Beyond the Double-Strand Breaks: The Role of DNA Repair Proteins in Cancer Stem-Cell Regulation
Abstract
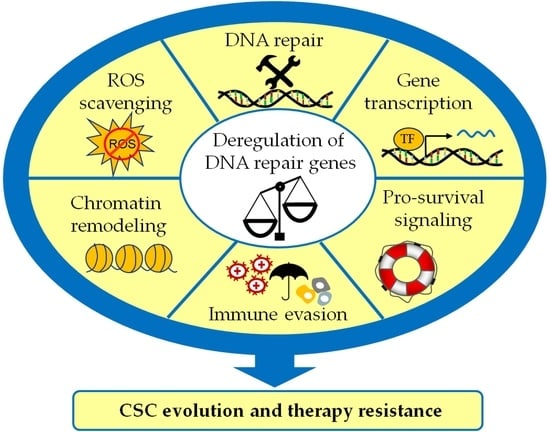
:Simple Summary
Abstract
1. Introduction
2. Adaptation of CSCs to Replication Stress
2.1. Endogenous Causes of Replication Stress
2.2. How CSCs Deal with RS and Consequences for Treatment Resistance
3. The Role of DNA Repair Proteins in the CSC Induction and Maintenance
3.1. Co-Regulation of Gene Transcription
3.2. Regulation of EMT, Survival, and Transformation by Interference with CSC-Related Signaling
3.2.1. ATM- and ATR-Related Signaling
3.2.2. PI3K/Akt Signaling and DNA Repair Proteins
3.2.3. TGF-ß/SMAD Signaling and FA/BRCA Proteins
3.2.4. p53-Related Signaling and FA/BRCA Proteins
3.2.5. p63 Signaling and FANCD2/BRCA1
3.2.6. WNT Signaling, Intrinsic Apoptotic Pathway, and Mismatch Repair (MMR)
3.3. ROS Detoxification and Redox Signaling
4. The Role of DNA Repair Signaling in the Immune Response against Cancer Cells
5. DNA Repair in Cancer Stem Cells as a Therapeutic Target
5.1. Targeting of ATM and ATR Signaling for CSC Eradication
5.2. Mutation of DNA Repair-Related Genes as a Sweet Point for CSC Targeting
5.3. Current Controversy Regarding the Role of DNA Repair Genes as Tumor Regulators and Defenders of Genome Integrity
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Baumann, M.; Krause, M.; Overgaard, J.; Debus, J.; Bentzen, S.M.; Daartz, J.; Richter, C.; Zips, D.; Bortfeld, T. Radiation oncology in the era of precision medicine. Nat. Rev. Cancer 2016, 16, 234–249. [Google Scholar] [CrossRef]
- Jackson, S.E.; Chester, J.D. Personalised cancer medicine. Int. J. Cancer 2015, 137, 262–266. [Google Scholar] [CrossRef]
- Ganesh, K.; Massague, J. Targeting metastatic cancer. Nat. Med. 2021, 27, 34–44. [Google Scholar] [CrossRef]
- Klein, C.A. Cancer progression and the invisible phase of metastatic colonization. Nat. Rev. Cancer 2020, 20, 681–694. [Google Scholar] [CrossRef]
- Mitra, A.; Mishra, L.; Li, S. EMT, CTCs and CSCs in tumor relapse and drug-resistance. Oncotarget 2015, 6, 10697–10711. [Google Scholar] [CrossRef] [Green Version]
- Schulz, A.; Meyer, F.; Dubrovska, A.; Borgmann, K. Cancer Stem Cells and Radioresistance: DNA Repair and Beyond. Cancers 2019, 11, 862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peitzsch, C.; Tyutyunnykova, A.; Pantel, K.; Dubrovska, A. Cancer stem cells: The root of tumor recurrence and metastases. Semin. Cancer Biol. 2017, 44, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Kreso, A.; Dick, J.E. Evolution of the cancer stem cell model. Cell Stem Cell 2014, 14, 275–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desai, A.; Yan, Y.; Gerson, S.L. Concise Reviews: Cancer Stem Cell Targeted Therapies: Toward Clinical Success. Stem Cells Transl. Med. 2019, 8, 75–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapidot, T.; Sirard, C.; Vormoor, J.; Murdoch, B.; Hoang, T.; Caceres-Cortes, J.; Minden, M.; Paterson, B.; Caligiuri, M.A.; Dick, J.E. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 1994, 367, 645–648. [Google Scholar] [CrossRef] [PubMed]
- Vitale, I.; Manic, G.; De Maria, R.; Kroemer, G.; Galluzzi, L. DNA Damage in Stem Cells. Mol. Cell 2017, 66, 306–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desai, A.; Webb, B.; Gerson, S.L. CD133+ cells contribute to radioresistance via altered regulation of DNA repair genes in human lung cancer cells. Radiother. Oncol. 2014, 110, 538–545. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.H.; Zhang, M.; Rajapakshe, K.; Coarfa, C.; Edwards, D.; Huang, S.; Rosen, J.M. Mammary Stem Cells and Tumor-Initiating Cells Are More Resistant to Apoptosis and Exhibit Increased DNA Repair Activity in Response to DNA Damage. Stem Cell Rep. 2015, 5, 378–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurth, I.; Hein, L.; Mabert, K.; Peitzsch, C.; Koi, L.; Cojoc, M.; Kunz-Schughart, L.; Baumann, M.; Dubrovska, A. Cancer stem cell related markers of radioresistance in head and neck squamous cell carcinoma. Oncotarget 2015, 6, 34494–34509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carruthers, R.; Ahmed, S.U.; Strathdee, K.; Gomez-Roman, N.; Amoah-Buahin, E.; Watts, C.; Chalmers, A.J. Abrogation of radioresistance in glioblastoma stem-like cells by inhibition of ATM kinase. Mol. Oncol. 2015, 9, 192–203. [Google Scholar] [CrossRef]
- Obara, E.A.A.; Aguilar-Morante, D.; Rasmussen, R.D.; Frias, A.; Vitting-Serup, K.; Lim, Y.C.; Elbaek, K.J.; Pedersen, H.; Vardouli, L.; Jensen, K.E.; et al. SPT6-driven error-free DNA repair safeguards genomic stability of glioblastoma cancer stem-like cells. Nat. Commun. 2020, 11, 4709. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Xiong, Y.; Wang, R.; Xiang, L.; Zhou, H.; Gu, H. Identification of a subpopulation of long-term tumor-initiating cells in colon cancer. Biosci. Rep. 2020, 40, BSR20200437. [Google Scholar] [CrossRef]
- Karimi-Busheri, F.; Rasouli-Nia, A.; Mackey, J.R.; Weinfeld, M. Senescence evasion by MCF-7 human breast tumor-initiating cells. Breast Cancer Res. 2010, 12, R31. [Google Scholar] [CrossRef] [Green Version]
- Lim, Y.C.; Roberts, T.L.; Day, B.W.; Harding, A.; Kozlov, S.; Kijas, A.W.; Ensbey, K.S.; Walker, D.G.; Lavin, M.F. A role for homologous recombination and abnormal cell-cycle progression in radioresistance of glioma-initiating cells. Mol. Cancer Ther. 2012, 11, 1863–1872. [Google Scholar] [CrossRef] [Green Version]
- Ropolo, M.; Daga, A.; Griffero, F.; Foresta, M.; Casartelli, G.; Zunino, A.; Poggi, A.; Cappelli, E.; Zona, G.; Spaziante, R.; et al. Comparative analysis of DNA repair in stem and nonstem glioma cell cultures. Mol. Cancer Res. 2009, 7, 383–392. [Google Scholar] [CrossRef] [Green Version]
- Xie, A.; Puget, N.; Shim, I.; Odate, S.; Jarzyna, I.; Bassing, C.H.; Alt, F.W.; Scully, R. Control of sister chromatid recombination by histone H2AX. Mol. Cell 2004, 16, 1017–1025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundholm, L.; Haag, P.; Zong, D.; Juntti, T.; Mork, B.; Lewensohn, R.; Viktorsson, K. Resistance to DNA-damaging treatment in non-small cell lung cancer tumor-initiating cells involves reduced DNA-PK/ATM activation and diminished cell cycle arrest. Cell Death Dis. 2013, 4, e478. [Google Scholar] [CrossRef] [PubMed]
- Gorodetska, I.; Kozeretska, I.; Dubrovska, A. BRCA Genes: The Role in Genome Stability, Cancer Stemness and Therapy Resistance. J. Cancer 2019, 10, 2109–2127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagasse, E. Cancer stem cells with genetic instability: The best vehicle with the best engine for cancer. Gene Ther. 2008, 15, 136–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nalepa, G.; Clapp, D.W. Fanconi anaemia and cancer: An intricate relationship. Nat. Rev. Cancer 2018, 18, 168–185. [Google Scholar] [CrossRef]
- Wu, J.; Mu, Q.; Thiviyanathan, V.; Annapragada, A.; Vigneswaran, N. Cancer stem cells are enriched in Fanconi anemia head and neck squamous cell carcinomas. Int. J. Oncol. 2014, 45, 2365–2372. [Google Scholar] [CrossRef] [Green Version]
- Roos, W.P.; Thomas, A.D.; Kaina, B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer 2016, 16, 20–33. [Google Scholar] [CrossRef]
- Zeman, M.K.; Cimprich, K.A. Causes and consequences of replication stress. Nat. Cell Biol. 2014, 16, 2–9. [Google Scholar] [CrossRef] [Green Version]
- Birkbak, N.J.; Eklund, A.C.; Li, Q.; McClelland, S.E.; Endesfelder, D.; Tan, P.; Tan, I.B.; Richardson, A.L.; Szallasi, Z.; Swanton, C. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011, 71, 3447–3452. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Li, D.; Wang, D.; Liu, X.; Yin, N.; Song, Y.; Lu, S.H.; Ju, Z.; Zhan, Q. Quiescence and attenuated DNA damage response promote survival of esophageal cancer stem cells. J. Cell Biochem. 2012, 113, 3643–3652. [Google Scholar] [CrossRef]
- Maugeri-Sacca, M.; Bartucci, M.; De Maria, R. DNA damage repair pathways in cancer stem cells. Mol. Cancer Ther. 2012, 11, 1627–1636. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.S.; O’Carrigan, B.; Jackson, S.P.; Yap, T.A. Targeting DNA Repair in Cancer: Beyond PARP Inhibitors. Cancer Discov. 2017, 7, 20–37. [Google Scholar] [CrossRef] [Green Version]
- Li, L.Y.; Guan, Y.D.; Chen, X.S.; Yang, J.M.; Cheng, Y. DNA Repair Pathways in Cancer Therapy and Resistance. Front. Pharmacol. 2020, 11, 629266. [Google Scholar] [CrossRef] [PubMed]
- Techer, H.; Koundrioukoff, S.; Nicolas, A.; Debatisse, M. The impact of replication stress on replication dynamics and DNA damage in vertebrate cells. Nat. Rev. Genet. 2017, 18, 535–550. [Google Scholar] [CrossRef] [PubMed]
- Techer, H.; Pasero, P. The Replication Stress Response on a Narrow Path Between Genomic Instability and Inflammation. Front. Cell Dev. Biol. 2021, 9, 702584. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Kustatscher, G.; Alabert, C.; Hodl, M.; Forne, I.; Volker-Albert, M.; Satpathy, S.; Beyer, T.E.; Mailand, N.; Choudhary, C.; et al. Proteome dynamics at broken replication forks reveal a distinct ATM-directed repair response suppressing DNA double-strand break ubiquitination. Mol. Cell 2021, 81, 1084–1099.e6. [Google Scholar] [CrossRef] [PubMed]
