cardiovascular disease and cancer: An update. Drug News Perspect. 2007, 20, 171–181. [Google Scholar] [CrossRef]
- Massudi, H.; Grant, R.; Guillemin, G.J.; Braidy, N. NAD+ metabolism and oxidative stress: The golden nucleotide on a crown of thorns. Redox Rep. 2012, 17, 28–46. [Google Scholar] [CrossRef]
- Chinopoulos, C. Acute sources of mitochondrial NAD+ during respiratory chain dysfunction. Exp. Neurol. 2020, 327, 113218. [Google Scholar] [CrossRef] [PubMed]
- Yin, F.; Sancheti, H.; Cadenas, E. Silencing of nicotinamide nucleotide transhydrogenase impairs cellular redox homeostasis and energy metabolism in PC12 cells. Biochim. Biophys. Acta 2012, 1817, 401–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheeran, F.L.; Rydström, J.; Shakhparonov, M.I.; Pestov, N.B.; Pepe, S. Diminished NADPH transhydrogenase activity and mitochondrial redox regulation in human failing myocardium. Biochim. Biophys. Acta 2010, 1797, 1138–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bainbridge, M.N.; Davis, E.E.; Choi, W.Y.; Dickson, A.; Martinez, H.R.; Wang, M.; Dinh, H.; Muzny, D.M.; Pignatelli, R.; Katsanis, N.; et al. Loss of function mutations in NNT are associated with left ventricular noncompaction. Circ. Cardiovasc. Genet. 2015, 8, 544–552. [Google Scholar] [CrossRef] [Green Version]
- Weinberg-Shukron, A.; Abu-Libdeh, A.; Zhadeh, F.; Carmel, L.; Kogot-Levin, A.; Kamal, L.; Kanaan, M.; Zeligson, S.; Renbaum, P.; Levy-Lahad, E. Combined mineralocorticoid and glucocorticoid deficiency is caused by a novel founder nicotinamide nucleotide transhydrogenase mutation that alters mitochondrial morphology and increases oxidative stress. J. Med. Genet. 2015, 52, 636–641. [Google Scholar] [CrossRef]
- Nickel, A.G.; Von Hardenberg, A.; Hohl, M.; Löffler, J.R.; Kohlhaas, M.; Becker, J.; Reil, J.C.; Kazakov, A.; Bonnekoh, J.; Stadelmaier, M. Reversal of Mitochondrial Transhydrogenase Causes Oxidative Stress in Heart Failure. Cell Metab. 2015, 22, 472–484. [Google Scholar] [CrossRef] [Green Version]
- Go, A.S.; Hylek, E.M.; Phillips, K.A.; Chang, Y.; Henault, L.E.; Selby, J.V.; Singer, D.E. Prevalence of diagnosed atrial fibrillation in adults: National implications for rhythm management and stroke prevention: The AnTicoagulation and risk factors in atrial fibrillation (ATRIA) study. JAMA 2001, 285, 2370–2375. [Google Scholar] [CrossRef] [PubMed]
- Lip, G.Y.; Brechin, C.M.; Lane, D.A. The global burden of atrial fibrillation and stroke: A systematic review of the epidemiology of atrial fibrillation in regions outside North America and Europe. Chest 2012, 142, 1489–1498. [Google Scholar] [CrossRef] [PubMed]
- Naccarelli, G.V.; Varker, H.; Lin, J.; Schulman, K.L. Increasing prevalence of atrial fibrillation and flutter in the United States. Am. J. Cardiol. 2009, 104, 1534–1539. [Google Scholar] [CrossRef]
- Hong, K.L.; Borges, J.; Glover, B. Catheter ablation for the management of atrial fibrillation: Current technical perspectives. Open Heart 2020, 7, e001207. [Google Scholar] [CrossRef]
- Roberts, S.A.; Diaz, C.; Nolan, P.E.; Salerno, D.M.; Stapczynski, J.S.; Zbrozek, A.S.; Ritz, E.G.; Bauman, J.L.; Vlasses, P.H. Effectiveness and costs of digoxin treatment for atrial fibrillation and flutter. Am. J. Cardiol. 1993, 72, 567–573. [Google Scholar] [CrossRef]
