Secondary Somatic Mutations in G-Protein-Related Pathways and Mutation Signatures in Uveal Melanoma
Abstract
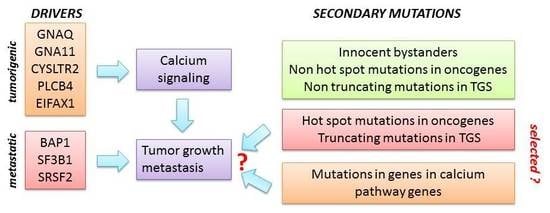
:1. Introduction
2. Results
2.1. Enrichment of Secondary Mutations in G-Protein Related Pathways
2.2. Expression Analysis
2.3. PTK2B as a Secondary Driver Mutation
2.4. Signatures of Somatic Mutations in UM
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Coupland, S.E.; Lake, S.L.; Zeschnigk, M.; Damato, B.E. Molecular pathology of uveal melanoma. Eye (Lond.) 2013, 27, 230–242. [Google Scholar] [CrossRef]
- Zeschnigk, M.; Lohmann, D.R. Prognostic testing in uveal melanoma. In Cancer Genomics: Molecular Classification, Prognosis and Response Prediction; Pfeffer, U., Ed.; Springer Science and Business Media: Dordrecht, The Netherlands, 2013; pp. 79–96. [Google Scholar]
- Amaro, A.; Gangemi, R.; Piaggio, F.; Angelini, G.; Barisione, G.; Ferrini, S.; Pfeffer, U. The biology of uveal melanoma. Cancer Metastasis Rev. 2017, 36, 109–140. [Google Scholar] [CrossRef]
- Zhang, T.; Dutton-Regester, K.; Brown, K.M.; Hayward, N.K. The genomic landscape of cutaneous melanoma. Pigment Cell Melanoma Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Van Raamsdonk, C.D.; Bezrookove, V.; Green, G.; Bauer, J.; Gaugler, L.; O’Brien, J.M.; Simpson, E.M.; Barsh, G.S.; Bastian, B.C. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 2009, 457, 599–602. [Google Scholar] [CrossRef] [PubMed]
- Van Raamsdonk, C.D.; Griewank, K.G.; Crosby, M.B.; Garrido, M.C.; Vemula, S.; Wiesner, T.; Obenauf, A.C.; Wackernagel, W.; Green, G.; Bouvier, N.; et al. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med. 2010, 363, 2191–2199. [Google Scholar] [CrossRef] [PubMed]
- Johansson, P.; Aoude, L.G.; Wadt, K.; Glasson, W.J.; Warrier, S.K.; Hewitt, A.W.; Kiilgaard, J.F.; Heegaard, S.; Isaacs, T.; Franchina, M.; et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget 2016, 7, 4624–4631. [Google Scholar] [CrossRef]
- Harbour, J.W.; Onken, M.D.; Roberson, E.D.; Duan, S.; Cao, L.; Worley, L.A.; Council, M.L.; Matatall, K.A.; Helms, C.; Bowcock, A.M. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 2010, 330, 1410–1413. [Google Scholar] [CrossRef]
- Harbour, J.W.; Roberson, E.D.; Anbunathan, H.; Onken, M.D.; Worley, L.A.; Bowcock, A.M. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat. Genet. 2013, 45, 133–135. [Google Scholar] [CrossRef]
- Field, M.G.; Durante, M.A.; Anbunathan, H.; Cai, L.Z.; Decatur, C.L.; Bowcock, A.M.; Kurtenbach, S.; Harbour, J.W. Punctuated evolution of canonical genomic aberrations in uveal melanoma. Nat. Commun. 2018, 9, 116. [Google Scholar] [CrossRef]
- Martin, M.; Masshofer, L.; Temming, P.; Rahmann, S.; Metz, C.; Bornfeld, N.; van de Nes, J.; Klein-Hitpass, L.; Hinnebusch, A.G.; Horsthemke, B.; et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet. 2013, 45, 933–936. [Google Scholar] [CrossRef] [Green Version]