- McGrail, D.J.; Lin, C.C.; Dai, H.; Mo, W.; Li, Y.; Stephan, C.; Davies, P.; Lu, Z.; Mills, G.B.; Lee, J.S.; et al. Defective Replication Stress Response Is Inherently Linked to the Cancer Stem Cell Phenotype. Cell Rep. 2018, 23, 2095–2106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef] [PubMed]
- Venere, M.; Hamerlik, P.; Wu, Q.; Rasmussen, R.D.; Song, L.A.; Vasanji, A.; Tenley, N.; Flavahan, W.A.; Hjelmeland, A.B.; Bartek, J.; et al. Therapeutic targeting of constitutive PARP activation compromises stem cell phenotype and survival of glioblastoma-initiating cells. Cell Death Differ. 2014, 21, 258–269. [Google Scholar] [CrossRef] [Green Version]
- Berti, M.; Vindigni, A. Replication stress: Getting back on track. Nat. Struct. Mol. Biol. 2016, 23, 103–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Muse, T.; Aguilera, A. Transcription-replication conflicts: How they occur and how they are resolved. Nat. Rev. Mol. Cell Biol. 2016, 17, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Santos-Pereira, J.M.; Aguilera, A. R loops: New modulators of genome dynamics and function. Nat. Rev. Genet. 2015, 16, 583–597. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, V.; Barroso, S.I.; Garcia-Rubio, M.L.; Tumini, E.; Herrera-Moyano, E.; Aguilera, A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 2014, 511, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rubio, M.L.; Perez-Calero, C.; Barroso, S.I.; Tumini, E.; Herrera-Moyano, E.; Rosado, I.V.; Aguilera, A. The Fanconi Anemia Pathway Protects Genome Integrity from R-loops. PLoS Genet. 2015, 11, e1005674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zhu, P.; Luo, J.; Wang, J.; Liu, Z.; Wu, W.; Du, Y.; Ye, B.; Wang, D.; He, L.; et al. LncRNA HAND2-AS1 promotes liver cancer stem cell self-renewal via BMP signaling. EMBO J. 2019, 38, e101110. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.U.; Carruthers, R.; Gilmour, L.; Yildirim, S.; Watts, C.; Chalmers, A.J. Selective Inhibition of Parallel DNA Damage Response Pathways Optimizes Radiosensitization of Glioblastoma Stem-like Cells. Cancer Res. 2015, 75, 4416–4428. [Google Scholar] [CrossRef] [Green Version]
- Carruthers, R.D.; Ahmed, S.U.; Ramachandran, S.; Strathdee, K.; Kurian, K.M.; Hedley, A.; Gomez-Roman, N.; Kalna, G.; Neilson, M.; Gilmour, L.; et al. Replication Stress Drives Constitutive Activation of the DNA Damage Response and Radioresistance in Glioblastoma Stem-like Cells. Cancer Res. 2018, 78, 5060–5071. [Google Scholar] [CrossRef] [Green Version]
- Murayama, T.; Takeuchi, Y.; Yamawaki, K.; Natsume, T.; Li, M.; Marcela, R.N.; Nishimura, T.; Kogure, Y.; Nakata, A.; Tominaga, K.; et al. MCM10 compensates for Myc-induced DNA replication stress in breast cancer stem-like cells. Cancer Sci. 2021, 112, 1209–1224. [Google Scholar] [CrossRef]
- Manic, G.; Musella, M.; Corradi, F.; Sistigu, A.; Vitale, S.; Soliman Abdel Rehim, S.; Mattiello, L.; Malacaria, E.; Galassi, C.; Signore, M.; et al. Control of replication stress and mitosis in colorectal cancer stem cells through the interplay of PARP1, MRE11 and RAD51. Cell Death Differ. 2021, 28, 2060–2082. [Google Scholar] [CrossRef]
- Mattiello, L.; Soliman Abdel Rehim, S.; Musella, M.; Sistigu, A.; Guarracino, A.; Vitale, S.; Corradi, F.; Galassi, C.; Sperati, F.; Manic, G.; et al. The Targeting of MRE11 or RAD51 Sensitizes Colorectal Cancer Stem Cells to CHK1 Inhibition. Cancers 2021, 13, 1957. [Google Scholar] [CrossRef]
- Blakemore, D.; Vilaplana-Lopera, N.; Almaghrabi, R.; Gonzalez, E.; Moya, M.; Ward, C.; Murphy, G.; Gambus, A.; Petermann, E.; Stewart, G.S.; et al. MYBL2 and ATM suppress replication stress in pluripotent stem cells. EMBO Rep. 2021, 22, e51120. [Google Scholar] [CrossRef]
- Sun, L.L.; Yang, R.Y.; Li, C.W.; Chen, M.K.; Shao, B.; Hsu, J.M.; Chan, L.C.; Yang, Y.; Hsu, J.L.; Lai, Y.J.; et al. Inhibition of ATR downregulates PD-L1 and sensitizes tumor cells to T cell-mediated killing. Am. J. Cancer Res. 2018, 8, 1307–1316. [Google Scholar]
- Tang, Z.; Pilie, P.G.; Geng, C.; Manyam, G.C.; Yang, G.; Park, S.; Wang, D.; Peng, S.; Wu, C.; Peng, G.; et al. ATR Inhibition Induces CDK1-SPOP Signaling and Enhances Anti-PD-L1 Cytotoxicity in Prostate Cancer. Clin. Cancer Res. 2021, 27, 4898–4909. [Google Scholar] [CrossRef]
- Cheung-Ong, K.; Giaever, G.; Nislow, C. DNA-damaging agents in cancer chemotherapy: Serendipity and chemical biology. Chem. Biol. 2013, 20, 648–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atun, R.; Jaffray, D.A.; Barton, M.B.; Bray, F.; Baumann, M.; Vikram, B.; Hanna, T.P.; Knaul, F.M.; Lievens, Y.; Lui, T.Y.; et al. Expanding global access to radiotherapy. Lancet Oncol. 2015, 16, 1153–1186. [Google Scholar] [CrossRef]
- Zheng, H.C. The molecular mechanisms of chemoresistance in cancers. Oncotarget 2017, 8, 59950–59964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorolla, M.A.; Parisi, E.; Sorolla, A. Determinants of Sensitivity to Radiotherapy in Endometrial Cancer. Cancers 2020, 12, 1906. [Google Scholar] [CrossRef]
- Seshacharyulu, P.; Baine, M.J.; Souchek, J.J.; Menning, M.; Kaur, S.; Yan, Y.; Ouellette, M.M.; Jain, M.; Lin, C.; Batra, S.K. Biological determinants of radioresistance and their remediation in pancreatic cancer. Biochim. Biophys. Acta Rev. Cancer 2017, 1868, 69–92. [Google Scholar] [CrossRef]
- Yoshida, G.J. Emerging roles of Myc in stem cell biology and novel tumor therapies. J. Exp. Clin. Cancer Res. 2018, 37, 173. [Google Scholar] [CrossRef] [Green Version]
- Le Grand, M.; Mukha, A.; Puschel, J.; Valli, E.; Kamili, A.; Vittorio, O.; Dubrovska, A.; Kavallaris, M. Interplay between MycN and c-Myc regulates radioresistance and cancer stem cell phenotype in neuroblastoma upon glutamine deprivation. Theranostics 2020, 10, 6411–6429. [Google Scholar] [CrossRef]
- Karagonlar, Z.F.; Akbari, S.; Karabicici, M.; Sahin, E.; Avci, S.T.; Ersoy, N.; Ates, K.E.; Balli, T.; Karacicek, B.; Kaplan, K.N.; et al. A Novel Function for KLF4 in Modulating the De-differentiation of EpCAM(-)/CD133(-) nonStem Cells into EpCAM(+)/CD133(+) Liver Cancer Stem Cells in HCC Cell Line HuH7. Cells 2020, 9, 1198. [Google Scholar] [CrossRef]
- Saha, A.; Roy, S.; Kar, M.; Roy, S.; Thakur, S.; Padhi, S.; Akhter, Y.; Banerjee, B. Role of Telomeric TRF2 in Orosphere Formation and CSC Phenotype Maintenance Through Efficient DNA Repair Pathway and its Correlation with Recurrence in OSCC. Stem Cell Rev. Rep. 2018, 14, 871–887. [Google Scholar] [CrossRef]
- Mao, Z.; Seluanov, A.; Jiang, Y.; Gorbunova, V. TRF2 is required for repair of nontelomeric DNA double-strand breaks by homologous recombination. Proc. Natl. Acad. Sci. USA 2007, 104, 13068–13073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Steensel, B.; Smogorzewska, A.; de Lange, T. TRF2 protects human telomeres from end-to-end fusions. Cell 1998, 92, 401–413. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Xian, M.; Yang, B.; Ying, M.; He, Q. Inhibition of KLF4 by Statins Reverses Adriamycin-Induced Metastasis and Cancer Stemness in Osteosarcoma Cells. Stem Cell Rep. 2017, 8, 1617–1629. [Google Scholar] [CrossRef] [Green Version]
- Rowland, B.D.; Peeper, D.S. KLF4, p21 and context-dependent opposing forces in cancer. Nat. Rev. Cancer 2006, 6, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Pazin, M.J.; Schwartz, C.M.; Becker, K.G.; Wersto, R.P.; Dilley, C.M.; Mattson, M.P. Nontelomeric TRF2-REST interaction modulates neuronal gene silencing and fate of tumor and stem cells. Curr. Biol. 2008, 18, 1489–1494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Lin, T.; Lian, N.; Tao, H.; Li, C.; Li, L.; Yang, X. Hop2 Interacts with ATF4 to Promote Osteoblast Differentiation. J. Bone Miner. Res. 2019, 34, 2287–2300. [Google Scholar] [CrossRef]
- Pezza, R.J.; Voloshin, O.N.; Volodin, A.A.; Boateng, K.A.; Bellani, M.A.; Mazin, A.V.; Camerini-Otero, R.D. The dual role of HOP2 in mammalian meiotic homologous recombination. Nucleic Acids Res. 2014, 42, 2346–2357. [Google Scholar] [CrossRef]
- Wei, L.; Lin, Q.; Lu, Y.; Li, G.; Huang, L.; Fu, Z.; Chen, R.; Zhou, Q. Cancer-associated fibroblasts-mediated ATF4 expression promotes malignancy and gemcitabine resistance in pancreatic cancer via the TGF-β1/SMAD2/3 pathway and ABCC1 transactivation. Cell Death Dis. 2021, 12, 334. [Google Scholar] [CrossRef]
- Gonzalez-Gonzalez, A.; Munoz-Muela, E.; Marchal, J.A.; Cara, F.E.; Molina, M.P.; Cruz-Lozano, M.; Jimenez, G.; Verma, A.; Ramirez, A.; Qian, W.; et al. Activating Transcription Factor 4 Modulates TGFβ-Induced Aggressiveness in Triple-Negative Breast Cancer via SMAD2/3/4 and mTORC2 Signaling. Clin. Cancer Res. 2018, 24, 5697–5709. [Google Scholar] [CrossRef] [Green Version]