- D’Oria, R.; Schipani, R.; Leonardini, A.; Natalicchio, A.; Perrini, S.; Cignarelli, A.; Laviola, L.; Giorgino, F. The role of oxidative stress in cardiac disease: From physiological response to injury factor. Oxid. Med. Cell. Longev. 2020, 2020, 5732956. [Google Scholar] [CrossRef] [PubMed]
- Sovari, A.A.; Dudley, S.C., Jr. Reactive oxygen species-targeted therapeutic interventions for atrial fibrillation. Front. Physiol. 2012, 3, 311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carnes, C.A.; Chung, M.K.; Nakayama, T.; Nakayama, H.; Baliga, R.S.; Piao, S.; Kanderian, A.; Pavia, S.; Hamlin, R.L.; McCarthy, P.M. Ascorbate attenuates atrial pacing-induced peroxynitrite formation and electrical remodeling and decreases the incidence of postoperative atrial fibrillation. Circ. Res. 2001, 89, e32–e38. [Google Scholar] [CrossRef] [PubMed]
- Eren, N.; Çakir, Ö.; Oruc, A.; Kaya, Z.; Erdinc, L. Effects of N-acetylcysteine on pulmonary function in patients undergoing coronary artery bypass surgery with cardiopulmonary bypass. Perfusion 2003, 18, 345–350. [Google Scholar] [CrossRef]
- Castillo, R.; Rodrigo, R.; Perez, F.; Cereceda, M.; Asenjo, R.; Zamorano, J.; Navarrete, R.; Villalabeitia, E.; Sanz, J.; Baeza, C. Antioxidant therapy reduces oxidative and inflammatory tissue damage in patients subjected to cardiac surgery with extracorporeal circulation. Basic Clin. Pharmacol. Toxicol. 2011, 108, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo, R.; Vinay, J.; Castillo, R.; Cereceda, M.; Asenjo, R.; Zamorano, J.; Araya, J.; Castillo-Koch, R.; Espinoza, J.; Larraín, E. Use of vitamins C and E as a prophylactic therapy to prevent postoperative atrial fibrillation. Int. J. Cardiol. 2010, 138, 221–228. [Google Scholar] [CrossRef]
- Hemilä, H.; Suonsyrjä, T. Vitamin C for preventing atrial fibrillation in high risk patients: A systematic review and meta-analysis. BMC Cardiovasc. Disord. 2017, 17, 49. [Google Scholar] [CrossRef] [Green Version]
- Panagiotou, O.A.; Contopoulos-Ioannidis, D.G.; Ioannidis, J.P. Comparative effect sizes in randomised trials from less developed and more developed countries: Meta-epidemiological assessment. BMJ 2013, 346, f707. [Google Scholar] [CrossRef] [Green Version]
- Basili, S.; Tanzilli, G.; Mangieri, E.; Raparelli, V.; Di Santo, S.; Pignatelli, P.; Violi, F. Intravenous ascorbic acid infusion improves myocardial perfusion grade during elective percutaneous coronary intervention: Relationship with oxidative stress markers. JACC Cardiovasc. Interv. 2010, 3, 221–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oktar, G.; Sinci, V.; Kalaycioğlu, S.; Soncul, H.; Gökgöz, L.; Halit, V.; Ersöz, A. Biochemical and hemodynamic effects of ascorbic acid and alpha-tocopherol in coronary artery surgery. Scand. J. Clin. Lab. Investig. 2001, 61, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Sesso, H.D.; Buring, J.E.; Christen, W.G.; Kurth, T.; Belanger, C.; MacFadyen, J.; Bubes, V.; Manson, J.E.; Glynn, R.J.; Gaziano, J.M. Vitamins E and C in the prevention of cardiovascular disease in men: The physicians’ health study II randomized controlled trial. JAMA 2008, 300, 2123–2133. [Google Scholar] [CrossRef] [Green Version]
