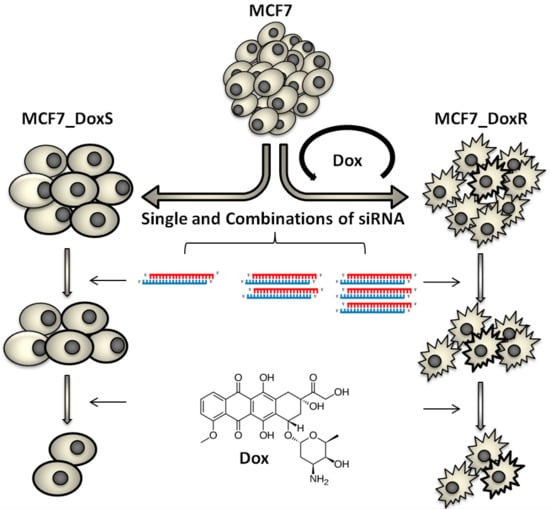
Downregulation of STAT3, β-Catenin, and Notch-1 by Single and Combinations of siRNA Treatment Enhance Chemosensitivity of Wild Type and Doxorubicin Resistant MCF7 Breast Cancer Cells to Doxorubicin
Abstract
:1. Introduction
2. Results and Discussion
2.1. Induction of Doxorubicin Resistant MCF7 Cells (MCF7_DoxR)
2.2. The Expression of Multidrug Resistant-Related Genes in MCF7_DoxR
2.3. Expression of Notch-1, STAT3, and β-Catenin in MCF7_DoxS and MCF7_DoxR Cells.
2.4. Expression of CD44/CD24 Surface Markers
2.5. Silencing of Notch-1, STAT3, and β-Catenin in MCF7_DoxS and MCF7_DoxR Cells Affects Cell Proliferation
2.6. Combination Index
2.7. The IC50 of Doxorubicin Decreased after siRNA Treatment
3. Materials and Methods
3.1. Oligonucleotides
3.2. Cell Culture
3.3. Development of MCF7 Doxorubicin Resistant Cells (MCF7_DoxR)
3.4. Array Q-PCR for Quantification of Multidrug Resistance Related-Genes Expression
3.5. Q-PCR for Quantification of STAT3, β-Catenin, and Notch-1 Expression
3.6. Western Blot
3.7. Detection of CD44 and CD24 Expression
3.8. Detection of Cell Viability by MTT Assay
3.8.1. IC50 of Doxorubicin before siRNA Treatment
3.8.2. IC50 of siRNA Treatment
3.8.3. IC50 of Doxorubicin after siRNA Treatment (Chemosensitivity)
3.9. Combination Index
3.10. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- McGuire, S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv. Nutr. 2016, 7, 418–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghoncheh, M.; Pournamdar, Z.; Salehiniya, H. Incidence and Mortality and Epidemiology of Breast Cancer in the World. Asian Pac. J. Cancer Prev. 2016, 17, 43–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug resistance in cancer: An overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef] [PubMed]
- Vasir, J.K.; Labhasetwar, V. Targeted drug delivery in cancer therapy. Technol. Cancer Res. Treat 2005, 4, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Baudino, T.A. Targeted Cancer Therapy: The Next Generation of Cancer Treatment. Curr. Drug Discov. Technol. 2015, 12, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Takebe, N.; Miele, L.; Harris, P.J.; Jeong, W.; Bando, H.; Kahn, M.; Yang, S.X.; Ivy, S.P. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: Clinical update. Nat. Rev. Clin. Oncol. 2015, 12, 445–464. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Zhu, Y.; Oupicky, D. Recent advances in delivery of drug-nucleic acid combinations for cancer treatment. J. Control Release 2013, 172, 589–600. [Google Scholar] [CrossRef]
- Kc, R.B.; Thapa, B.; Valencia-Serna, J.; Aliabadi, H.M.; Uludag, H. Nucleic acid combinations: A new frontier for cancer treatment. J. Control Release 2017, 256, 153–169. [Google Scholar] [Green Version]
- Cardoso, A.; Trabulo, S.; Moreira, J.N.; Düzgüneş, N.; de Lima, M.C.P. Targeted lipoplexes for siRNA delivery. Methods Enzymol. 2009, 465, 267–287. [Google Scholar]
