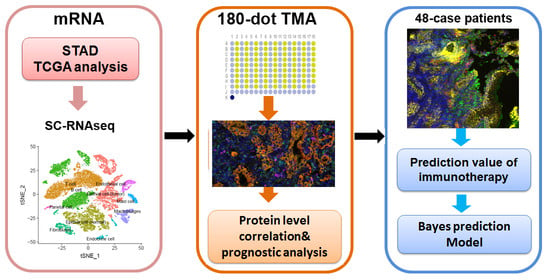
Co-Expression with Membrane CMTM6/4 on Tumor Epithelium Enhances the Prediction Value of PD-L1 on Anti-PD-1/L1 Therapeutic Efficacy in Gastric Adenocarcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Antibodies
2.2. Bioinformatics Analysis
2.3. Human Gastric Adenocarcinoma Tissue Array and GC Specimens
2.4. Multiplex Immunofluoresence Staining with Opal Fluorophores
2.5. Bayes Model
2.6. Statistical Analysis
3. Results
3.1. The Transcriptional Profile of CMTM6/4 and PD-L1 in Gastric Cancer Tissues
3.2. Membrane Co-Expression of CMTM6 and PD-L1 in Human Gastric Adenocarcinoma Tissues Array
3.3. Higher Levels of CMTM6/4 Correlate with Poor Prognosis in GC Patients
3.3.1. Co-Expression of CMTM6/4 and PD-L1 Is Associated with Better Response to Anti-PD-1/L1 Therapy in Gastric Adenocarcinoma Patients
3.3.2. Co-Expression of Membrane CMTM6/4 and PD-L1 Is Beneficial to Precision Patients Screening
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nakamura, Y.; Kawazoe, A.; Lordick, F.; Janjigian, Y.Y.; Shitara, K. Biomarker-targeted therapies for advanced-stage gastric and gastro-oesophageal junction cancers: An emerging paradigm. Nat. Rev. Clin. Oncol. 2021, 18, 473–487. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.K.; Boku, N.; Satoh, T.; Ryu, M.H.; Chao, Y.; Kato, K.; Chung, H.C.; Chen, J.S.; Muro, K.; Kang, W.K.; et al. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017, 390, 2461–2471. [Google Scholar] [CrossRef]
- Bang, Y.J.; Kang, Y.K.; Catenacci, D.V.; Muro, K.; Fuchs, C.S.; Geva, R.; Hara, H.; Golan, T.; Garrido, M.; Jalal, S.I.; et al. Pembrolizumab alone or in combination with chemotherapy as first-line therapy for patients with advanced gastric or gastroesophageal junction adenocarcinoma: Results from the phase II nonrandomized KEYNOTE-059 study. Gastric Cancer 2019, 22, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Shitara, K.; Ozguroglu, M.; Bang, Y.J.; Di Bartolomeo, M.; Mandala, M.; Ryu, M.H.; Fornaro, L.; Olesinski, T.; Caglevic, C.; Chung, H.C.; et al. Pembrolizumab versus paclitaxel for previously treated, advanced gastric or gastro-oesophageal junction cancer (KEYNOTE-061): A randomised, open-label, controlled, phase 3 trial. Lancet 2018, 392, 123–133. [Google Scholar] [CrossRef]
- Bang, Y.J.; Ruiz, E.Y.; Van Cutsem, E.; Lee, K.W.; Wyrwicz, L.; Schenker, M.; Alsina, M.; Ryu, M.H.; Chung, H.C.; Evesque, L.; et al. Phase III, randomised trial of avelumab versus physician’s choice of chemotherapy as third-line treatment of patients with advanced gastric or gastro-oesophageal junction cancer: Primary analysis of JAVELIN Gastric 300. Ann. Oncol. 2018, 29, 2052–2060. [Google Scholar] [CrossRef]
- Fuchs, C.S.; Doi, T.; Jang, R.W.; Muro, K.; Satoh, T.; Machado, M.; Sun, W.; Jalal, S.I.; Shah, M.A.; Metges, J.P.; et al. Safety and Efficacy of Pembrolizumab Monotherapy in Patients With Previously Treated Advanced Gastric and Gastroesophageal Junction Cancer: Phase 2 Clinical KEYNOTE-059 Trial. JAMA Oncol. 2018, 4, e180013. [Google Scholar] [CrossRef]
- Pietrantonio, F.; Randon, G.; Di Bartolomeo, M.; Luciani, A.; Chao, J.; Smyth, E.C.; Petrelli, F. Predictive role of microsatellite instability for PD-1 blockade in patients with advanced gastric cancer: A meta-analysis of randomized clinical trials. ESMO Open 2021, 6, 100036. [Google Scholar] [CrossRef]
- Kawazoe, A.; Kuwata, T.; Kuboki, Y.; Shitara, K.; Nagatsuma, A.K.; Aizawa, M.; Yoshino, T.; Doi, T.; Ohtsu, A.; Ochiai, A. Clinicopathological features of programmed death ligand 1 expression with tumor-infiltrating lymphocyte, mismatch repair, and Epstein-Barr virus status in a large cohort of gastric cancer patients. Gastric Cancer 2017, 20, 407–415. [Google Scholar] [CrossRef]