- Sovari, A.A.; Morita, N.; Karagueuzian, H.S. Apocynin: A potent NADPH oxidase inhibitor for the management of atrial fibrillation. Redox Rep. 2008, 13, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Budanov, A.V.; Lee, J.H.; Karin, M. Stressin’Sestrins take an aging fight. EMBO Mol. Med. 2010, 2, 388–400. [Google Scholar] [CrossRef]
- Kallenborn-Gerhardt, W.; Lu, R.; Syhr, K.M.; Heidler, J.; von Melchner, H.; Geisslinger, G.; Bangsow, T.; Schmidtko, A. Antioxidant activity of sestrin 2 controls neuropathic pain after peripheral nerve injury. Antioxid. Redox Signal. 2013, 19, 2013–2023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Z.; Lin, C.; Liu, Y.; Jin, H.; Wu, H.; Li, Z.; Sun, L.; Zhang, L.; Hu, X.; Wei, Y. Upregulation of sestrins protect atriums against oxidative damage and fibrosis in human and experimental atrial fibrillation. Sci. Rep. 2017, 7, 46307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajman, L.; Chwalek, K.; Sinclair, D.A. Therapeutic potential of NAD-boosting molecules: The in vivo evidence. Cell Metab. 2018, 27, 529–547. [Google Scholar] [CrossRef] [Green Version]
- Bogan, K.L.; Brenner, C. Nicotinic acid, nicotinamide, and nicotinamide riboside: A molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu. Rev. Nutr. 2008, 28, 115–130. [Google Scholar] [CrossRef] [Green Version]
- de Picciotto, N.E.; Gano, L.B.; Johnson, L.C.; Martens, C.R.; Sindler, A.L.; Mills, K.F.; Imai, S.I.; Seals, D.R. Nicotinamide mononucleotide supplementation reverses vascular dysfunction and oxidative stress with aging in mice. Aging Cell 2016, 15, 522–530. [Google Scholar] [CrossRef]
- Imai, S. The NAD World 2.0: The importance of the inter-tissue communication mediated by NAMPT/NAD+/SIRT1 in mammalian aging and longevity control. NPJ Syst. Biol. Appl. 2016, 2, 16018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mills, K.F.; Yoshida, S.; Stein, L.R.; Grozio, A.; Kubota, S.; Sasaki, Y.; Redpath, P.; Migaud, M.E.; Apte, R.S.; Uchida, K.; et al. Long-term administration of nicotinamide mononucleotide mitigates age-associated physiological decline in mice. Cell Metab. 2016, 24, 795–806. [Google Scholar] [CrossRef] [Green Version]
- Tarantini, S.; Valcarcel-Ares, M.N.; Toth, P.; Yabluchanskiy, A.; Tucsek, Z.; Kiss, T.; Hertelendy, P.; Kinter, M.; Ballabh, P.; Süle, Z.; et al. Nicotinamide mononucleotide (NMN) supplementation rescues cerebromicrovascular endothelial function and neurovascular coupling responses and improves cognitive function in aged mice. Redox Biol. 2019, 24, 101192. [Google Scholar] [CrossRef]
- Yoshino, J.; Mills, K.F.; Yoon, M.J.; Imai, S. Nicotinamide mononucleotide, a key NAD+ intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011, 14, 528–536. [Google Scholar] [CrossRef] [Green Version]
- Benyo, Z.; Gille, A.; Bennett, C.L.; Clausen, B.E.; Offermanns, S. Nicotinic acid-induced flushing is mediated by activation of epidermal langerhans cells. Mol. Pharmacol. 2006, 70, 1844–1849. [Google Scholar] [CrossRef]