- Ko, L.; Cardona, G.R.; Henrion-Caude, A.; Chin, W.W. Identification and characterization of a tissue-specific coactivator, GT198, that interacts with the DNA-binding domains of nuclear receptors. Mol. Cell. Biol. 2002, 22, 357–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wortel, I.M.N.; van der Meer, L.T.; Kilberg, M.S.; van Leeuwen, F.N. Surviving Stress: Modulation of ATF4-Mediated Stress Responses in Normal and Malignant Cells. Trends Endocrinol. Metab. 2017, 28, 794–806. [Google Scholar] [CrossRef] [PubMed]
- Bian, L.; Meng, Y.; Zhang, M.; Li, D. MRE11-RAD50-NBS1 complex alterations and DNA damage response: Implications for cancer treatment. Mol. Cancer 2019, 18, 169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Syed, A.; Tainer, J.A. The MRE11-RAD50-NBS1 Complex Conducts the Orchestration of Damage Signaling and Outcomes to Stress in DNA Replication and Repair. Annu. Rev. Biochem. 2018, 87, 263–294. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Canny, M.D.; Buschmann, T.A.; Latham, M.P. A Survey of Reported Disease-Related Mutations in the MRE11-RAD50-NBS1 Complex. Cells 2020, 9, 1678. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.W.; Tong, G.H.; Liu, Y. Cancer stem cells and hypoxia-inducible factors (Review). Int. J. Oncol. 2018, 53, 469–476. [Google Scholar] [CrossRef] [Green Version]
- MacLachlan, T.K.; Takimoto, R.; El-Deiry, W.S. BRCA1 directs a selective p53-dependent transcriptional response towards growth arrest and DNA repair targets. Mol. Cell. Biol. 2002, 22, 4280–4292. [Google Scholar] [CrossRef] [Green Version]
- Gorski, J.J.; James, C.R.; Quinn, J.E.; Stewart, G.E.; Staunton, K.C.; Buckley, N.E.; McDyer, F.A.; Kennedy, R.D.; Wilson, R.H.; Mullan, P.B.; et al. BRCA1 transcriptionally regulates genes associated with the basal-like phenotype in breast cancer. Breast Cancer Res. Treat. 2010, 122, 721–731. [Google Scholar] [CrossRef]
- Kalkat, M.; Resetca, D.; Lourenco, C.; Chan, P.K.; Wei, Y.; Shiah, Y.J.; Vitkin, N.; Tong, Y.; Sunnerhagen, M.; Done, S.J.; et al. MYC Protein Interactome Profiling Reveals Functionally Distinct Regions that Cooperate to Drive Tumorigenesis. Mol. Cell 2018, 72, 836–848.e7. [Google Scholar] [CrossRef] [Green Version]
- Arroyo, R.; Sune, G.; Zanzoni, A.; Duran-Frigola, M.; Alcalde, V.; Stracker, T.H.; Soler-Lopez, M.; Aloy, P. Systematic identification of molecular links between core and candidate genes in breast cancer. J. Mol. Biol. 2015, 427, 1436–1450. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Chen, Y.K.; Lo, S.; Chi, T.C.; Chen, Y.H.; Hu, S.C.; Chen, Y.W.; Jiang, S.S.; Tsai, F.Y.; Liu, W.; et al. MRE11 promotes oral cancer progression through RUNX2/CXCR4/AKT/FOXA2 signaling in a nuclease-independent manner. Oncogene 2021, 40, 3510–3532. [Google Scholar] [CrossRef]
- Kamarli, A.P.; Davliatkadamov, S. Hypodermin-chlorophos against warble fly larvae in yaks. Veterinariia 1975, 10, 64–65. [Google Scholar]
- Chen, Y.C.; Su, Y.N.; Chou, P.C.; Chiang, W.C.; Chang, M.C.; Wang, L.S.; Teng, S.C.; Wu, K.J. Overexpression of NBS1 contributes to transformation through the activation of phosphatidylinositol 3-kinase/Akt. J. Biol. Chem. 2005, 280, 32505–32511. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.H.; Chang, S.Y.; Chiou, S.H.; Liu, C.J.; Chi, C.W.; Chen, P.M.; Teng, S.C.; Wu, K.J. Overexpression of NBS1 induces epithelial-mesenchymal transition and co-expression of NBS1 and Snail predicts metastasis of head and neck cancer. Oncogene 2007, 26, 1459–1467. [Google Scholar] [CrossRef]
- Valencia-Gonzalez, H.A.; Ruiz, G.; Ortiz-Sanchez, E.; Garcia-Carranca, A. Cancer Stem Cells from Tumor Cell Lines Activate the DNA Damage Response Pathway after Ionizing Radiation More Efficiently Than Noncancer Stem Cells. Stem Cells Int. 2019, 2019, 7038953. [Google Scholar] [CrossRef]
- Gemenetzidis, E.; Gammon, L.; Biddle, A.; Emich, H.; Mackenzie, I.C. Invasive oral cancer stem cells display resistance to ionising radiation. Oncotarget 2015, 6, 43964–43977. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Hu, M.; Liu, P.; Jiao, M.; Zhou, M.; Lee, A.K.; Li, F.; Li, C.Y. ATM Paradoxically Promotes Oncogenic Transformation via Transcriptional Reprogramming. Cancer Res. 2020, 80, 1669–1680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, S.M.; Kauzlaric, A.; Quivy, J.P.; Pontis, J.; Rauwel, B.; Coluccio, A.; Offner, S.; Duc, J.; Turelli, P.; Almouzni, G.; et al. KAP1 facilitates reinstatement of heterochromatin after DNA replication. Nucleic Acids Res. 2018, 46, 8788–8802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziv, Y.; Bielopolski, D.; Galanty, Y.; Lukas, C.; Taya, Y.; Schultz, D.C.; Lukas, J.; Bekker-Jensen, S.; Bartek, J.; Shiloh, Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 2006, 8, 870–876. [Google Scholar] [CrossRef] [PubMed]
- Bernardino-Sgherri, J.; Siberchicot, C.; Auvre, F.; Busso, D.; Brocas, C.; El Masri, G.; Lioutsko, A.; Ferri, F.; Radicella, J.P.; Romeo, P.H.; et al. Tumor resistance to radiotherapy is triggered by an ATM/TAK1-dependent-increased expression of the cellular prion protein. Oncogene 2021, 40, 3460–3469. [Google Scholar] [CrossRef]
- Yang, D.; Peng, M.; Hou, Y.; Qin, Y.; Wan, X.; Zhu, P.; Liu, S.; Yang, L.; Zeng, H.; Jin, T.; et al. Oxidized ATM promotes breast cancer stem cell enrichment through energy metabolism reprogram-mediated acetyl-CoA accumulation. Cell Death Dis. 2020, 11, 508. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Sun, Y.; Ma, L. ZEB1: At the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle 2015, 14, 481–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, P.; Wei, Y.; Wang, L.; Debeb, B.G.; Yuan, Y.; Zhang, J.; Yuan, J.; Wang, M.; Chen, D.; Sun, Y.; et al. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat. Cell Biol. 2014, 16, 864–875. [Google Scholar] [CrossRef] [PubMed]
- Chaffer, C.L.; Marjanovic, N.D.; Lee, T.; Bell, G.; Kleer, C.G.; Reinhardt, F.; D’Alessio, A.C.; Young, R.A.; Weinberg, R.A. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 2013, 154, 61–74. [Google Scholar] [CrossRef] [Green Version]
- Antonelli, M.; Strappazzon, F.; Arisi, I.; Brandi, R.; D’Onofrio, M.; Sambucci, M.; Manic, G.; Vitale, I.; Barila, D.; Stagni, V. ATM kinase sustains breast cancer stem-like cells by promoting ATG4C expression and autophagy. Oncotarget 2017, 8, 21692–21709. [Google Scholar] [CrossRef]
- Song, M.; Bode, A.M.; Dong, Z.; Lee, M.H. AKT as a Therapeutic Target for Cancer. Cancer Res. 2019, 79, 1019–1031. [Google Scholar] [CrossRef] [Green Version]
- Zhou, B.P.; Deng, J.; Xia, W.; Xu, J.; Li, Y.M.; Gunduz, M.; Hung, M.C. Dual regulation of Snail by GSK-3β-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat. Cell Biol. 2004, 6, 931–940. [Google Scholar] [CrossRef]
- Korkaya, H.; Paulson, A.; Charafe-Jauffret, E.; Ginestier, C.; Brown, M.; Dutcher, J.; Clouthier, S.G.; Wicha, M.S. Regulation of mammary stem/progenitor cells by PTEN/Akt/β-catenin signaling. PLoS Biol. 2009, 7, e1000121. [Google Scholar] [CrossRef]
- Feng, J.; Liang, J.; Li, J.; Li, Y.; Liang, H.; Zhao, X.; McNutt, M.A.; Yin, Y. PTEN Controls the DNA Replication Process through MCM2 in Response to Replicative Stress. Cell Rep. 2015, 13, 1295–1303. [Google Scholar] [CrossRef] [Green Version]
- Brunet, A.; Bonni, A.; Zigmond, M.J.; Lin, M.Z.; Juo, P.; Hu, L.S.; Anderson, M.J.; Arden, K.C.; Blenis, J.; Greenberg, M.E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999, 96, 857–868. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.W.; Kirtay, M.; Schneble, N.; Yakoub, G.; Ding, M.; Rudiger, T.; Siniuk, K.; Lu, R.; Jiang, Y.N.; Li, T.L.; et al. NBS1 interacts with Notch signaling in neuronal homeostasis. Nucleic Acids Res. 2020, 48, 10924–10939. [Google Scholar] [CrossRef]
- Yuan, S.S.; Hou, M.F.; Hsieh, Y.C.; Huang, C.Y.; Lee, Y.C.; Chen, Y.J.; Lo, S. Role of MRE11 in cell proliferation, tumor invasion, and DNA repair in breast cancer. J. Natl. Cancer Inst. 2012, 104, 1485–1502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burrell, R.A.; McGranahan, N.; Bartek, J.; Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013, 501, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.W.; Kang, H.J.; Kim, H.J.; Hwang, J.S.; Wang, A.; Bae, I. Inhibition of constitutively activated phosphoinositide 3-kinase/AKT pathway enhances antitumor activity of chemotherapeutic agents in breast cancer susceptibility gene 1-defective breast cancer cells. Mol. Carcinog. 2013, 52, 667–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, T.; Jia, Y.; Sherris, D.; Li, S.; Wang, H.; Lu, D.; Yang, Q. Targeting the Akt/mTOR pathway in Brca1-deficient cancers. Oncogene 2011, 30, 2443–2450. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Hu, C.; Riegel, A.T.; Fan, S.; Rosen, E.M. Growth factor signaling pathways modulate BRCA1 repression of estrogen receptor-α activity. Mol. Endocrinol. 2007, 21, 1905–1923. [Google Scholar] [CrossRef] [Green Version]
- Xiang, T.; Ohashi, A.; Huang, Y.; Pandita, T.K.; Ludwig, T.; Powell, S.N.; Yang, Q. Negative Regulation of AKT Activation by BRCA1. Cancer Res. 2008, 68, 10040–10044. [Google Scholar] [CrossRef] [Green Version]