- Dono, M.; Angelini, G.; Cecconi, M.; Amaro, A.; Esposito, A.I.; Mirisola, V.; Maric, I.; Lanza, F.; Nasciuti, F.; Viaggi, S.; et al. Mutation frequencies of GNAQ, GNA11, BAP1, SF3B1, EIF1AX and TERT in uveal melanoma: Detection of an activating mutation in the TERT gene promoter in a single case of uveal melanoma. Br. J. Cancer 2014, 110, 1058–1065. [Google Scholar] [CrossRef] [PubMed]
- Jager, M.J.; Brouwer, N.J.; Esmaeli, B. The Cancer Genome Atlas Project: An Integrated Molecular View of Uveal Melanoma. Ophthalmology 2018, 125, 1139–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, A.R.; Ceraudo, E.; Sher, J.J.; Guan, Y.; Shoushtari, A.N.; Chang, M.T.; Zhang, J.Q.; Walczak, E.G.; Kazmi, M.A.; Taylor, B.S.; et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat. Genet. 2016, 48, 675–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, A.G.; Shih, J.; Yau, C.; Gibb, E.A.; Oba, J.; Mungall, K.L.; Hess, J.M.; Uzunangelov, V.; Walter, V.; Danilova, L.; et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell 2017, 32, 204–220.e15. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Borresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [Green Version]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Campbell, P.J.; Stratton, M.R. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013, 3, 246–259. [Google Scholar] [CrossRef]
- Shah, C.P.; Weis, E.; Lajous, M.; Shields, J.A.; Shields, C.L. Intermittent and chronic ultraviolet light exposure and uveal melanoma: A meta-analysis. Ophthalmology 2005, 112, 1599–1607. [Google Scholar] [CrossRef]
- Dolin, P.J.; Foss, A.J.; Hungerford, J.L. Uveal melanoma: Is solar ultraviolet radiation a risk factor? Ophthalmic Epidemiol. 1994, 1, 27–30. [Google Scholar] [CrossRef]
- Reynaud, C.; Billaud, M. The theory of punctuated equilibrium, a breakthrough in understanding cancer. Med. Sci. M/S 2011, 27, 921–923. [Google Scholar] [CrossRef]
- Amaro, A.; Chiara, S.; Pfeffer, U. Molecular evolution of colorectal cancer: From multistep carcinogenesis to the big bang. Cancer Metastasis Rev. 2016, 35, 63–74. [Google Scholar] [CrossRef]
- Bakhoum, M.F.; Esmaeli, B. Molecular Characteristics of Uveal Melanoma: Insights from the Cancer Genome Atlas (TCGA) Project. Cancers 2019, 11, 1061. [Google Scholar] [CrossRef] [PubMed]
- Vader, M.J.C.; Madigan, M.C.; Versluis, M.; Suleiman, H.M.; Gezgin, G.; Gruis, N.A.; Out-Luiting, J.J.; Bergman, W.; Verdijk, R.M.; Jager, M.J.; et al. GNAQ and GNA11 mutations and downstream YAP activation in choroidal nevi. Br. J. Cancer 2017, 117, 884–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenman, C.; Stephens, P.; Smith, R.; Dalgliesh, G.L.; Hunter, C.; Bignell, G.; Davies, H.; Teague, J.; Butler, A.; Stevens, C.; et al. Patterns of somatic mutation in human cancer genomes. Nature 2007, 446, 153–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef]
- Nieuwenhuis, B.; Assen-Bolt, A.J.V.; AWH Van Waarde-Verhagen, M.; Sijmons, R.H.; Hout, A.H.V.d.; Bauch, T.; Streffer, C.; Kampinga, H.H. BRCA1 and BRCA2 heterozygosity and repair of X-ray-induced DNA damage. Int. J. Radiat. Biol. 2002, 78, 285–295. [Google Scholar] [CrossRef]
- Cosmic–Catlogue of somatic Mutations in Cancer. Available online: https://cancer.sanger.ac.uk/cosmic (accessed on 9 October 2019).