- Canto, C.; Houtkooper, R.H.; Pirinen, E.; Youn, D.Y.; Oosterveer, M.H.; Cen, Y.; Fernandez-Marcos, P.J.; Yamamoto, H.; Andreux, P.A.; Cettour-Rose, P.; et al. The NAD+ precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012, 15, 838–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trammell, S.A.; Weidemann, B.J.; Chadda, A.; Yorek, M.S.; Holmes, A.; Coppey, L.J.; Obrosov, A.; Kardon, R.H.; Yorek, M.A.; Brenner, C. Nicotinamide riboside opposes type 2 diabetes and neuropathy in mice. Sci. Rep. 2016, 6, 26933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diguet, N.; Trammell, S.A.J.; Tannous, C.; Deloux, R.; Piquereau, J.; Mougenot, N.; Mougenot, N.; Gouge, A.; Gressette, M.; Manoury, B.; et al. Nicotinamide riboside preserves cardiac function in a mouse model of dilated cardiomyopathy. Circulation 2018, 137, 2256–2273. [Google Scholar] [CrossRef] [Green Version]
- Shi, W.; Hegeman, M.A.; van Dartel, D.A.M.; Tang, J.; Suarez, M.; Swarts, H.; van der Hee, B.; Arola, L.; Keijer, J. Effects of a wide range of dietary nicotinamide riboside (NR) concentrations on metabolic flexibility and white adipose tissue (WAT) of mice fed a mildly obesogenic diet. Mol. Nutr. Food Res. 2017, 61, 1600878. [Google Scholar] [CrossRef] [Green Version]
- Shi, W.; Hegeman, M.; Doncheva, A.; Bekkenkamp-Grovenstein, M.; de Boer, V.C.J.; Keijer, J. High Dose of Dietary Nicotinamide Riboside Induces Glucose Intolerance and White Adipose Tissue Dysfunction in Mice Fed a Mildly Obesogenic Diet. Nutrients 2019, 11, 2439. [Google Scholar] [CrossRef] [Green Version]
- Janssen, J.J.; Grefte, S.; Keijer, J.; De Boer, V.C. Mito-nuclear communication by mitochondrial metabolites and its regulation by B-vitamins. Front. Physiol. 2019, 10, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, H.; Li, X.; Gao, H.; Feng, Z.; Li, X.; Zhao, L.; Jia, X.; Zhang, H.; Liu, J. High doses of nicotinamide prevent oxidative mitochondrial dysfunction in a cellular model and improve motor deficit in a Drosophila model of Parkinson’s disease. J. Neurosci. Res. 2008, 86, 2083–2090. [Google Scholar] [CrossRef]
- Suzuki, E.; Okuda, H.; Nishida, K.; Fujimoto, S.; Nagasawa, K. Protective effect of nicotinamide against poly (ADP-ribose) polymerase-1-mediated astrocyte death depends on its transporter-mediated uptake. Life Sci. 2010, 86, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wu, C.T.; Qi, X.; Meijering, R.A.; Hoogstra-Berends, F.; Tadevosyan, A.; Cubukcuoglu, D.G.; Durdu, S.; Akar, A.R.; Sibon, O.C.; et al. Activation of histone deacetylase-6 induces contractile dysfunction through derailment of α-tubulin proteostasis in experimental and human atrial fibrillation. Circulation 2014, 29, 346–358. [Google Scholar] [CrossRef] [Green Version]
- Pillai, V.B.; Sundaresan, N.R.; Kim, G.; Gupta, M.; Rajamohan, S.B.; Pillai, J.B.; Samant, S.; Ravindra, P.; Isbatan, A.; Gupta, M.P. Exogenous NAD blocks cardiac hypertrophic response via activation of the SIRT3-LKB1-AMP-activated kinase pathway. J. Biol. Chem. 2010, 285, 3133–3144. [Google Scholar] [CrossRef] [Green Version]