- Tseng, Y.-C.; Mozumdar, S.; Huang, L. Lipid-based systemic delivery of siRNA. Adv. Drug Deliv. Rev. 2009, 61, 721–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eroles, P.; Bosch, A.; Perez-Fidalgo, J.A.; Lluch, A. Molecular biology in breast cancer: Intrinsic subtypes and signaling pathways. Cancer Treat. Rev. 2012, 38, 698–707. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zang, S.; Chen, F.; Dai, J.; Guo, D.; Tse, W.; Qu, X.; Ma, D.; Ji, C. RNAi-mediated knockdown of Notch-1 leads to cell growth inhibition and enhanced chemosensitivity in human breast cancer. Oncol. Rep. 2010, 23, 893–899. [Google Scholar] [PubMed]
- Yang, Z.; Cai, J.H.; Xie, S.J.; Li, G.X.; Song, W.Q.; Yan, Q.H.; Yan, L.; Zhang, F. Therapeutic effects of signal transducer and activator of transcription 3 siRNA on human breast cancer in xenograft mice. Chin. Med. J. 2011, 124, 1854–1861. [Google Scholar]
- Zhang, D.; Fei, F.; Li, S.; Zhao, Y.; Yang, Z.; Qu, J.; Zhang, X.; Yin, Y.; Zhang, S. The role of beta-catenin in the initiation and metastasis of TA2 mice spontaneous breast cancer. J. Cancer 2017, 8, 2114–2123. [Google Scholar] [CrossRef]
- Deng, X.; Apple, S.; Zhao, H.; Song, J.; Lee, M.; Luo, W.; Wu, X.; Chung, D.; Pietras, R.J.; Chang, H.R. CD24 Expression and differential resistance to chemotherapy in triple-negative breast cancer. Oncotarget 2017, 8, 38294–38308. [Google Scholar] [CrossRef] [Green Version]
- Van Phuc, P.; Nhan, P.L.; Nhung, T.H.; Tam, N.T.; Hoang, N.M.; Tue, V.G.; Thuy, D.T.; Ngoc, P.K. Downregulation of CD44 reduces doxorubicin resistance of CD44CD24 breast cancer cells. OncoTargetsTher. 2011, 4, 71–78. [Google Scholar]
- Minotti, G.; Menna, P.; Salvatorelli, E.; Cairo, G.; Gianni, L. Anthracyclines: Molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol. Rev. 2004, 56, 185–229. [Google Scholar] [CrossRef]
- Tassone, P.; Tagliaferri, P.; Perricelli, A.; Blotta, S.; Quaresima, B.; Martelli, M.; Goel, A.; Barbieri, V.; Costanzo, F.; Boland, C. BRCA1 expression modulates chemosensitivity of BRCA1-defective HCC1937 human breast cancer cells. Br. J. Cancer 2003, 88, 1285–1291. [Google Scholar] [CrossRef] [Green Version]
- Taylor, C.W.; Dalton, W.S.; Parrish, P.R.; Gleason, M.C.; Bellamy, W.T.; Thompson, F.H.; Roe, D.J.; Trent, J.M. Different mechanisms of decreased drug accumulation in doxorubicin and mitoxantrone resistant variants of the MCF7 human breast cancer cell line. Br. J. Cancer 1991, 63, 923–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AbuHammad, S.; Zihlif, M. Gene expression alterations in doxorubicin resistant MCF7 breast cancer cell line. Genomics 2013, 101, 213–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weihua, Z.; Lin, Q.; Ramoth, A.J.; Fan, D.; Fidler, I.J. Formation of solid tumors by a single multinucleated cancer cell. Cancer 2011, 117, 4092–4099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sansone, P.; Bromberg, J. Targeting the interleukin-6/Jak/stat pathway in human malignancies. J. Clin. Oncol. 2012, 30, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- VanKlompenberg, M.K.; Bedalov, C.O.; Soto, K.F.; Prosperi, J.R. APC selectively mediates response to chemotherapeutic agents in breast cancer. BMC Cancer 2015, 15, 457. [Google Scholar] [CrossRef] [PubMed]