- Luscan, A.; Just, P.A.; Briand, A.; Burin des Roziers, C.; Goussard, P.; Nitschke, P.; Vidaud, M.; Avril, M.F.; Terris, B.; Pasmant, E. Uveal melanoma hepatic metastases mutation spectrum analysis using targeted next-generation sequencing of 400 cancer genes. Br. J. Ophthalmol. 2015, 99, 437–439. [Google Scholar] [CrossRef]
- McCarthy, C.; Kalirai, H.; Lake, S.L.; Dodson, A.; Damato, B.E.; Coupland, S.E. Insights into genetic alterations of liver metastases from uveal melanoma. Pigment Cell Melanoma Res. 2015, 29, 60–67. [Google Scholar] [CrossRef]
- Drake, J.M.; Graham, N.A.; Stoyanova, T.; Sedghi, A.; Goldstein, A.S.; Cai, H.; Smith, D.A.; Zhang, H.; Komisopoulou, E.; Huang, J.; et al. Oncogene-specific activation of tyrosine kinase networks during prostate cancer progression. Proc. Natl. Acad. Sci. USA 2012. [Google Scholar] [CrossRef]
- Dutton-Regester, K.; Irwin, D.; Hunt, P.; Aoude, L.G.; Tembe, V.; Pupo, G.M.; Lanagan, C.; Carter, C.D.; O’Connor, L.; O’Rourke, M.; et al. A high-throughput panel for identifying clinically relevant mutation profiles in melanoma. Mol. Cancer Ther. 2012, 11, 888–897. [Google Scholar] [CrossRef]
- Prickett, T.D.; Agrawal, N.S.; Wei, X.; Yates, K.E.; Lin, J.C.; Wunderlich, J.R.; Cronin, J.C.; Cruz, P.; Rosenberg, S.A.; Samuels, Y. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat. Genet. 2009, 41, 1127–1132. [Google Scholar] [CrossRef]
- Jones, A.M.; Ferguson, P.; Gardner, J.; Rooker, S.; Sutton, T.; Ahn, A.; Chatterjee, A.; Bickley, V.M.; Sarwar, M.; Emanuel, P.; et al. NRAS and EPHB6 mutation rates differ in metastatic melanomas of patients in the North Island versus South Island of New Zealand. Oncotarget 2016, 7, 41017–41030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting Functional Effect of Human Missense Mutations Using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013. [Google Scholar] [CrossRef] [PubMed]
- Vaser, R.; Adusumalli, S.; Leng, S.N.; Sikic, M.; Ng, P.C. SIFT missense predictions for genomes. Nat. Protoc. 2016, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Sims, G.E.; Murphy, S.; Miller, J.R.; Chan, A.P. Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLoS ONE 2012, 7, e46688. [Google Scholar] [CrossRef]
- Shihab, H.A.; Gough, J.; Cooper, D.N.; Day, I.N.M.; Gaunt, T.R. Predicting the Functional Consequences of Cancer-Associated Amino Acid Substitutions. Bioinformatics 2013, 29, 1504–1510. [Google Scholar] [CrossRef] [PubMed]
- Tozzo, V.; Barla, A. Cancer Mutational Signatures Identification with Sparse Dictionary Learning; Springer: Cham, Switzerland, 2017; pp. 32–41. [Google Scholar]
- McFarland, C.D.; Yaglom, J.A.; Wojtkowiak, J.W.; Scott, J.G.; Morse, D.L.; Sherman, M.Y.; Mirny, L.A. The Damaging Effect of Passenger Mutations on Cancer Progression. Cancer Res. 2017, 77, 4763–4772. [Google Scholar] [CrossRef] [Green Version]
- McFarland, C.D.; Korolev, K.S.; Kryukov, G.V.; Sunyaev, S.R.; Mirny, L.A. Impact of deleterious passenger mutations on cancer progression. Proc. Natl. Acad. Sci. USA 2013, 110, 2910–2915. [Google Scholar] [CrossRef] [Green Version]
- Pon, J.R.; Marra, M.A. Driver and passenger mutations in cancer. Annu. Rev. Pathol. 2015, 10, 25–50. [Google Scholar] [CrossRef]
- Bozic, I.; Antal, T.; Ohtsuki, H.; Carter, H.; Kim, D.; Chen, S.; Karchin, R.; Kinzler, K.W.; Vogelstein, B.; Nowak, M.A. Accumulation of driver and passenger mutations during tumor progression. Proc. Natl. Acad. Sci. USA 2010, 107, 18545–18550. [Google Scholar] [CrossRef] [Green Version]