- Airhart, S.E.; Shireman, L.M.; Risler, L.J.; Anderson, G.D.; Nagana Gowda, G.; Raftery, D.; Tian, R.; Shen, D.D.; O’Brien, K.D. An open-label, non-randomized study of the pharmacokinetics of the nutritional supplement nicotinamide riboside (NR) and its effects on blood NAD+ levels in healthy volunteers. PLoS ONE 2017, 12, e0186459. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Hashim, M.J.; Mustafa, H.; Baniyas, M.Y.; Al Suwaidi, S.; AlKatheeri, R.; Alblooshi, F.M.K.; Almatrooshi, M.; Alzaabi, M.E.H.; Al Darmaki, R.S.; et al. Global epidemiology of ischemic heart disease: Results from the global burden of disease study. Cureus 2020, 12, e9349. [Google Scholar] [CrossRef]
- Kalogeris, T.; Baines, C.P.; Krenz, M.; Korthuis, R.J. Cell biology of ischemia/reperfusion injury. Int. Rev. Cell Mol. Biol. 2012, 298, 229–317. [Google Scholar] [CrossRef] [Green Version]
- Tran, D.H.; Wang, Z.V. Glucose metabolism in cardiac hypertrophy and heart failure. J. Am. Heart Assoc. 2019, 8, e012673. [Google Scholar] [CrossRef]
- Badalzadeh, R.; Azimi, A.; Alihemmati, A.; Yousefi, B. Chronic type-I diabetes could not impede the anti-inflammatory and anti-apoptotic effects of combined postconditioning with ischemia and cyclosporine A in myocardial reperfusion injury. J. Physiol. Biochem. 2017, 73, 111–120. [Google Scholar] [CrossRef]
- Feyzizadeh, S.; Javadi, A.; Badalzadeh, R.; Vafaee, M.S. Signaling mediators modulated by cardioprotective interventions in healthy and diabetic myocardium with ischaemia–reperfusion injury. Eur. J. Prev. Cardiol. 2018, 25, 1463–1481. [Google Scholar] [CrossRef]
- Gao, Q.; Deng, H.; Li, H.; Sun, C.; Sun, Y.; Wei, B.; Guo, M.; Jiang, X. Glycolysis and fatty acid β-oxidation, which one is the culprit of ischemic reperfusion injury. Int. J. Clin. Exp. Med. 2018, 11, 59–68. [Google Scholar]
- Yellon, D.M.; Hausenloy, D.J. Myocardial reperfusion injury. N. Engl. J. Med. 2007, 357, 1121–1135. [Google Scholar] [CrossRef]
- Baxter, P.; Chen, Y.; Xu, Y.; Swanson, R.A. Mitochondrial dysfunction induced by nuclear poly(ADP-ribose)polymerase-1: A treatable cause of cell death in stroke. Transl. Stroke Res. 2014, 5, 136–144. [Google Scholar] [CrossRef] [Green Version]
- Di Lisa, F.; Menabò, R.; Canton, M.; Barile, M.; Bernardi, P. Opening of the mitochondrial permeability transition pore causes depletion of mitochondrial and cytosolic NAD+ and is a causative event in the death of myocytes in postischemic reperfusion of the heart. J. Biol. Chem. 2001, 276, 2571–2575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, C.K.; Chhabra, G.; Ndiaye, M.A.; Garcia-Peterson, L.M.; Mack, N.J.; Ahmad, N. The role of sirtuins in antioxidant and redox signaling. Antioxid. Redox Signal. 2018, 28, 643–661. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, S.; Sadoshima, J. The role of sirtuins in cardiac disease. Am. J. Physiol. Heart Circ. Physiol. 2015, 309, H1375–H1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becatti, M.; Taddei, N.; Cecchi, C.; Nassi, N.; Nassi, P.A.; Fiorillo, C. SIRT1 modulates MAPK pathways in ischemic–reperfused cardiomyocytes. Cell. Mol. Life Sci. 2012, 69, 2245–2260. [Google Scholar] [CrossRef]
- Zhang, J.; Ren, D.; Fedorova, J.; He, Z.; Li, J. SIRT1/SIRT3 Modulates redox homeostasis during ischemia/reperfusion in the aging heart. Antioxidants 2020, 9, 858. [Google Scholar] [CrossRef]
- Hosseini, L.; Vafaee, M.S.; Mahmoudi, J.; Badalzadeh, R. Nicotinamide adenine dinucleotide emerges as a therapeutic target in aging and ischemic conditions. Biogerontology 2019, 20, 381–395. [Google Scholar] [CrossRef]