- VanKlompenberg, M.K.; Leyden, E.; Arnason, A.H.; Zhang, J.T.; Stefanski, C.D.; Prosperi, J.R. APC loss in breast cancer leads to doxorubicin resistance via STAT3 activation. Oncotarget 2017, 8, 102868–102879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumawat, K.; Koopmans, T.; Gosens, R. β-catenin as a regulator and therapeutic target for asthmatic airway remodeling. Expert Opin. Ther. Targets 2014, 18, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.; Li, N.; Qi, J.; Yin, P.; Zhao, C.; Wang, L.; Li, Z.; Zha, X. Wnt/β-catenin signaling induces the transcription of cystathionine-γ-lyase, a stimulator of tumor in colon cancer. Cell. Signal. 2014, 26, 2801–2808. [Google Scholar] [CrossRef]
- Teng, Y.; Wang, X.; Wang, Y.; Ma, D. Wnt/β-catenin signaling regulates cancer stem cells in lung cancer A549 cells. Biochem. Biophys. Res. Commun. 2010, 392, 373–379. [Google Scholar] [CrossRef]
- Deng, Y.; Pu, X.; Huang, M.; Xiao, J.; Zhou, J.; Lin, T.; Lin, E.H. 5-Fluorouracil upregulates the activity of Wnt signaling pathway in CD133-positive colon cancer stem-like cells. Chin. J. Cancer 2010, 29, 810–815. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.; Xia, W.; Wong, G. Hyaluronan-mediated CD44 interaction with p300 and SIRT1 regulates beta-catenin signaling and NFkappaB-specific transcription activity leading to MDR1 and Bcl-xL gene expression and chemoresistance in breast tumor cells. J. Biol. Chem. 2009, 284, 2657–2671. [Google Scholar] [CrossRef]
- Reedijk, M.; Odorcic, S.; Zhang, H.; Chetty, R.; Tennert, C.; Dickson, B.C.; Lockwood, G.; Gallinger, S.; Egan, S.E. Activation of Notch signaling in human colon adenocarcinoma. Int. J. Oncol. 2008, 33, 1223–1229. [Google Scholar]
- Wang, Z.; Banerjee, S.; Li, Y.; Rahman, K.W.; Zhang, Y.; Sarkar, F.H. Down-regulation of Notch-1 inhibits invasion by inactivation of nuclear factor-κB, vascular endothelial growth factor, and matrix metalloproteinase-9 in pancreatic cancer cells. Cancer Res. 2006, 66, 2778–2784. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Y.; Banerjee, S.; Kong, D.; Ahmad, A.; Nogueira, V.; Hay, N.; Sarkar, F.H. Down-regulation of Notch-1 and Jagged-1 inhibits prostate cancer cell growth, migration and invasion, and induces apoptosis via inactivation of Akt, mTOR, and NF-κB signaling pathways. J. Cell. Biochem. 2010, 109, 726–736. [Google Scholar] [CrossRef]
- Politi, K.; Feirt, N.; Kitajewski, J. Notch in Mammary Gland Development and Breast Cancer. Seminars in Cancer Biology. 2004, 14, 341–347. [Google Scholar] [CrossRef]