- Janjigian, Y.Y.; Bendell, J.; Calvo, E.; Kim, J.W.; Ascierto, P.A.; Sharma, P.; Ott, P.A.; Peltola, K.; Jaeger, D.; Evans, J.; et al. CheckMate-032 Study: Efficacy and Safety of Nivolumab and Nivolumab Plus Ipilimumab in Patients With Metastatic Esophagogastric Cancer. J. Clin. Oncol. 2018, 36, 2836–2844. [Google Scholar] [CrossRef]
- Kelly, R.J. Immunotherapy for Esophageal and Gastric Cancer. Am. Soc. Clin. Oncol. Educ. Book 2017, 37, 292–300. [Google Scholar] [CrossRef]
- Sanchez-Pulido, L.; Martin-Belmonte, F.; Valencia, A.; Alonso, M.A. MARVEL: A conserved domain involved in membrane apposition events. Trends Biochem. Sci. 2002, 27, 599–601. [Google Scholar] [CrossRef]
- Han, W.; Ding, P.; Xu, M.; Wang, L.; Rui, M.; Shi, S.; Liu, Y.; Zheng, Y.; Chen, Y.; Yang, T.; et al. Identification of eight genes encoding chemokine-like factor superfamily members 1-8 (CKLFSF1-8) by in silico cloning and experimental validation. Genomics 2003, 81, 609–617. [Google Scholar] [CrossRef]
- Li, H.; Liu, Y.T.; Chen, L.; Zhou, J.J.; Chen, D.R.; Li, S.J.; Sun, Z.J. CMTM4 regulates epithelial-mesenchymal transition and PD-L1 expression in head and neck squamous cell carcinoma. Mol. Carcinog. 2021. [Google Scholar] [CrossRef]
- Mezzadra, R.; Sun, C.; Jae, L.T.; Gomez-Eerland, R.; de Vries, E.; Wu, W.; Logtenberg, M.E.W.; Slagter, M.; Rozeman, E.A.; Hofland, I.; et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 2017, 549, 106–110. [Google Scholar] [CrossRef]
- Burr, M.L.; Sparbier, C.E.; Chan, Y.C.; Williamson, J.C.; Woods, K.; Beavis, P.A.; Lam, E.Y.N.; Henderson, M.A.; Bell, C.C.; Stolzenburg, S.; et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 2017, 549, 101–105. [Google Scholar] [CrossRef] [Green Version]
- Guan, X.; Zhang, C.; Zhao, J.; Sun, G.; Song, Q.; Jia, W. CMTM6 overexpression is associated with molecular and clinical characteristics of malignancy and predicts poor prognosis in gliomas. EBioMedicine 2018, 35, 233–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Yang, Q.C.; Li, Y.C.; Yang, L.L.; Liu, J.F.; Li, H.; Xiao, Y.; Bu, L.L.; Zhang, W.F.; Sun, Z.J. Targeting CMTM6 Suppresses Stem Cell-Like Properties and Enhances Antitumor Immunity in Head and Neck Squamous Cell Carcinoma. Cancer Immunol. Res. 2020, 8, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.L.; Zhang, S.W.; Chao, X.; Wang, C.H.; Yang, X.; Zhang, X.K.; Wen, Y.L.; Yun, J.P.; Luo, R.Z. Coexpression of CMTM6 and PD-L1 as a predictor of poor prognosis in macrotrabecular-massive hepatocellular carcinoma. Cancer Immunol. Immunother. 2021, 70, 417–429. [Google Scholar] [CrossRef]
- Mamessier, E.; Birnbaum, D.J.; Finetti, P.; Birnbaum, D.; Bertucci, F. CMTM6 stabilizes PD-L1 expression and refines its prognostic value in tumors. Ann. Transl. Med. 2018, 6, 54. [Google Scholar] [CrossRef]
- Liang, Z.; Xie, J.; Huang, L.; Huang, Y.; Zhang, Y.; Ma, R.; Zheng, Z.; Wang, Q.; Li, X. Comprehensive analysis of the prognostic value of the chemokine-like factor-like MARVEL transmembrane domain-containing family in gastric cancer. J. Gastrointest. Oncol. 2021, 12, 388–406. [Google Scholar] [CrossRef]
- Zhang, C.; Zhao, S.; Wang, X. Co-expression of CMTM6 and PD-L1: A novel prognostic indicator of gastric cancer. Cancer Cell Int. 2021, 21, 78. [Google Scholar] [CrossRef] [PubMed]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehar, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Zhang, M.; Hu, S.; Min, M.; Ni, Y.; Lu, Z.; Sun, X.; Wu, J.; Liu, B.; Ying, X.; Liu, Y. Dissecting transcriptional heterogeneity in primary gastric adenocarcinoma by single cell RNA sequencing. Gut 2021, 70, 464–475. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., 3rd; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef] [PubMed]