- Bliksøen, M.; Baysa, A.; Eide, L.; Bjørås, M.; Suganthan, R.; Vaage, J.; Stensløkken, K.; Valen, G. Mitochondrial DNA damage and repair during ischemia–reperfusion injury of the heart. J. Mol. Cell. Cardiol. 2015, 78, 9–22. [Google Scholar] [CrossRef]
- Inafuku, H.; Kuniyoshi, Y.; Yamashiro, S.; Arakaki, K.; Nagano, T.; Morishima, Y.; Kise, Y. Determination of oxidative stress and cardiac dysfunction after ischemia/reperfusion injury in isolated rat hearts. Ann. Thorac. Cardiovasc. Surg. 2013, 19, 186–194. [Google Scholar] [CrossRef] [Green Version]
- Cordis, G.A.; Maulik, G.; Bagchi, D.; Riedel, W.; Das, D.K. Detection of oxidative DNA damage to ischemic reperfused rat hearts by 8-hydroxydeoxyguanosine formation. J. Mol. Cell. Cardiol. 1998, 30, 1939–1944. [Google Scholar] [CrossRef]
- Mercer, J.R.; Cheng, K.K.; Figg, N.; Gorenne, I.; Mahmoudi, M.; Griffin, J.; Vidal-Puig, A.; Logan, A.; Murphy, M.P.; Bennett, M. DNA damage links mitochondrial dysfunction to atherosclerosis and the metabolic syndrome. Circ. Res. 2010, 107, 1021–1031. [Google Scholar] [CrossRef]
- Shukla, P.C.; Singh, K.K.; Yanagawa, B.; Teoh, H.; Verma, S. DNA damage repair and cardiovascular diseases. Can. J. Cardiol. 2010, 26, 13A–16A. [Google Scholar] [CrossRef]
- Powers, S.K.; Murlasits, Z.; Wu, M.; Kavazis, A.N. Ischemia-reperfusion-induced cardiac injury: A brief review. Med. Sci. Sports Exerc. 2007, 39, 1529–1536. [Google Scholar] [CrossRef] [PubMed]
- Karahalil, B.; Kesimci, E.; Emerce, E.; Gumus, T.; Kanbak, O. The impact of OGG1, MTH1 and MnSOD gene polymorphisms on 8-hydroxy-2′-deoxyguanosine and cellular superoxide dismutase activity in myocardial ischemia–reperfusion. Mol. Biol. Rep. 2011, 38, 2427–2435. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.Q.; Zhou, H.Z.; Cecchini, G.; Gray, M.O.; Karliner, J.S. MnSOD in mouse heart: Acute responses to ischemic preconditioning and ischemia-reperfusion injury. Am. J. Physiol. Heart Circ. Physiol. 2005, 288, H2986–H2994. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Siu, B.; Ho, Y.-S.; Vincent, R.; Chua, C.C.; Hamdy, R.C.; Chua, B.H. Overexpression of MnSOD protects against myocardial ischemia/reperfusion injury in transgenic mice. J. Mol. Cell. Cardiol. 1998, 30, 2281–2289. [Google Scholar] [CrossRef] [PubMed]
- Dare, A.J.; Logan, A.; Prime, T.A.; Rogatti, S.; Goddard, M.; Bolton, E.M.; Bradley, J.A.; Pettigrew, G.J.; Murphy, M.P.; Saeb-Parsy, K. The mitochondria-targeted anti-oxidant MitoQ decreases ischemia-reperfusion injury in a murine syngeneic heart transplant model. J. Heart Lung Transplant. 2015, 34, 1471–1480. [Google Scholar] [CrossRef] [Green Version]
- Pei, H.F.; Hou, J.N.; Wei, F.P.; Xue, Q.; Zhang, F.; Peng, C.F.; Yang, Y.; Tian, Y.; Feng, J.; Du, J. Melatonin attenuates postmyocardial infarction injury via increasing Tom70 expression. J. Pineal Res. 2017, 62, e12371. [Google Scholar] [CrossRef]
- Zhou, H.; Ma, Q.; Zhu, P.; Ren, J.; Reiter, R.J.; Chen, Y. Protective role of melatonin in cardiac ischemia-reperfusion injury: From pathogenesis to targeted therapy. J. Pineal Res. 2018, 64, e12471. [Google Scholar] [CrossRef] [Green Version]