- Weijzen, S.; Rizzo, P.; Braid, M.; Vaishnav, R.; Jonkheer, S.M.; Zlobin, A.; Osborne, B.A.; Gottipati, S.; Aster, J.C.; Hahn, W.C. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat. Med. 2002, 8, 979. [Google Scholar] [CrossRef]
- Diévart, A.; Beaulieu, N.; Jolicoeur, P. Involvement of Notch1 in the development of mouse mammary tumors. Oncogene 1999, 18, 5973. [Google Scholar] [CrossRef]
- Cho, S.; Lu, M.; He, X.; Ee, P.L.; Bhat, U.; Schneider, E.; Miele, L.; Beck, W.T. Notch1 regulates the expression of the multidrug resistance gene ABCC1/MRP1 in cultured cancer cells. Proc. Natl. Acad. Sci. USA 2011, 108, 20778–20783. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Ma, H.; Zhang, J.; Zhu, L.; Wang, C.; Yang, Y. Unraveling the roles of CD44/CD24 and ALDH1 as cancer stem cell markers in tumorigenesis and metastasis. Sci. Rep. 2017, 7, 13856. [Google Scholar] [CrossRef]
- Ricardo, S.; Vieira, A.F.; Gerhard, R.; Leitao, D.; Pinto, R.; Cameselle-Teijeiro, J.F.; Milanezi, F.; Schmitt, F.; Paredes, J. Breast cancer stem cell markers CD44, CD24 and ALDH1: Expression distribution within intrinsic molecular subtype. J. Clin. Pathol. 2011, 64, 937–946. [Google Scholar] [CrossRef]
- Mylona, E.; Giannopoulou, I.; Fasomytakis, E.; Nomikos, A.; Magkou, C.; Bakarakos, P.; Nakopoulou, L. The clinicopathologic and prognostic significance of CD44+/CD24(-/low) and CD44-/CD24+ tumor cells in invasive breast carcinomas. Hum. Pathol. 2008, 39, 1096–1102. [Google Scholar] [CrossRef]
- Ahmed, M.A.; Aleskandarany, M.A.; Rakha, E.A.; Moustafa, R.Z.; Benhasouna, A.; Nolan, C.; Green, A.R.; Ilyas, M.; Ellis, I.O. A CD44(-)/CD24(+) phenotype is a poor prognostic marker in early invasive breast cancer. Breast Cancer Res. Treat. 2012, 133, 979–995. [Google Scholar] [CrossRef]
- Marotta, L.L.; Almendro, V.; Marusyk, A.; Shipitsin, M.; Schemme, J.; Walker, S.R.; Bloushtain-Qimron, N.; Kim, J.J.; Choudhury, S.A.; Maruyama, R.; et al. The JAK2/STAT3 signaling pathway is required for growth of CD44(+)CD24(-) stem cell-like breast cancer cells in human tumors. J. Clin. Investig. 2011, 121, 2723–2735. [Google Scholar] [CrossRef]
- Tsou, S.-H.; Chen, T.-M.; Hsiao, H.-T.; Chen, Y.-H. A critical dose of doxorubicin is required to alter the gene expression profiles in MCF-7 cells acquiring multidrug resistance. PLoS ONE 2015, 10, e0116747. [Google Scholar] [CrossRef]
- Wang, X.; Crowe, P.J.; Goldstein, D.; Yang, J.L. STAT3 inhibition, a novel approach to enhancing targeted therapy in human cancers (review). Int. J. Oncol. 2012, 41, 1181–1191. [Google Scholar] [CrossRef]
- Deng, J.; Miller, S.A.; Wang, H.Y.; Xia, W.; Wen, Y.; Zhou, B.P.; Li, Y.; Lin, S.Y.; Hung, M.C. beta-catenin interacts with and inhibits NF-kappa B in human colon and breast cancer. Cancer Cell 2002, 2, 323–334. [Google Scholar] [CrossRef]
- Armanious, H.; Gelebart, P.; Mackey, J.; Ma, Y.; Lai, R. STAT3 upregulates the protein expression and transcriptional activity of beta-catenin in breast cancer. Int. J. Clin. Exp. Pathol. 2010, 3, 654–664. [Google Scholar]