- Ceccaldi, R.; Parmar, K.; Mouly, E.; Delord, M.; Kim, J.M.; Regairaz, M.; Pla, M.; Vasquez, N.; Zhang, Q.S.; Pondarre, C.; et al. Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell 2012, 11, 36–49. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Kozono, D.E.; O’Connor, K.W.; Vidal-Cardenas, S.; Rousseau, A.; Hamilton, A.; Moreau, L.; Gaudiano, E.F.; Greenberger, J.; Bagby, G.; et al. TGF-β Inhibition Rescues Hematopoietic Stem Cell Defects and Bone Marrow Failure in Fanconi Anemia. Cell Stem Cell 2016, 18, 668–681. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Zhou, W.; Cheng, C.T.; Ren, X.; Somlo, G.; Fong, M.Y.; Chin, A.R.; Li, H.; Yu, Y.; Xu, Y.; et al. TGFβ induces “BRCAness” and sensitivity to PARP inhibition in breast cancer by regulating DNA-repair genes. Mol. Cancer Res. 2014, 12, 1597–1609. [Google Scholar] [CrossRef] [Green Version]
- Dubrovska, A.; Kanamoto, T.; Lomnytska, M.; Heldin, C.H.; Volodko, N.; Souchelnytskyi, S. TGFβ1/Smad3 counteracts BRCA1-dependent repair of DNA damage. Oncogene 2005, 24, 2289–2297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, S.; Liu, M.; Shen, D.; Zhang, W.; Wang, T.; Bai, Y. TGF-β/Smads Signaling Affects Radiation Response and Prolongs Survival by Regulating DNA Repair Genes in Malignant Glioma. DNA Cell Biol. 2018, 37, 909–916. [Google Scholar] [CrossRef]
- Levinsky, H.; Smirnoff, P.; Khalef, S.; Birk, Y.; Applebaum, S.W. Proceedings: Trypsin-like enzymes and trypsin inhibitors. Isr. J. Med. Sci. 1975, 11, 1170. [Google Scholar] [PubMed]
- Mantovani, F.; Collavin, L.; Del Sal, G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019, 26, 199–212. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Xu, Y. p53 and stem cells: New developments and new concerns. Trends Cell Biol. 2010, 20, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.H.; Ro, E.J.; Yoon, J.S.; Mizutani, T.; Kang, D.W.; Park, J.C.; Il Kim, T.; Clevers, H.; Choi, K.Y. 5-FU promotes stemness of colorectal cancer via p53-mediated WNT/beta-catenin pathway activation. Nat. Commun. 2020, 11, 5321. [Google Scholar] [CrossRef]
- Shetzer, Y.; Solomon, H.; Koifman, G.; Molchadsky, A.; Horesh, S.; Rotter, V. The paradigm of mutant p53-expressing cancer stem cells and drug resistance. Carcinogenesis 2014, 35, 1196–1208. [Google Scholar] [CrossRef] [Green Version]
- Ceccaldi, R.; Briot, D.; Larghero, J.; Vasquez, N.; Dubois d’Enghien, C.; Chamousset, D.; Noguera, M.E.; Waisfisz, Q.; Hermine, O.; Pondarre, C.; et al. Spontaneous abrogation of the G(2)DNA damage checkpoint has clinical benefits but promotes leukemogenesis in Fanconi anemia patients. J. Clin. Investig. 2011, 121, 184–194. [Google Scholar] [CrossRef] [Green Version]
- Houghtaling, S.; Granville, L.; Akkari, Y.; Torimaru, Y.; Olson, S.; Finegold, M.; Grompe, M. Heterozygosity for p53 (Trp53+/−) accelerates epithelial tumor formation in fanconi anemia complementation group D2 (Fancd2) knockout mice. Cancer Res. 2005, 65, 85–91. [Google Scholar]
- Holstege, H.; Joosse, S.A.; van Oostrom, C.T.; Nederlof, P.M.; de Vries, A.; Jonkers, J. High incidence of protein-truncating TP53 mutations in BRCA1-related breast cancer. Cancer Res. 2009, 69, 3625–3633. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Luo, J.; Chen, F.; Yang, F.; Song, W.; Zhu, A.; Guan, X. BRCA1 regulates PIG3-mediated apoptosis in a p53-dependent manner. Oncotarget 2015, 6, 7608–7618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Z.; Kachnic, L.; Zhang, J.; Powell, S.N.; Xia, F. DNA damage induces p53-dependent BRCA1 nuclear export. J. Biol. Chem. 2004, 279, 28574–28584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, J.; Yang, E.S.; Jiang, G.; Nowsheen, S.; Wang, H.; Wang, T.; Wang, Y.; Billheimer, D.; Chakravarthy, A.B.; Brown, M.; et al. p53-dependent BRCA1 nuclear export controls cellular susceptibility to DNA damage. Cancer Res. 2011, 71, 5546–5557. [Google Scholar] [CrossRef] [Green Version]
- Buckley, N.E.; D’Costa, Z.; Kaminska, M.; Mullan, P.B. S100A2 is a BRCA1/p63 coregulated tumour suppressor gene with roles in the regulation of mutant p53 stability. Cell Death Dis. 2014, 5, e1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valenti, F.; Ganci, F.; Fontemaggi, G.; Sacconi, A.; Strano, S.; Blandino, G.; Di Agostino, S. Gain of function mutant p53 proteins cooperate with E2F4 to transcriptionally downregulate RAD17 and BRCA1 gene expression. Oncotarget 2015, 6, 5547–5566. [Google Scholar] [CrossRef] [Green Version]
- Panneerselvam, J.; Pickering, A.; Zhang, J.; Wang, H.; Tian, H.; Zheng, J.; Fei, P. A hidden role of the inactivated FANCD2: Upregulating DeltaNp63. Oncotarget 2013, 4, 1416–1426. [Google Scholar] [CrossRef]
- Gatti, V.; Bongiorno-Borbone, L.; Fierro, C.; Annicchiarico-Petruzzelli, M.; Melino, G.; Peschiaroli, A. p63 at the Crossroads between Stemness and Metastasis in Breast Cancer. Int. J. Mol. Sci. 2019, 20, 2683. [Google Scholar] [CrossRef] [Green Version]
- Su, X.; Napoli, M.; Abbas, H.A.; Venkatanarayan, A.; Bui, N.H.B.; Coarfa, C.; Gi, Y.J.; Kittrell, F.; Gunaratne, P.H.; Medina, D.; et al. TAp63 suppresses mammary tumorigenesis through regulation of the Hippo pathway. Oncogene 2017, 36, 2377–2393. [Google Scholar] [CrossRef]
- Park, E.; Kim, H.; Kim, J.M.; Primack, B.; Vidal-Cardenas, S.; Xu, Y.; Price, B.D.; Mills, A.A.; D’Andrea, A.D. FANCD2 activates transcription of TAp63 and suppresses tumorigenesis. Mol. Cell 2013, 50, 908–918. [Google Scholar] [CrossRef] [Green Version]
- Du, Z.; Li, J.; Wang, L.; Bian, C.; Wang, Q.; Liao, L.; Dou, X.; Bian, X.; Zhao, R.C. Overexpression of DeltaNp63alpha induces a stem cell phenotype in MCF7 breast carcinoma cell line through the Notch pathway. Cancer Sci. 2010, 101, 2417–2424. [Google Scholar] [CrossRef] [PubMed]
- Memmi, E.M.; Sanarico, A.G.; Giacobbe, A.; Peschiaroli, A.; Frezza, V.; Cicalese, A.; Pisati, F.; Tosoni, D.; Zhou, H.; Tonon, G.; et al. p63 Sustains self-renewal of mammary cancer stem cells through regulation of Sonic Hedgehog signaling. Proc. Natl. Acad. Sci. USA 2015, 112, 3499–3504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakrabarti, R.; Wei, Y.; Hwang, J.; Hang, X.; Andres Blanco, M.; Choudhury, A.; Tiede, B.; Romano, R.A.; DeCoste, C.; Mercatali, L.; et al. DeltaNp63 promotes stem cell activity in mammary gland development and basal-like breast cancer by enhancing Fzd7 expression and Wnt signalling. Nat. Cell Biol. 2014, 16, 1004–1015. [Google Scholar] [CrossRef] [PubMed]
- Buckley, N.E.; Conlon, S.J.; Jirstrom, K.; Kay, E.W.; Crawford, N.T.; O’Grady, A.; Sheehan, K.; McDade, S.S.; Wang, C.W.; McCance, D.J.; et al. The DeltaNp63 proteins are key allies of BRCA1 in the prevention of basal-like breast cancer. Cancer Res. 2011, 71, 1933–1944. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Bierie, B.; Li, A.G.; Pathania, S.; Toomire, K.; Dimitrov, S.D.; Liu, B.; Gelman, R.; Giobbie-Hurder, A.; Feunteun, J.; et al. BRCA1/FANCD2/BRG1-Driven DNA Repair Stabilizes the Differentiation State of Human Mammary Epithelial Cells. Mol. Cell 2016, 63, 277–292. [Google Scholar] [CrossRef] [Green Version]
- Buckley, N.E.; Nic An tSaoir, C.B.; Blayney, J.K.; Oram, L.C.; Crawford, N.T.; D’Costa, Z.C.; Quinn, J.E.; Kennedy, R.D.; Harkin, D.P.; Mullan, P.B. BRCA1 is a key regulator of breast differentiation through activation of Notch signalling with implications for anti-endocrine treatment of breast cancers. Nucleic Acids Res. 2013, 41, 8601–8614. [Google Scholar] [CrossRef]
- Peltomaki, P. Deficient DNA mismatch repair: A common etiologic factor for colon cancer. Hum. Mol. Genet. 2001, 10, 735–740. [Google Scholar] [CrossRef] [Green Version]
- Poulogiannis, G.; Frayling, I.M.; Arends, M.J. DNA mismatch repair deficiency in sporadic colorectal cancer and Lynch syndrome. Histopathology 2010, 56, 167–179. [Google Scholar] [CrossRef] [Green Version]
- Norgaard, K.; Muller, C.; Christensen, N.; Chiloeches, M.L.; Madsen, C.L.; Nielsen, S.S.; Thingholm, T.E.; Belcheva, A. Loss of mismatch repair signaling impairs the WNT-bone morphogenetic protein crosstalk and the colonic homeostasis. J. Mol. Cell Biol. 2020, 12, 410–423. [Google Scholar] [CrossRef] [Green Version]
- Medema, J.P.; Vermeulen, L. Microenvironmental regulation of stem cells in intestinal homeostasis and cancer. Nature 2011, 474, 318–326. [Google Scholar] [CrossRef]
- Vermeulen, L.; De Sousa, E.M.F.; van der Heijden, M.; Cameron, K.; de Jong, J.H.; Borovski, T.; Tuynman, J.B.; Todaro, M.; Merz, C.; Rodermond, H.; et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 2010, 12, 468–476. [Google Scholar] [CrossRef]
- Lombardo, Y.; Scopelliti, A.; Cammareri, P.; Todaro, M.; Iovino, F.; Ricci-Vitiani, L.; Gulotta, G.; Dieli, F.; de Maria, R.; Stassi, G. Bone morphogenetic protein 4 induces differentiation of colorectal cancer stem cells and increases their response to chemotherapy in mice. Gastroenterology 2011, 140, 297–309.e6. [Google Scholar] [CrossRef] [PubMed]
- Fink, D.; Aebi, S.; Howell, S.B. The role of DNA mismatch repair in drug resistance. Clin. Cancer Res. 1998, 4, 1–6. [Google Scholar]