- Koronowski, K.B.; Perez-Pinzon, M.A. Sirt1 in cerebral ischemia. Brain Circ. 2015, 1, 69–78. [Google Scholar] [CrossRef] [Green Version]
- Brunet, A.; Sweeney, L.B.; Sturgill, J.F.; Chua, K.F.; Greer, P.L.; Lin, Y.; Tran, H.; Ross, S.E.; Mostoslavsky, R.; Cohen, H.Y. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 2004, 303, 2011–2015. [Google Scholar] [CrossRef] [Green Version]
- Hsu, C.P.; Zhai, P.; Yamamoto, T.; Maejima, Y.; Matsushima, S.; Hariharan, N.; Shao, D.; Takagi, H.; Oka, S.; Sadoshima, J. Silent information regulator 1 protects the heart from ischemia/reperfusion. Circulation 2010, 122, 2170–2182. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, T.; Byun, J.; Zhai, P.; Ikeda, Y.; Oka, S.; Sadoshima, J. Nicotinamide mononucleotide, an intermediate of NAD+ synthesis, protects the heart from ischemia and reperfusion. PLoS ONE 2014, 9, e98972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Wang, Q.; Zhao, B.; Wu, Q.; Wang, P. Exogenous nicotinamide adenine dinucleotide administration alleviates ischemia/reperfusion-induced oxidative injury in isolated rat hearts via Sirt5-SDH-succinate pathway. Eur. J. Pharmacol. 2019, 858, 172520. [Google Scholar] [CrossRef] [PubMed]
- Lynn, E.G.; McLeod, C.J.; Gordon, J.P.; Bao, J.; Sack, M.N. SIRT2 is a negative regulator of anoxia-reoxygenation tolerance via regulation of 14-3-3 zeta and BAD in H9c2 cells. FEBS Lett. 2008, 582, 2857–2862. [Google Scholar] [CrossRef] [Green Version]
- Zeng, G.; Liu, H.; Wang, H. Amelioration of myocardial ischemia-reperfusion injury by SIRT4 involves mitochondrial protection and reduced apoptosis. Biochem. Biophys. Res. Commun. 2018, 502, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, B.; Fu, X.; Guan, S.; Han, W.; Zhang, J.; Gan, Q.; Fang, W.; Ying, W.; Qu, X. Exogenous NAD+ administration significantly protects against myocardial ischemia/reperfusion injury in rat model. Am. J. Transl. Res. 2016, 8, 3342–3350. [Google Scholar]
- Zhai, X.; Han, W.; Wang, M.; Guan, S.; Qu, X. Exogenous supplemental NAD+ protect myocardium against myocardial ischemic/reperfusion injury in swine model. Am. J. Transl. Res. 2019, 11, 6066–6074. [Google Scholar] [PubMed]
- Ping, L.; Yin, Z.; Li, Y. GW27-e0632 NR reduced myocardial ischemia-reperfusion injury by improving mitochondrial biogenesis and reducing excessive autophagy via Sirt3-PGC-1α/P53 pathway. J. Am. Coll. Cardiol. 2016, 68, C24–C25. [Google Scholar] [CrossRef]
- Nadtochiy, S.M.; Wang, Y.T.; Nehrke, K.; Munger, J.; Brookes, P.S. Cardioprotection by nicotinamide mononucleotide (NMN): Involvement of glycolysis and acidic pH. J. Mol. Cell. Cardiol. 2018, 121, 155–162. [Google Scholar] [CrossRef]
- Baines, C.P. The mitochondrial permeability transition pore and ischemia-reperfusion injury. Basic Res. Cardiol. 2009, 104, 181–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, M.V.; Yang, X.M.; Downey, J.M. The pH hypothesis of postconditioning: Staccato reperfusion reintroduces oxygen and perpetuates myocardial acidosis. Circulation 2007, 115, 1895–1903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wang, Q.; Watson, L.J.; Jones, S.P.; Epstein, P.N. Cardiac overexpression of 8-oxoguanine DNA glycosylase 1 protects mitochondrial DNA and reduces cardiac fibrosis following transaortic constriction. Am. J. Physiol. Heart Circ. Physiol. 2011, 301, H2073–H2080. [Google Scholar] [CrossRef] [Green Version]