- Jin, Y.H.; Kim, H.; Ki, H.; Yang, I.; Yang, N.; Lee, K.Y.; Kim, N.; Park, H.S.; Kim, K. Beta-catenin modulates the level and transcriptional activity of Notch1/NICD through its direct interaction. Biochim. Biophys. Acta 2009, 1793, 290–299. [Google Scholar] [CrossRef] [Green Version]
- Hayward, P.; Brennan, K.; Sanders, P.; Balayo, T.; DasGupta, R.; Perrimon, N.; Martinez Arias, A. Notch modulates Wntsignalling by associating with Armadillo/beta-catenin and regulating its transcriptional activity. Development 2005, 132, 1819–1830. [Google Scholar] [CrossRef]
- Jin, S.; Mutvei, A.P.; Chivukula, I.V.; Andersson, E.R.; Ramskold, D.; Sandberg, R.; Lee, K.L.; Kronqvist, P.; Mamaeva, V.; Ostling, P.; et al. Non-canonical Notch signaling activates IL-6/JAK/STAT signaling in breast tumor cells and is controlled by p53 and IKKalpha/IKKbeta. Oncogene 2013, 32, 4892–4902. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, M.; Liu, W.; Wang, C.; Zhang, Q.; Li, W. beta-Catenin knockdown inhibits pituitary adenoma cell proliferation and invasion via interfering with AKT and gelatinases expression. Int. J. Oncol. 2015, 46, 1643–1650. [Google Scholar] [CrossRef]
- Ayyanan, A.; Civenni, G.; Ciarloni, L.; Morel, C.; Mueller, N.; Lefort, K.; Mandinova, A.; Raffoul, W.; Fiche, M.; Dotto, G.P.; et al. Increased Wnt signaling triggers oncogenic conversion of human breast epithelial cells by a Notch-dependent mechanism. Proc. Natl. Acad. Sci. USA 2006, 103, 3799–3804. [Google Scholar] [CrossRef] [Green Version]
- Gariboldi, M.B.; Ravizza, R.; Molteni, R.; Osella, D.; Gabano, E.; Monti, E. Inhibition of Stat3 increases doxorubicin sensitivity in a human metastatic breast cancer cell line. Cancer Lett. 2007, 258, 181–188. [Google Scholar] [CrossRef]
- Xu, J.; Prosperi, J.R.; Choudhury, N.; Olopade, O.I.; Goss, K.H. β-Catenin is required for the tumorigenic behavior of triple-negative breast cancer cells. PLoS ONE 2015, 10, e0117097. [Google Scholar] [CrossRef]
- Jaganathan, H.; Mitra, S.; Srinivasan, S.; Dave, B.; Godin, B. Design and in vitro evaluation of layer by layer siRNAnanovectors targeting breast tumor initiating cells. PLoS ONE 2014, 9, e91986. [Google Scholar] [CrossRef]
- Sakamoto, K.; Fujii, T.; Kawachi, H.; Miki, Y.; Omura, K.; Morita, K.; Kayamori, K.; Katsube, K.; Yamaguchi, A. Reduction of NOTCH1 expression pertains to maturation abnormalities of keratinocytes in squamous neoplasms. Lab. Investig. 2012, 92, 688–702. [Google Scholar] [CrossRef] [Green Version]
- Jiang, G.; Xiao, X.; Zeng, Y.; Nagabhushanam, K.; Majeed, M.; Xiao, D. Targeting beta-catenin signaling to induce apoptosis in human breast cancer cells by z-guggulsterone and Gugulipid extract of Ayurvedic medicine plant Commiphoramukul. BMC Complement. Altern. Med. 2013, 13, 203. [Google Scholar] [CrossRef]