- Knudson, A.G. Mutation and Cancer: Statistical Study of Retinoblastoma. Proc. Natl. Acad. Sci. USA 1971, 68, 820–823. [Google Scholar] [CrossRef] [Green Version]
- Moolgavkar, S.H.; Knudson, A.G. Mutation and Cancer: A Model for Human Carcinogenesis. J. Natl. Cancer Inst. 1981, 66, 1037–1052. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Kinzler, K.W. The multistep nature of cancer. Trends Genet. 1993, 9, 138–141. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Sottoriva, A.; Kang, H.; Ma, Z.; Graham, T.A.; Salomon, M.P.; Zhao, J.; Marjoram, P.; Siegmund, K.; Press, M.F.; Shibata, D.; et al. A Big Bang model of human colorectal tumor growth. Nat. Genet. 2015, 47, 209–216. [Google Scholar] [CrossRef]
- Dankort, D.; Curley, D.P.; Cartlidge, R.A.; Nelson, B.; Karnezis, A.N.; Damsky, W.E., Jr.; You, M.J.; DePinho, R.A.; McMahon, M.; Bosenberg, M. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat. Genet. 2009, 41, 544–552. [Google Scholar] [CrossRef]
- Ben-David, U.; Beroukhim, R.; Golub, T.R. Genomic evolution of cancer models: Perils and opportunities. Nat. Rev. Cancer 2019, 19, 97–109. [Google Scholar] [CrossRef]
- Durante, M.A.; Field, M.G.; Sanchez, M.I.; Covington, K.R.; Decatur, C.L.; Dubovy, S.R.; Harbour, J.W. Genomic evolution of uveal melanoma arising in ocular melanocytosis. Mol. Case Stud. 2019. [Google Scholar] [CrossRef]
- Feng, X.; Arang, N.; Rigiracciolo, D.C.; Lee, J.S.; Yeerna, H.; Wang, Z.; Lubrano, S.; Kishore, A.; Pachter, J.A.; Konig, G.M.; et al. A Platform of Synthetic Lethal Gene Interaction Networks Reveals That the GNAQ Uveal Melanoma Oncogene Controls the Hippo Pathway through FAK. Cancer Cell 2019. [Google Scholar] [CrossRef]
- Naylor, T.L.; Greshock, J.; Wang, Y.; Colligon, T.; Yu, Q.C.; Clemmer, V.; Zaks, T.Z.; Weber, B.L. High resolution genomic analysis of sporadic breast cancer using array-based comparative genomic hybridization. Breast Cancer Res. 2005, 7, R1186–R1198. [Google Scholar] [CrossRef]
- Sun, C.K.; Man, K.; Ng, K.T.; Ho, J.W.; Lim, Z.X.; Cheng, Q.; Lo, C.-M.; Poon, R.T.; Fan, S.-T. Proline-rich tyrosine kinase 2 (Pyk2) promotes proliferation and invasiveness of hepatocellular carcinoma cells through c-Src/ERK activation. Carcinogenesis 2008, 29, 2096–2105. [Google Scholar] [CrossRef] [Green Version]
- Tozzo, V.; D’Amerio, V.; Barla, A. Hey there’s DALILA: A DictionAry LearnIng LibrAry. Dagstuhl Res. Online Publ. Serv. 2017. [Google Scholar] [CrossRef]
- De Lange, M.J.; Razzaq, L.; Versluis, M.; Verlinde, S.; Dogrusoz, M.; Bohringer, S.; Marinkovic, M.; Luyten, G.P.; de Keizer, R.J.; de Gruijl, F.R.; et al. Distribution of GNAQ and GNA11 Mutation Signatures in Uveal Melanoma Points to a Light Dependent Mutation Mechanism. PLoS ONE 2015, 10, e0138002. [Google Scholar] [CrossRef]
- Balazs, E.A. Studies on the structure of the vitreous body. I. The absorption of ultraviolet light. Am. J. Ophthalmol. 1954, 38, 21–28. [Google Scholar] [CrossRef]
- Boru, G.; Cebulla, C.M.; Sample, K.M.; Massengill, J.B.; Davidorf, F.H.; Abdel-Rahman, M.H. Heterogeneity in Mitogen-Activated Protein Kinase (MAPK) Pathway Activation in Uveal Melanoma with Somatic GNAQ and GNA11 Mutations. Investig. Ophthalmol. Vis. Sci. 2019, 60, 2474–2480. [Google Scholar] [CrossRef]
- EnrichR. Available online: http://amp.pharm.mssm.edu/Enrichr/ (accessed on 10 September 2019).