- Hickman, M.J.; Samson, L.D. Role of DNA mismatch repair and p53 in signaling induction of apoptosis by alkylating agents. Proc. Natl. Acad. Sci. USA 1999, 96, 10764–10769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hickman, M.J.; Samson, L.D. Apoptotic signaling in response to a single type of DNA lesion, O(6)-methylguanine. Mol. Cell 2004, 14, 105–116. [Google Scholar] [CrossRef]
- Tobar, N.; Villar, V.; Santibanez, J.F. ROS-NFkappaB mediates TGF-beta1-induced expression of urokinase-type plasminogen activator, matrix metalloproteinase-9 and cell invasion. Mol. Cell. Biochem. 2010, 340, 195–202. [Google Scholar] [CrossRef]
- Mori, K.; Shibanuma, M.; Nose, K. Invasive potential induced under long-term oxidative stress in mammary epithelial cells. Cancer Res. 2004, 64, 7464–7472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kambach, D.M.; Sodi, V.L.; Lelkes, P.I.; Azizkhan-Clifford, J.; Reginato, M.J. ErbB2, FoxM1 and 14-3-3zeta prime breast cancer cells for invasion in response to ionizing radiation. Oncogene 2014, 33, 589–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahya, U.; Koseer, A.S.; Dubrovska, A. Amino Acid Transporters on the Guard of Cell Genome and Epigenome. Cancers 2021, 13, 125. [Google Scholar] [CrossRef]
- Evans, A.R.; Limp-Foster, M.; Kelley, M.R. Going APE over ref-1. Mutat. Res. 2000, 461, 83–108. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, T.; Dong, Q.; Nice, E.C.; Huang, C.; Wei, Y. Redox homeostasis: The linchpin in stem cell self-renewal and differentiation. Cell Death Dis. 2013, 4, e537. [Google Scholar] [CrossRef] [Green Version]
- Diehn, M.; Cho, R.W.; Lobo, N.A.; Kalisky, T.; Dorie, M.J.; Kulp, A.N.; Qian, D.; Lam, J.S.; Ailles, L.E.; Wong, M.; et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 2009, 458, 780–783. [Google Scholar] [CrossRef]
- Herault, O.; Hope, K.J.; Deneault, E.; Mayotte, N.; Chagraoui, J.; Wilhelm, B.T.; Cellot, S.; Sauvageau, M.; Andrade-Navarro, M.A.; Hebert, J.; et al. A role for GPx3 in activity of normal and leukemia stem cells. J. Exp. Med. 2012, 209, 895–901. [Google Scholar] [CrossRef]
- Srinivas, U.S.; Tan, B.W.Q.; Vellayappan, B.A.; Jeyasekharan, A.D. ROS and the DNA damage response in cancer. Redox Biol. 2019, 25, 101084. [Google Scholar] [CrossRef]
- Kaur, G.; Cholia, R.P.; Mantha, A.K.; Kumar, R. DNA repair and redox activities and inhibitors of apurinic/apyrimidinic endonuclease 1/redox effector factor 1 (APE1/Ref-1): A comparative analysis and their scope and limitations toward anticancer drug development. J. Med. Chem. 2014, 57, 10241–10256. [Google Scholar] [CrossRef] [PubMed]
- Li, M.X.; Shan, J.L.; Wang, D.; He, Y.; Zhou, Q.; Xia, L.; Zeng, L.L.; Li, Z.P.; Wang, G.; Yang, Z.Z. Human apurinic/apyrimidinic endonuclease 1 translocalizes to mitochondria after photodynamic therapy and protects cells from apoptosis. Cancer Sci. 2012, 103, 882–888. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, L.; Murthy, K.G.; Zhu, C.; Curran, T.; Xanthoudakis, S.; Prives, C. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev. 1997, 11, 558–570. [Google Scholar] [CrossRef] [Green Version]
- Zaky, A.; Busso, C.; Izumi, T.; Chattopadhyay, R.; Bassiouny, A.; Mitra, S.; Bhakat, K.K. Regulation of the human AP-endonuclease (APE1/Ref-1) expression by the tumor suppressor p53 in response to DNA damage. Nucleic Acids Res. 2008, 36, 1555–1566. [Google Scholar] [CrossRef]
- Sengupta, S.; Mitra, S.; Bhakat, K.K. Dual regulatory roles of human AP-endonuclease (APE1/Ref-1) in CDKN1A/p21 expression. PLoS ONE 2013, 8, e68467. [Google Scholar] [CrossRef] [Green Version]
- Skvortsova, I.; Skvortsov, S.; Stasyk, T.; Raju, U.; Popper, B.A.; Schiestl, B.; von Guggenberg, E.; Neher, A.; Bonn, G.K.; Huber, L.A.; et al. Intracellular signaling pathways regulating radioresistance of human prostate carcinoma cells. Proteomics 2008, 8, 4521–4533. [Google Scholar] [CrossRef] [PubMed]
- Lou, D.; Zhu, L.; Ding, H.; Dai, H.Y.; Zou, G.M. Aberrant expression of redox protein Ape1 in colon cancer stem cells. Oncol. Lett. 2014, 7, 1078–1082. [Google Scholar] [CrossRef] [Green Version]
- Fishel, M.L.; Jiang, Y.; Rajeshkumar, N.V.; Scandura, G.; Sinn, A.L.; He, Y.; Shen, C.; Jones, D.R.; Pollok, K.E.; Ivan, M.; et al. Impact of APE1/Ref-1 redox inhibition on pancreatic tumor growth. Mol. Cancer Ther. 2011, 10, 1698–1708. [Google Scholar] [CrossRef] [Green Version]
- Skvortsov, S.; Debbage, P.; Lukas, P.; Skvortsova, I. Crosstalk between DNA repair and cancer stem cell (CSC) associated intracellular pathways. Semin. Cancer Biol. 2015, 31, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Saha, T.; Rih, J.K.; Rosen, E.M. BRCA1 down-regulates cellular levels of reactive oxygen species. FEBS Lett. 2009, 583, 1535–1543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorrini, C.; Baniasadi, P.S.; Harris, I.S.; Silvester, J.; Inoue, S.; Snow, B.; Joshi, P.A.; Wakeham, A.; Molyneux, S.D.; Martin, B.; et al. BRCA1 interacts with Nrf2 to regulate antioxidant signaling and cell survival. J. Exp. Med. 2013, 210, 1529–1544. [Google Scholar] [CrossRef] [Green Version]
- Bae, I.; Fan, S.; Meng, Q.; Rih, J.K.; Kim, H.J.; Kang, H.J.; Xu, J.; Goldberg, I.D.; Jaiswal, A.K.; Rosen, E.M. BRCA1 induces antioxidant gene expression and resistance to oxidative stress. Cancer Res. 2004, 64, 7893–7909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosey, A.M.; Gorski, J.J.; Murray, M.M.; Quinn, J.E.; Chung, W.Y.; Stewart, G.E.; James, C.R.; Farragher, S.M.; Mulligan, J.M.; Scott, A.N.; et al. Molecular basis for estrogen receptor alpha deficiency in BRCA1-linked breast cancer. J. Natl. Cancer Inst. 2007, 99, 1683–1694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curtis, C.D.; Thorngren, D.L.; Ziegler, Y.S.; Sarkeshik, A.; Yates, J.R.; Nardulli, A.M. Apurinic/apyrimidinic endonuclease 1 alters estrogen receptor activity and estrogen-responsive gene expression. Mol. Endocrinol. 2009, 23, 1346–1359. [Google Scholar] [CrossRef] [Green Version]
- Du, W.; Rani, R.; Sipple, J.; Schick, J.; Myers, K.C.; Mehta, P.; Andreassen, P.R.; Davies, S.M.; Pang, Q. The FA pathway counteracts oxidative stress through selective protection of antioxidant defense gene promoters. Blood 2012, 119, 4142–4151. [Google Scholar] [CrossRef]
- Pagano, G.; Talamanca, A.A.; Castello, G.; d’Ischia, M.; Pallardo, F.V.; Petrovic, S.; Porto, B.; Tiano, L.; Zatterale, A. Bone marrow cell transcripts from Fanconi anaemia patients reveal in vivo alterations in mitochondrial, redox and DNA repair pathways. Eur. J. Haematol. 2013, 91, 141–151. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.S.; Leung, K.S.; Hicks, M.J.; Hastings, P.J.; Youssoufian, H.; Plon, S.E. Defective mitochondrial peroxiredoxin-3 results in sensitivity to oxidative stress in Fanconi anemia. J. Cell Biol. 2006, 175, 225–235. [Google Scholar] [CrossRef] [Green Version]
- Willers, H.; Kachnic, L.A.; Luo, C.M.; Li, L.; Purschke, M.; Borgmann, K.; Held, K.D.; Powell, S.N. Biomarkers and mechanisms of FANCD2 function. J. Biomed. Biotechnol. 2008, 2008, 821529. [Google Scholar] [CrossRef] [PubMed]
- Zdzalik, D.; Vagbo, C.B.; Kirpekar, F.; Davydova, E.; Puscian, A.; Maciejewska, A.M.; Krokan, H.E.; Klungland, A.; Tudek, B.; van den Born, E.; et al. Protozoan ALKBH8 oxygenases display both DNA repair and tRNA modification activities. PLoS ONE 2014, 9, e98729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ougland, R.; Rognes, T.; Klungland, A.; Larsen, E. Non-homologous functions of the AlkB homologs. J. Mol. Cell Biol. 2015, 7, 494–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimada, K.; Nakamura, M.; Anai, S.; De Velasco, M.; Tanaka, M.; Tsujikawa, K.; Ouji, Y.; Konishi, N. A novel human AlkB homologue, ALKBH8, contributes to human bladder cancer progression. Cancer Res. 2009, 69, 3157–3164. [Google Scholar] [CrossRef] [Green Version]
- Ke, B.; Ye, K.; Cheng, S. ALKBH2 inhibition alleviates malignancy in colorectal cancer by regulating BMI1-mediated activation of NF-kappaB pathway. World J. Surg. Oncol. 2020, 18, 328. [Google Scholar] [CrossRef]
- Ohshio, I.; Kawakami, R.; Tsukada, Y.; Nakajima, K.; Kitae, K.; Shimanoe, T.; Saigo, Y.; Hase, H.; Ueda, Y.; Jingushi, K.; et al. ALKBH8 promotes bladder cancer growth and progression through regulating the expression of survivin. Biochem. Biophys. Res. Commun. 2016, 477, 413–418. [Google Scholar] [CrossRef] [Green Version]
- Pilzys, T.; Marcinkowski, M.; Kukwa, W.; Garbicz, D.; Dylewska, M.; Ferenc, K.; Mieczkowski, A.; Kukwa, A.; Migacz, E.; Wolosz, D.; et al. ALKBH overexpression in head and neck cancer: Potential target for novel anticancer therapy. Sci. Rep. 2019, 9, 13249. [Google Scholar] [CrossRef] [Green Version]
- Fu, D.; Jordan, J.J.; Samson, L.D. Human ALKBH7 is required for alkylation and oxidation-induced programmed necrosis. Genes Dev. 2013, 27, 1089–1100. [Google Scholar] [CrossRef] [Green Version]
- Baritaud, M.; Boujrad, H.; Lorenzo, H.K.; Krantic, S.; Susin, S.A. Histone H2AX: The missing link in AIF-mediated caspase-independent programmed necrosis. Cell Cycle 2010, 9, 3166–3173. [Google Scholar] [CrossRef]