- Liaudet, L.; Szabó, É.; Timashpolsky, L.; Virág, L.; Cziráki, A.; Szabó, C. Suppression of poly(ADP-ribose)polymerase activation by 3-aminobenzamide in a rat model of myocardial infarction: Long-term morphological and functional consequences. Br. J. Pharmacol. 2001, 133, 1424–1430. [Google Scholar] [CrossRef] [Green Version]
- Szabó, G.; Soós, P.; Mandera, S.; Heger, U.; Flechtenmacher, C.; Bährle, S.; Seres, L.; Cziráki, A.; Gries, A.; Zsengellér, Z. INO-1001 a novel poly(ADP-ribose)polymerase (PARP) inhibitor improves cardiac and pulmonary function after crystalloid cardioplegia and extracorporal circulation. Shock 2004, 21, 426–432. [Google Scholar] [CrossRef]
- Faro, R.; Toyoda, Y.; McCully, J.D.; Jagtap, P.; Szabo, E.; Virag, L.; Bianchi, C.; Levitsky, S.; Szabo, C.; Sellke, F.W. Myocardial protection by PJ34, a novel potent poly(ADP-ribose)synthetase inhibitor. Ann. Thorac. Surg. 2002, 73, 575–581. [Google Scholar] [CrossRef]
- Thiemermann, C.; Bowes, J.; Myint, F.P.; Vane, J.R. Inhibition of the activity of poly (ADP ribose) synthetase reduces ischemia–reperfusion injury in the heart and skeletal muscle. Proc. Natl. Acad. Sci. USA 1997, 94, 679–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zingarelli, B.; Cuzzocrea, S.; Zsengeller, Z.; Salzman, A.; Szabó, C. Inhibition of poly (ADP ribose) synthetase protects against myocardial ischemia and reperfusion injury. Cardiovasc. Res. 1997, 36, 205–212. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Wang, D.F.; Zhang, Z.; Han, D.; Yang, K. EGb 761 protects cardiac microvascular endothelial cells against hypoxia/reoxygenation injury and exerts inhibitory effect on the ATM pathway. J. Microbiol. Biotechnol. 2017, 27, 584–590. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Wei, Q.; Dong, G.; Huo, Y.; Dong, Z. DNA damage response in renal ischemia–reperfusion and ATP-depletion injury of renal tubular cells. J. Microbiol. Biotechnol. 2014, 1842, 1088–1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Literature 1 | Subjects | Measurements | Results |
|---|---|---|---|
| Montaigne et al. [123] |
|
|
|
| Xie et al. [124] |
|
| AF patients’ samples:
|
| Jeganathan et al. [125] |
|
|
|
| Wiersma et al. [126] |
|
|
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, L.; Wang, Z.; Carmone, C.; Keijer, J.; Zhang, D. Role of Oxidative DNA Damage and Repair in Atrial Fibrillation and Ischemic Heart Disease. Int. J. Mol. Sci. 2021, 22, 3838. https://doi.org/10.3390/ijms22083838
Hu L, Wang Z, Carmone C, Keijer J, Zhang D. Role of Oxidative DNA Damage and Repair in Atrial Fibrillation and Ischemic Heart Disease. International Journal of Molecular Sciences. 2021; 22(8):3838. https://doi.org/10.3390/ijms22083838
Chicago/Turabian StyleHu, Liangyu, Zhengkun Wang, Claudia Carmone, Jaap Keijer, and Deli Zhang. 2021. "Role of Oxidative DNA Damage and Repair in Atrial Fibrillation and Ischemic Heart Disease" International Journal of Molecular Sciences 22, no. 8: 3838. https://doi.org/10.3390/ijms22083838