- Meserve, J.; Facciorusso, A.; Holmer, A.K.; Annese, V.; Sandborn, W.J.; Singh, S. Systematic review with meta-analysis: Safety and tolerability of immune checkpoint inhibitors in patients with pre-existing inflammatory bowel diseases. Aliment. Pharmacol. Ther. 2021, 53, 374–382. [Google Scholar] [CrossRef]
- Abd El Aziz, M.A.; Facciorusso, A.; Nayfeh, T.; Saadi, S.; Elnaggar, M.; Cotsoglou, C.; Sacco, R. Immune Checkpoint Inhibitors for Unresectable Hepatocellular Carcinoma. Vaccines 2020, 8, 616. [Google Scholar] [CrossRef]
- Wu, X.; Lan, X.; Hu, W.; Zhang, W.; Lai, X.; Xu, S.; Li, J.; Qiu, W.; Wang, W.; Xiao, J.; et al. CMTM6 expression in M2 macrophages is a potential predictor of PD-1/PD-L1 inhibitor response in colorectal cancer. Cancer Immunol. Immunother. 2021. [Google Scholar] [CrossRef]
- Zugazagoitia, J.; Liu, Y.; Toki, M.; McGuire, J.; Ahmed, F.S.; Henick, B.S.; Gupta, R.; Gettinger, S.N.; Herbst, R.S.; Schalper, K.A.; et al. Quantitative Assessment of CMTM6 in the Tumor Microenvironment and Association with Response to PD-1 Pathway Blockade in Advanced-Stage Non–Small Cell Lung Cancer. J. Thorac. Oncol. 2019, 14, 2084–2096. [Google Scholar] [CrossRef]
- Peng, Q.H.; Wang, C.H.; Chen, H.M.; Zhang, R.X.; Pan, Z.Z.; Lu, Z.H.; Wang, G.Y.; Yue, X.; Huang, W.; Liu, R.Y. CMTM6 and PD-L1 coexpression is associated with an active immune microenvironment and a favorable prognosis in colorectal cancer. J. Immunother. Cancer 2021, 9, e001638. [Google Scholar] [CrossRef]
- Li, X.; Chen, L.; Gu, C.; Sun, Q.; Li, J. CMTM6 significantly relates to PD-L1 and predicts the prognosis of gastric cancer patients. PeerJ 2020, 8, e9536. [Google Scholar] [CrossRef] [PubMed]
- Yugawa, K.; Itoh, S.; Yoshizumi, T.; Iseda, N.; Tomiyama, T.; Morinaga, A.; Toshima, T.; Harada, N.; Kohashi, K.; Oda, Y.; et al. CMTM6 Stabilizes PD-L1 Expression and Is a New Prognostic Impact Factor in Hepatocellular Carcinoma. Hepatol. Commun. 2021, 5, 334–348. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Xia, X.; Hu, J.; Fowlkes, N.W.; Li, S. WSX1 act as a tumor suppressor in hepatocellular carcinoma by downregulating neoplastic PD-L1 expression. Nat. Commun. 2021, 12, 3500. [Google Scholar] [CrossRef] [PubMed]
- Long, L.; Chen, M.; Yuan, Y.; Ming, A.L.; Guo, W.; Wu, K.; Chen, H. High expression of PKM2 synergizes with PD-L1 in tumor cells and immune cells to predict worse survival in human lung adenocarcinoma. J. Cancer 2020, 11, 4442–4452. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Peng, Z.; Liu, Q.; Guo, Z.; Menatola, M.; Su, J.; Li, T.; Ge, Q.; Wang, P.; Shen, L.; et al. Co-Expression with Membrane CMTM6/4 on Tumor Epithelium Enhances the Prediction Value of PD-L1 on Anti-PD-1/L1 Therapeutic Efficacy in Gastric Adenocarcinoma. Cancers 2021, 13, 5175. https://doi.org/10.3390/cancers13205175
Wang Z, Peng Z, Liu Q, Guo Z, Menatola M, Su J, Li T, Ge Q, Wang P, Shen L, et al. Co-Expression with Membrane CMTM6/4 on Tumor Epithelium Enhances the Prediction Value of PD-L1 on Anti-PD-1/L1 Therapeutic Efficacy in Gastric Adenocarcinoma. Cancers. 2021; 13(20):5175. https://doi.org/10.3390/cancers13205175
Chicago/Turabian StyleWang, Ziqi, Zhi Peng, Qiyao Liu, Zixia Guo, Merey Menatola, Jing Su, Ting Li, Qing Ge, Pingzhang Wang, Lin Shen, and et al. 2021. "Co-Expression with Membrane CMTM6/4 on Tumor Epithelium Enhances the Prediction Value of PD-L1 on Anti-PD-1/L1 Therapeutic Efficacy in Gastric Adenocarcinoma" Cancers 13, no. 20: 5175. https://doi.org/10.3390/cancers13205175