- Ho, S.S.; Zhang, W.Y.; Tan, N.Y.; Khatoo, M.; Suter, M.A.; Tripathi, S.; Cheung, F.S.; Lim, W.K.; Tan, P.H.; Ngeow, J.; et al. The DNA Structure-Specific Endonuclease MUS81 Mediates DNA Sensor STING-Dependent Host Rejection of Prostate Cancer Cells. Immunity 2016, 44, 1177–1189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ablasser, A.; Bauernfeind, F.; Hartmann, G.; Latz, E.; Fitzgerald, K.A.; Hornung, V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 2009, 10, 1065–1072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burdette, D.L.; Monroe, K.M.; Sotelo-Troha, K.; Iwig, J.S.; Eckert, B.; Hyodo, M.; Hayakawa, Y.; Vance, R.E. STING is a direct innate immune sensor of cyclic di-GMP. Nature 2011, 478, 515–518. [Google Scholar] [CrossRef] [PubMed]
- Wan, D.; Jiang, W.; Hao, J. Research Advances in How the cGAS-STING Pathway Controls the Cellular Inflammatory Response. Front. Immunol. 2020, 11, 615. [Google Scholar] [CrossRef]
- Zitvogel, L.; Galluzzi, L.; Kepp, O.; Smyth, M.J.; Kroemer, G. Type I interferons in anticancer immunity. Nat. Rev. Immunol. 2015, 15, 405–414. [Google Scholar] [CrossRef]
- Konno, H.; Yamauchi, S.; Berglund, A.; Putney, R.M.; Mule, J.J.; Barber, G.N. Suppression of STING signaling through epigenetic silencing and missense mutation impedes DNA damage mediated cytokine production. Oncogene 2018, 37, 2037–2051. [Google Scholar] [CrossRef]
- Suter, M.A.; Tan, N.Y.; Thiam, C.H.; Khatoo, M.; MacAry, P.A.; Angeli, V.; Gasser, S.; Zhang, Y.L. cGAS-STING cytosolic DNA sensing pathway is suppressed by JAK2-STAT3 in tumor cells. Sci. Rep. 2021, 11, 7243. [Google Scholar] [CrossRef]
- Galoczova, M.; Coates, P.; Vojtesek, B. STAT3, stem cells, cancer stem cells and p63. Cell Mol. Biol. Lett. 2018, 23, 12. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Jin, C.; Li, H.B.; Tong, J.; Ouyang, X.; Cetinbas, N.M.; Zhu, S.; Strowig, T.; Lam, F.C.; Zhao, C.; et al. The DNA-sensing AIM2 inflammasome controls radiation-induced cell death and tissue injury. Science 2016, 354, 765–768. [Google Scholar] [CrossRef] [Green Version]
- Fernandes-Alnemri, T.; Yu, J.W.; Juliana, C.; Solorzano, L.; Kang, S.; Wu, J.; Datta, P.; McCormick, M.; Huang, L.; McDermott, E.; et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat. Immunol. 2010, 11, 385–393. [Google Scholar] [CrossRef] [Green Version]
- Karpova, A.Y.; Trost, M.; Murray, J.M.; Cantley, L.C.; Howley, P.M. Interferon regulatory factor-3 is an in vivo target of DNA-PK. Proc. Natl. Acad. Sci. USA 2002, 99, 2818–2823. [Google Scholar] [CrossRef] [Green Version]
- Huang, T.T.; Wuerzberger-Davis, S.M.; Wu, Z.H.; Miyamoto, S. Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell 2003, 115, 565–576. [Google Scholar] [CrossRef] [Green Version]
- Federico, A.; Morgillo, F.; Tuccillo, C.; Ciardiello, F.; Loguercio, C. Chronic inflammation and oxidative stress in human carcinogenesis. Int. J. Cancer 2007, 121, 2381–2386. [Google Scholar] [CrossRef]
- Wolf, C.; Rapp, A.; Berndt, N.; Staroske, W.; Schuster, M.; Dobrick-Mattheuer, M.; Kretschmer, S.; Konig, N.; Kurth, T.; Wieczorek, D.; et al. RPA and Rad51 constitute a cell intrinsic mechanism to protect the cytosol from self DNA. Nat. Commun. 2016, 7, 11752. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Srinivasan, K.; Abdisalaam, S.; Su, F.; Raj, P.; Dozmorov, I.; Mishra, R.; Wakeland, E.K.; Ghose, S.; Mukherjee, S.; et al. RAD51 interconnects between DNA replication, DNA repair and immunity. Nucleic Acids Res. 2017, 45, 4590–4605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Burness, M.L.; Martin-Trevino, R.; Guy, J.; Bai, S.; Harouaka, R.; Brooks, M.D.; Shang, L.; Fox, A.; Luther, T.K.; et al. RAD51 Mediates Resistance of Cancer Stem Cells to PARP Inhibition in Triple-Negative Breast Cancer. Clin. Cancer Res. 2017, 23, 514–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, H.O.; Brend, T.; Payne, H.L.; Wright, A.; Ward, T.A.; Patel, K.; Egnuni, T.; Stead, L.F.; Patel, A.; Wurdak, H.; et al. RAD51 Is a Selective DNA Repair Target to Radiosensitize Glioma Stem Cells. Stem Cell Rep. 2017, 8, 125–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reislander, T.; Lombardi, E.P.; Groelly, F.J.; Miar, A.; Porru, M.; Di Vito, S.; Wright, B.; Lockstone, H.; Biroccio, A.; Harris, A.; et al. BRCA2 abrogation triggers innate immune responses potentiated by treatment with PARP inhibitors. Nat. Commun. 2019, 10, 3143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanpouille-Box, C.; Alard, A.; Aryankalayil, M.J.; Sarfraz, Y.; Diamond, J.M.; Schneider, R.J.; Inghirami, G.; Coleman, C.N.; Formenti, S.C.; Demaria, S. DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity. Nat. Commun. 2017, 8, 15618. [Google Scholar] [CrossRef] [PubMed]
- Parkes, E.E.; Walker, S.M.; Taggart, L.E.; McCabe, N.; Knight, L.A.; Wilkinson, R.; McCloskey, K.D.; Buckley, N.E.; Savage, K.I.; Salto-Tellez, M.; et al. Activation of STING-Dependent Innate Immune Signaling By S-Phase-Specific DNA Damage in Breast Cancer. J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Behbod, F.; Atkinson, R.L.; Landis, M.D.; Kittrell, F.; Edwards, D.; Medina, D.; Tsimelzon, A.; Hilsenbeck, S.; Green, J.E.; et al. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res. 2008, 68, 4674–4682. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Xu, H.; Liu, T.; Huang, M.; Butter, P.P.; Li, C.; Zhang, L.; Kao, G.D.; Gong, Y.; Maity, A.; et al. Temporal DNA-PK activation drives genomic instability and therapy resistance in glioma stem cells. JCI Insight 2018, 3. [Google Scholar] [CrossRef] [Green Version]
- Abad, E.; Civit, L.; Potesil, D.; Zdrahal, Z.; Lyakhovich, A. Enhanced DNA damage response through RAD50 in triple negative breast cancer resistant and cancer stem-like cells contributes to chemoresistance. FEBS J. 2021, 288, 2184–2202. [Google Scholar] [CrossRef] [PubMed]
- Borgmann, K.; Kocher, S.; Kriegs, M.; Mansour, W.Y.; Parplys, A.C.; Rieckmann, T.; Rothkamm, K. DNA Repair. Recent. Results Cancer Res. 2016, 198, 1–24. [Google Scholar] [CrossRef]
- Kim, H.; Lin, Q.; Yun, Z. BRCA1 regulates the cancer stem cell fate of breast cancer cells in the context of hypoxia and histone deacetylase inhibitors. Sci. Rep. 2019, 9, 9702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorodetska, I.; Lukiyanchuk, V.; Peitzsch, C.; Kozeretska, I.; Dubrovska, A. BRCA1 and EZH2 cooperate in regulation of prostate cancer stem cell phenotype. Int. J. Cancer 2019, 145, 2974–2985. [Google Scholar] [CrossRef] [PubMed]
- Anuja, K.; Chowdhury, A.R.; Saha, A.; Roy, S.; Rath, A.K.; Kar, M.; Banerjee, B. Radiation-induced DNA damage response and resistance in colorectal cancer stem-like cells. Int. J. Radiat. Biol. 2019, 95, 667–679. [Google Scholar] [CrossRef] [Green Version]
- Nathansen, J.; Lukiyanchuk, V.; Hein, L.; Stolte, M.I.; Borgmann, K.; Lock, S.; Kurth, I.; Baumann, M.; Krause, M.; Linge, A.; et al. Oct4 confers stemness and radioresistance to head and neck squamous cell carcinoma by regulating the homologous recombination factors PSMC3IP and RAD54L. Oncogene 2021, 40, 4214–4228. [Google Scholar] [CrossRef]
- Cheng, L.; Wu, Q.; Huang, Z.; Guryanova, O.A.; Huang, Q.; Shou, W.; Rich, J.N.; Bao, S. L1CAM regulates DNA damage checkpoint response of glioblastoma stem cells through NBS1. EMBO J. 2011, 30, 800–813. [Google Scholar] [CrossRef]
- Pedersen, H.; Anne Adanma Obara, E.; Elbaek, K.J.; Vitting-Serup, K.; Hamerlik, P. Replication Protein A (RPA) Mediates Radio-Resistance of Glioblastoma Cancer Stem-Like Cells. Int. J. Mol. Sci. 2020, 21, 1388. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.L.; Xing, B.C.; Han, H.B.; Zhao, W.; Hu, M.H.; Xu, Z.L.; Li, J.Y.; Xie, Y.; Gu, J.; Wang, Y.; et al. The properties of tumor-initiating cells from a hepatocellular carcinoma patient’s primary and recurrent tumor. Carcinogenesis 2010, 31, 167–174. [Google Scholar] [CrossRef] [Green Version]
- Tse, A.N.; Carvajal, R.; Schwartz, G.K. Targeting checkpoint kinase 1 in cancer therapeutics. Clin. Cancer Res. 2007, 13, 1955–1960. [Google Scholar] [CrossRef] [Green Version]
- Soriani, A.; Zingoni, A.; Cerboni, C.; Iannitto, M.L.; Ricciardi, M.R.; Di Gialleonardo, V.; Cippitelli, M.; Fionda, C.; Petrucci, M.T.; Guarini, A.; et al. ATM-ATR-dependent up-regulation of DNAM-1 and NKG2D ligands on multiple myeloma cells by therapeutic agents results in enhanced NK-cell susceptibility and is associated with a senescent phenotype. Blood 2009, 113, 3503–3511. [Google Scholar] [CrossRef] [Green Version]
- Gasser, S.; Orsulic, S.; Brown, E.J.; Raulet, D.H. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature 2005, 436, 1186–1190. [Google Scholar] [CrossRef] [Green Version]
- Rodier, F.; Coppe, J.P.; Patil, C.K.; Hoeijmakers, W.A.; Munoz, D.P.; Raza, S.R.; Freund, A.; Campeau, E.; Davalos, A.R.; Campisi, J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 2009, 11, 973–979. [Google Scholar] [CrossRef]