- Qiagen. Available online: www.SABiosciences.com/pcrarraydataanalysis.php (accessed on 23 January 2019).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta DeltaC(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef]
- Nishimura, M.; Jung, E.-J.; Shah, M.Y.; Lu, C.; Spizzo, R.; Shimizu, M.; Han, H.D.; Ivan, C.; Rossi, S.; Zhang, X. Therapeutic synergy between microRNA and siRNA in ovarian cancer treatment. Cancer Discov. 2013, 3, 1302–1315. [Google Scholar] [CrossRef]







| Mechanism | Fold Change | |
|---|---|---|
| Upregulated (≥2-folds) | ||
| Drug efflux | ABCC1 | 2.4 |
| ABCC2 | 3.0 | |
| ABCC3 | 5.4 | |
| MVP | 2.5 | |
| Drug inactivation | CYP1A1 | 7.1 |
| CYP3A5 | 3.4 | |
| AhR | 4.1 | |
| ARNT1 | 2.3 | |
| SULT1E1 | 2.5 | |
| EPHX1 | 2.4 | |
| Cell cycle and cell death inhibition | PPARG | 14.5 |
| MET | 6.8 | |
| HIF1A | 6.2 | |
| CDKN1A | 3.6 | |
| RXRB | 2.8 | |
| IGF1R | 2.6 | |
| PPARD | 2.2 | |
| IGF1R | 2.6 | |
| XPA | 4.6 | |
| Growth factors receptors axes | ERBB4 | 21.5 |
| EGFR | 11.4 | |
| ERBB2 | 3.4 | |
| ERBB3 | 2.9 | |
| Myc | 2.1 | |
| Downregulated (≥2-folds) | ||
| Apoptotic regulator | Bcl-2 | −13.9 |
| Cell Type | siRNA Combination | CI/Fa = 0.5 | CI/Fa = 0.75 | CI/Fa = 0.9 | CI/Fa = 0.95 | CI/Fa = 0.97 |
|---|---|---|---|---|---|---|
| MCF7_DoxS | STAT3 + β-catenin | 1.19 | 1.20 | 1.21 | 1.21 | 1.22 |
| STAT3 + Notch-1 | 0.58 | 0.59 | 0.61 | 0.62 | 0.62 | |
| β-Catenin + Notch-1 | 1.07 | 1.06 | 1.05 | 1.05 | 1.04 | |
| STAT3 + β-catenin + Notch-1 | 0.69 | 0.68 | 0.67 | 0.67 | 0.66 | |
| MCF7_DoxR | STAT3 + β-catenin | 1.11 | 1.08 | 1.06 | 1.06 | 1.06 |
| STAT3 + Notch-1 | 0.53 | 0.54 | 0.56 | 0.58 | 0.59 | |
| β-Catenin + Notch-1 | 1.07 | 1.07 | 1.08 | 1.08 | 1.08 | |
| STAT3 + β-catenin + Notch-1 | 0.50 | 0.52 | 0.53 | 0.54 | 0.55 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alshaer, W.; Alqudah, D.A.; Wehaibi, S.; Abuarqoub, D.; Zihlif, M.; Hatmal, M.M.; Awidi, A. Downregulation of STAT3, β-Catenin, and Notch-1 by Single and Combinations of siRNA Treatment Enhance Chemosensitivity of Wild Type and Doxorubicin Resistant MCF7 Breast Cancer Cells to Doxorubicin. Int. J. Mol. Sci. 2019, 20, 3696. https://doi.org/10.3390/ijms20153696
Alshaer W, Alqudah DA, Wehaibi S, Abuarqoub D, Zihlif M, Hatmal MM, Awidi A. Downregulation of STAT3, β-Catenin, and Notch-1 by Single and Combinations of siRNA Treatment Enhance Chemosensitivity of Wild Type and Doxorubicin Resistant MCF7 Breast Cancer Cells to Doxorubicin. International Journal of Molecular Sciences. 2019; 20(15):3696. https://doi.org/10.3390/ijms20153696
Chicago/Turabian StyleAlshaer, Walhan, Dana A. Alqudah, Suha Wehaibi, Duaa Abuarqoub, Malek Zihlif, Ma’mon M. Hatmal, and Abdalla Awidi. 2019. "Downregulation of STAT3, β-Catenin, and Notch-1 by Single and Combinations of siRNA Treatment Enhance Chemosensitivity of Wild Type and Doxorubicin Resistant MCF7 Breast Cancer Cells to Doxorubicin" International Journal of Molecular Sciences 20, no. 15: 3696. https://doi.org/10.3390/ijms20153696