- Milanovic, M.; Fan, D.N.Y.; Belenki, D.; Dabritz, J.H.M.; Zhao, Z.; Yu, Y.; Dorr, J.R.; Dimitrova, L.; Lenze, D.; Monteiro Barbosa, I.A.; et al. Senescence-associated reprogramming promotes cancer stemness. Nature 2018, 553, 96–100. [Google Scholar] [CrossRef] [Green Version]
- Sato, H.; Niimi, A.; Yasuhara, T.; Permata, T.B.M.; Hagiwara, Y.; Isono, M.; Nuryadi, E.; Sekine, R.; Oike, T.; Kakoti, S.; et al. DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat. Commun. 2017, 8, 1751. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Lin, A.; Cao, M.; Ding, W.; Mou, W.; Guo, N.; Chen, Z.; Zhang, J.; Luo, P. Activation of the DDR Pathway Leads to the Down-Regulation of the TGFbeta Pathway and a Better Response to ICIs in Patients With Metastatic Urothelial Carcinoma. Front. Immunol. 2021, 12, 634741. [Google Scholar] [CrossRef]
- Song, Y.; Huang, J.; Liang, D.; Hu, Y.; Mao, B.; Li, Q.; Sun, H.; Yang, Y.; Zhang, J.; Zhang, H.; et al. DNA Damage Repair Gene Mutations Are Indicative of a Favorable Prognosis in Colorectal Cancer Treated With Immune Checkpoint Inhibitors. Front. Oncol. 2020, 10, 549777. [Google Scholar] [CrossRef]
- Muller, L.; Tunger, A.; Plesca, I.; Wehner, R.; Temme, A.; Westphal, D.; Meier, F.; Bachmann, M.; Schmitz, M. Bidirectional Crosstalk Between Cancer Stem Cells and Immune Cell Subsets. Front. Immunol. 2020, 11, 140. [Google Scholar] [CrossRef]
- Wei, Q.; Frazier, M.L.; Levin, B. DNA repair: A double-edged sword. J. Natl. Cancer Inst. 2000, 92, 440–441. [Google Scholar] [CrossRef] [Green Version]
- Santos, M.A.; Faryabi, R.B.; Ergen, A.V.; Day, A.M.; Malhowski, A.; Canela, A.; Onozawa, M.; Lee, J.E.; Callen, E.; Gutierrez-Martinez, P.; et al. DNA-damage-induced differentiation of leukaemic cells as an anti-cancer barrier. Nature 2014, 514, 107–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vecchio, D.; Daga, A.; Carra, E.; Marubbi, D.; Baio, G.; Neumaier, C.E.; Vagge, S.; Corvo, R.; Pia Brisigotti, M.; Louis Ravetti, J.; et al. Predictability, efficacy and safety of radiosensitization of glioblastoma-initiating cells by the ATM inhibitor KU-60019. Int. J. Cancer 2014, 135, 479–491. [Google Scholar] [CrossRef]
- Raso, A.; Vecchio, D.; Cappelli, E.; Ropolo, M.; Poggi, A.; Nozza, P.; Biassoni, R.; Mascelli, S.; Capra, V.; Kalfas, F.; et al. Characterization of glioma stem cells through multiple stem cell markers and their specific sensitization to double-strand break-inducing agents by pharmacological inhibition of ataxia telangiectasia mutated protein. Brain Pathol. 2012, 22, 677–688. [Google Scholar] [CrossRef] [PubMed]
- Tachon, G.; Cortes, U.; Guichet, P.O.; Rivet, P.; Balbous, A.; Masliantsev, K.; Berger, A.; Boissonnade, O.; Wager, M.; Karayan-Tapon, L. Cell Cycle Changes after Glioblastoma Stem Cell Irradiation: The Major Role of RAD51. Int. J. Mol. Sci. 2018, 19, 3018. [Google Scholar] [CrossRef] [Green Version]
- Manic, G.; Signore, M.; Sistigu, A.; Russo, G.; Corradi, F.; Siteni, S.; Musella, M.; Vitale, S.; De Angelis, M.L.; Pallocca, M.; et al. CHK1-targeted therapy to deplete DNA replication-stressed, p53-deficient, hyperdiploid colorectal cancer stem cells. Gut 2018, 67, 903–917. [Google Scholar] [CrossRef]
- Di Franco, S.; Parrino, B.; Gaggianesi, M.; Pantina, V.D.; Bianca, P.; Nicotra, A.; Mangiapane, L.R.; Lo Iacono, M.; Ganduscio, G.; Veschi, V.; et al. CHK1 inhibitor sensitizes resistant colorectal cancer stem cells to nortopsentin. iScience 2021, 24, 102664. [Google Scholar] [CrossRef]
- Gallmeier, E.; Hermann, P.C.; Mueller, M.T.; Machado, J.G.; Ziesch, A.; De Toni, E.N.; Palagyi, A.; Eisen, C.; Ellwart, J.W.; Rivera, J.; et al. Inhibition of ataxia telangiectasia- and Rad3-related function abrogates the in vitro and in vivo tumorigenicity of human colon cancer cells through depletion of the CD133(+) tumor-initiating cell fraction. Stem Cells 2011, 29, 418–429. [Google Scholar] [CrossRef]
- Bartucci, M.; Svensson, S.; Romania, P.; Dattilo, R.; Patrizii, M.; Signore, M.; Navarra, S.; Lotti, F.; Biffoni, M.; Pilozzi, E.; et al. Therapeutic targeting of Chk1 in NSCLC stem cells during chemotherapy. Cell Death Differ. 2012, 19, 768–778. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Z.; Sperl, B.; Ullrich, A.; Knyazev, P. Metformin and salinomycin as the best combination for the eradication of NSCLC monolayer cells and their alveospheres (cancer stem cells) irrespective of EGFR, KRAS, EML4/ALK and LKB1 status. Oncotarget 2014, 5, 12877–12890. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Dong, J.; Haiech, J.; Kilhoffer, M.C.; Zeniou, M. Cancer Stem Cell Quiescence and Plasticity as Major Challenges in Cancer Therapy. Stem Cells Int. 2016, 2016, 1740936. [Google Scholar] [CrossRef] [Green Version]
- Hutcherson, R.J.; Kemp, M.G. ATR kinase inhibition sensitizes quiescent human cells to the lethal effects of cisplatin but increases mutagenesis. Mutat. Res. 2019, 816–818, 111678. [Google Scholar] [CrossRef]
- Turgeon, M.O.; Perry, N.J.S.; Poulogiannis, G. DNA Damage, Repair, and Cancer Metabolism. Front. Oncol. 2018, 8, 15. [Google Scholar] [CrossRef] [Green Version]
- Na, H.J.; Akan, I.; Abramowitz, L.K.; Hanover, J.A. Nutrient-Driven O-GlcNAcylation Controls DNA Damage Repair Signaling and Stem/Progenitor Cell Homeostasis. Cell Rep. 2020, 31, 107632. [Google Scholar] [CrossRef]
- Smith, H.L.; Southgate, H.; Tweddle, D.A.; Curtin, N.J. DNA damage checkpoint kinases in cancer. Expert Rev. Mol. Med. 2020, 22, e2. [Google Scholar] [CrossRef]
- Dent, P. Investigational CHK1 inhibitors in early phase clinical trials for the treatment of cancer. Expert Opin. Investig. Drugs 2019, 28, 1095–1100. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, Y.; Meng, L.; Ji, B.; Yang, D. Synergistic Antitumor Effect of Sorafenib in Combination with ATM Inhibitor in Hepatocellular Carcinoma Cells. Int. J. Med. Sci. 2017, 14, 523–529. [Google Scholar] [CrossRef] [Green Version]
- Hintelmann, K.; Berenz, T.; Kriegs, M.; Christiansen, S.; Gatzemeier, F.; Struve, N.; Petersen, C.; Betz, C.; Rothkamm, K.; Oetting, A.; et al. Dual Inhibition of PARP and the Intra-S/G2 Cell Cycle Checkpoints Results in Highly Effective Radiosensitization of HPV-Positive HNSCC Cells. Front. Oncol. 2021, 11, 683688. [Google Scholar] [CrossRef] [PubMed]
- Booth, L.; Cruickshanks, N.; Ridder, T.; Dai, Y.; Grant, S.; Dent, P. PARP and CHK inhibitors interact to cause DNA damage and cell death in mammary carcinoma cells. Cancer Biol. Ther. 2013, 14, 458–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, A.R.; Jin, M.H.; Bang, J.H.; Oh, K.S.; Seo, H.R.; Oh, D.Y.; Bang, Y.J. Inhibition of ATR Increases the Sensitivity to WEE1 Inhibitor in Biliary Tract Cancer. Cancer Res. Treat. 2020, 52, 945–956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanjiv, K.; Hagenkort, A.; Calderon-Montano, J.M.; Koolmeister, T.; Reaper, P.M.; Mortusewicz, O.; Jacques, S.A.; Kuiper, R.V.; Schultz, N.; Scobie, M.; et al. Cancer-Specific Synthetic Lethality between ATR and CHK1 Kinase Activities. Cell Rep. 2016, 14, 298–309. [Google Scholar] [CrossRef]
- Jin, J.; Fang, H.; Yang, F.; Ji, W.; Guan, N.; Sun, Z.; Shi, Y.; Zhou, G.; Guan, X. Combined Inhibition of ATR and WEE1 as a Novel Therapeutic Strategy in Triple-Negative Breast Cancer. Neoplasia 2018, 20, 478–488. [Google Scholar] [CrossRef]
- Zhang, X.; Chiang, H.C.; Wang, Y.; Zhang, C.; Smith, S.; Zhao, X.; Nair, S.J.; Michalek, J.; Jatoi, I.; Lautner, M.; et al. Attenuation of RNA polymerase II pausing mitigates BRCA1-associated R-loop accumulation and tumorigenesis. Nat. Commun. 2017, 8, 15908. [Google Scholar] [CrossRef] [PubMed]
- Chiang, H.C.; Zhang, X.; Li, J.; Zhao, X.; Chen, J.; Wang, H.T.; Jatoi, I.; Brenner, A.; Hu, Y.; Li, R. BRCA1-associated R-loop affects transcription and differentiation in breast luminal epithelial cells. Nucleic Acids Res. 2019, 47, 5086–5099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Vlerken, L.E.; Kiefer, C.M.; Morehouse, C.; Li, Y.; Groves, C.; Wilson, S.D.; Yao, Y.; Hollingsworth, R.E.; Hurt, E.M. EZH2 is required for breast and pancreatic cancer stem cell maintenance and can be used as a functional cancer stem cell reporter. Stem Cells Transl. Med. 2013, 2, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Peitzsch, C.; Cojoc, M.; Hein, L.; Kurth, I.; Mabert, K.; Trautmann, F.; Klink, B.; Schrock, E.; Wirth, M.P.; Krause, M.; et al. An Epigenetic Reprogramming Strategy to Resensitize Radioresistant Prostate Cancer Cells. Cancer Res. 2016, 76, 2637–2651. [Google Scholar] [CrossRef] [Green Version]
- Witt, A.E.; Lee, C.W.; Lee, T.I.; Azzam, D.J.; Wang, B.; Caslini, C.; Petrocca, F.; Grosso, J.; Jones, M.; Cohick, E.B.; et al. Identification of a cancer stem cell-specific function for the histone deacetylases, HDAC1 and HDAC7, in breast and ovarian cancer. Oncogene 2017, 36, 1707–1720. [Google Scholar] [CrossRef] [PubMed]
- Nagel, R.; Semenova, E.A.; Berns, A. Drugging the addict: Non-oncogene addiction as a target for cancer therapy. EMBO Rep. 2016, 17, 1516–1531. [Google Scholar] [CrossRef]
- Hutchinson, M.N.D.; Mierzwa, M.; D’Silva, N.J. Radiation resistance in head and neck squamous cell carcinoma: Dire need for an appropriate sensitizer. Oncogene 2020, 39, 3638–3649. [Google Scholar] [CrossRef] [Green Version]
- Chen, J. The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression. Cold Spring Harb. Perspect. Med. 2016, 6, a026104. [Google Scholar] [CrossRef]
- Huang, J. Current developments of targeting the p53 signaling pathway for cancer treatment. Pharmacol. Ther. 2021, 220, 107720. [Google Scholar] [CrossRef]
- Kang, J.; D’Andrea, A.D.; Kozono, D. A DNA repair pathway-focused score for prediction of outcomes in ovarian cancer treated with platinum-based chemotherapy. J. Natl. Cancer Inst. 2012, 104, 670–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitroda, S.P.; Pashtan, I.M.; Logan, H.L.; Budke, B.; Darga, T.E.; Weichselbaum, R.R.; Connell, P.P. DNA repair pathway gene expression score correlates with repair proficiency and tumor sensitivity to chemotherapy. Sci. Transl. Med. 2014, 6, 229ra242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bold, I.T.; Specht, A.K.; Droste, C.F.; Zielinski, A.; Meyer, F.; Clauditz, T.S.; Munscher, A.; Werner, S.; Rothkamm, K.; Petersen, C.; et al. DNA Damage Response during Replication Correlates with CIN70 Score and Determines Survival in HNSCC Patients. Cancers 2021, 13, 1194. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.; Perkhofer, L.; Liebau, S.; Lin, Q.; Lechel, A.; Feld, F.M.; Hessmann, E.; Gaedcke, J.; Guthle, M.; Zenke, M.; et al. Loss of ATM accelerates pancreatic cancer formation and epithelial-mesenchymal transition. Nat. Commun. 2015, 6, 7677. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Tumor Type | Chemical Inhibitor | CSC Enrichment and Analysis | Biological Effect | Tumor Model | Clinical Trial (Selected) | Refs |
|---|---|---|---|---|---|---|
| GBM | ATM inhibitor KU55933 | Stem-cell-enriching conditions; -Sphere formation | Inhibition of DNA DSB repair after RT; CSC sensitization to RT In Vitro | Patient-derived cell cultures | -NCT03423628: Study to test ATM inhibitor, AZD1390 in combination with radiation therapy for the treatment of brain tumors, Phase 1 -NCT04550104: Study to determine the recommended dose and safety profiles of different DDRis when given in combination with curative intent RT in patients with stage IIB/IIIA/IIIB NSCLC, Phase 1 -NCT02588105: Study will determine the best dose and safety of AZD0156 when given alone or in combination with other agents e.g., Olaparib, irinotecan, Fluorouracil, Folinic Acid in patients with locally advanced/metastatic tumors, Phase 1 -NCT03225105: Study to evaluate the safety, tolerability, PK, PD, antitumor activity of ATM inhibitor M3541 in combination with fractionated palliative RT in patients with solid tumors with malignant lesions e.g., in the thorax, abdominal cavity, head and neck region, Phase 1 | [15] |
| GBM | ATM inhibitor KU60019 KU55933 | Stem-cell-enriching conditions | Sensitization of tumor cells to radiotherapy In Vitro; -Down-regulation of Prom1, Bmi1, CD15, Msi1, Msi2, Nanog, Nestin, Oct4 and Sox2 in response to combination of KU60019 and RT | Established cell lines, in vitro sphere formation; Orthotopic mouse tumor models | [223,224] | |
| Mouse model of GBM | ATM inhibitor KU60019 DNA-PK inhibitor NU7441 | IHC analysis of CD133+ cells | Sensitization of CSCs to RT in vivo in GEMM | GEMM: primary GBM in Ntv-a;Ink4a–/–;Ptenfl/fl;LSL-Luc donor mice induced by RCAS-mediated somatic gene transfer | [202] | |
| Mouse model of AML | ATM inhibitor KU55933 | -Cell morphology | Inducing differentiation of leukemic blast cells | The frequency of blasts and differentiated cells was identified in vitro by morphology | [222] | |
| GBM | CHK1/CHK2 inhibitor DBH | CD133 expression | Sensitization of CSCs to RT In Vitro and in mouse xenograft model | Patient-derived cell cultures; Orthotopic mouse tumor model | n.a. | [38] |
| GBM | RAD51 inhibitor RI-1 | Stem-cell-enriching conditions | Combination of RI-1 and RT increased apoptosis and delayed G2 arrest in radioresistant CSC cultures | Patient-derived cell cultures | n.a. | [225] |
| GBM | ATR inhibitor VE821 in combination with PARP1/ PARP2 inhibitor Olaparib | Stem-cell-enriching conditions | Inhibition of DNA DSB repair after RT; Inhibition of sphere formation after RT; CSC sensitization to RT In Vitro | Patient-derived cell cultures; In Vitro sphere formation | -NCT03967938: Study to evaluate efficacy of Olaparib in advanced cancers in patients with germline mutations or somatic tumor mutations in homologous recombination genes (1-2018 BSMO), phase 2 -NCT03742895: Study to assess efficacy and safety of Olaparib in patients with previously treated, HR repair mutation (HRm) or HR deficiency (HRD) positive advanced cancers (MK-7339-002/LYNK-002), phase 2 -NCT03212742: Study to investigate the toxicity and efficacy of olaparib and TMZ concomitantly with radiotherapy in first line treatment of unresectable high risk HGG (OLA-TMZ-RTE-01), phase ½ -NCT03167619: A randomized multicenter study to explore the efficacy of olaparib or olaparib in combination with durvalumab in platinum-treated TNBC, Phase 2 | [47] |
| GBM | -CHK1 inhibitor SCH900776 (SCH); -ATM inhibitor KU55933 in combination with PARP1/ PARP2 inhibitor Olaparib -ATM inhibitor VE821 in combination with PARP1/ PARP2 inhibitor Olaparib | Stem-cell-enriching conditions | Inhibition of DNA DSB repair after or G2/M checkpoint activation after RT; CSC sensitization to RT In Vitro | Patient-derived cell cultures; In Vitro sphere formation | [46] | |
| GBM | PARP1/ PARP2 inhibitor Olaparib | In Vitro limiting dilution assays In Vitro limiting dilution assays | Sensitization of tumor cells to RT In Vitro in In Vivo; Inhibition CSC self-renewal In Vitro in In Vivo; Inhibition CSC viability in vitro and tumor growth In Vivo | Patient-derived cell cultures; S.c. xenograft mouse model | [39] | |
| TNBC | PARP1/ PARP2 inhibitor Olaparib | ALDEFLUOR assay | Sensitization to PARP inhibition mediated by RAD51 KD | Established BRCA1-mutant cell lines with or without RAD51 KD | [196] | |
| CRC | CHK1 inhibitor LY2606368 | Sphere formation | Inhibition of tumor growth In Vitro and In Vivo | Patient-derived cell cultures Mouse xenograft models | -NCT02203513: Single arm study in BRCA1/2 mutation associated BC or OC, TNBC, high grade serous OC, and mCRPC, phase 2; -NCT04032080: Study to evaluate efficacy of LY3023414 and prexasertib in patients with metastatic TNBC, phase 2; -NCT02555644: Combination with chemotherapy and radiation in participants with HNSCC, phase 1 | [226] |
| CRC | CHK1 inhibitor LY2603618 (Rebusertib) | Sphere formation | Apoptosis induction (Annexin V positivity); -Inhibition of tumor cell growth In Vitro | Patient-derived cell cultures with or without RAD51 KD | -NCT00988858: Study to evaluate efficacy and safety of LY2603618 in combination with pemetrexed in participants with advanced or metastatic NSCLC, phase 2; -NCT00839332: A study to evaluate the dose and efficacy of LY2603618 in combination with Gemcitabine for participants with PC, phase 1/2 | [227] |
| CRC | CHK1 inhibitor SB218078 | CD133 expression | Inhibition of CD133+ cell population; Sensitization of CSCs to cisplatin In Vitro | Established cell lines | n.a. | [228] |
| NSCLC | CHK1 inhibitor SB218078 AZD7762 | Sphere formation | Inhibition of tumor growth In Vitro and In Vivo in combination with conventional chemotherapeutic drugs | Patient-derived cell cultures; Mouse xenograft models | -NCT00413686: Study to assess safety of AZD7762 administered alone and in combination with gemcitabine in patients with advanced solid malignancies, phase 1 | [229] |
| NSCLC | Combination of antidiabetic drug metformin and antibiotic salinomycin | Sphere formation | Inhibition of CHK2 (T68) phosphorylation; Inhibition of sphere formation | Established cell lines, in vitro sphere formation | -NCT01579812: Targeting CSC for prevention of relapse in gynecologic patients, phase 2 -NCT02437656: Combination of metformin to neoadjuvant radiochemotherapy in locally advanced RC (METCAP), phase 2 -NCT04387630: Neoadjuvant chemotherapy with or without metformin in early BC, phase 2/3 | [230] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nathansen, J.; Meyer, F.; Müller, L.; Schmitz, M.; Borgmann, K.; Dubrovska, A. Beyond the Double-Strand Breaks: The Role of DNA Repair Proteins in Cancer Stem-Cell Regulation. Cancers 2021, 13, 4818. https://doi.org/10.3390/cancers13194818
Nathansen J, Meyer F, Müller L, Schmitz M, Borgmann K, Dubrovska A. Beyond the Double-Strand Breaks: The Role of DNA Repair Proteins in Cancer Stem-Cell Regulation. Cancers. 2021; 13(19):4818. https://doi.org/10.3390/cancers13194818
Chicago/Turabian StyleNathansen, Jacqueline, Felix Meyer, Luise Müller, Marc Schmitz, Kerstin Borgmann, and Anna Dubrovska. 2021. "Beyond the Double-Strand Breaks: The Role of DNA Repair Proteins in Cancer Stem-Cell Regulation" Cancers 13, no. 19: 4818. https://doi.org/10.3390/cancers13194818