- Amaro, A.; Parodi, F.; Diedrich, K.; Angelini, G.; Gotz, C.; Viaggi, S.; Maric, I.; Coviello, D.; Pistillo, M.P.; Morabito, A.; et al. Analysis of the Expression and Single-Nucleotide Variant Frequencies of the Butyrophilin-Like 2 Gene in Patients with Uveal Melanoma. JAMA Ophthalmol. 2016, 134, 1125–1133. [Google Scholar] [CrossRef]
- Tusher, V.G.; Tibshirani, R.; Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 2001, 98, 5116–5121. [Google Scholar] [CrossRef] [Green Version]
- Amaro, A.; Mirisola, V.; Angelini, G.; Musso, A.; Tosetti, F.; Esposito, A.I.; Perri, P.; Lanza, F.; Nasciuti, F.; Mosci, C.; et al. Evidence of epidermal growth factor receptor expression in uveal melanoma: Inhibition of epidermal growth factor-mediated signalling by Gefitinib and Cetuximab triggered antibody-dependent cellular cytotoxicity. Eur. J. Cancer 2013, 49, 3353–3365. [Google Scholar] [CrossRef]
- Herlihy, N.; Dogrusoz, M.; van Essen, T.H.; Harbour, J.W.; van der Velden, P.A.; van Eggermond, M.C.; Haasnoot, G.W.; van den Elsen, P.J.; Jager, M.J. Skewed expression of the genes encoding epigenetic modifiers in high-risk uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2015, 56, 1447–1458. [Google Scholar] [CrossRef]
- Laurent, C.; Valet, F.; Planque, N.; Silveri, L.; Maacha, S.; Anezo, O.; Hupe, P.; Plancher, C.; Reyes, C.; Albaud, B.; et al. High PTP4A3 phosphatase expression correlates with metastatic risk in uveal melanoma patients. Cancer Res. 2011, 71, 666–674. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef]
- Taminau, J.; Meganck, S.; Lazar, C.; Steenhoff, D.; Coletta, A.; Molter, C.; Duque, R.; de Schaetzen, V.; Weiss Solis, D.Y.; Bersini, H.; et al. Unlocking the potential of publicly available microarray data using inSilicoDb and inSilicoMerging R/Bioconductor packages. BMC Bioinform. 2012, 13, 335. [Google Scholar] [CrossRef]
- Nannya, Y.; Sanada, M.; Nakazaki, K.; Hosoya, N.; Wang, L.; Hangaishi, A.; Kurokawa, M.; Chiba, S.; Bailey, D.K.; Kennedy, G.C.; et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005, 65, 6071–6079. [Google Scholar] [CrossRef]
- Xie, X.-J.; Whitehurst, A.; White, M. A practical efficient approach in high throughput screening: Using FDR and fold change. Nat. Protoc. 2007. [Google Scholar] [CrossRef]
- Gangemi, R.; Amaro, A.; Gino, A.; Barisione, G.; Fabbi, M.; Pfeffer, U.; Brizzolara, A.; Queirolo, P.; Salvi, S.; Boccardo, S.; et al. ADAM10 correlates with uveal melanoma metastasis and promotes In Vitro invasion. Pigment Cell Melanoma Res. 2014, 27, 1138–1148. [Google Scholar] [CrossRef]
- Krüger, S.; Piro, R. Identification of mutational signatures active in individual tumors. PeerJ 2017, 5. [Google Scholar] [CrossRef]
- Qiao, H. New SVD based initialization strategy for non-negative matrix factorization. Pattern Recognit. Lett. 2015, 63, 71–77. [Google Scholar] [CrossRef] [Green Version]





| KEGG Pathway | # of Query Genes | # Total Annotated | p-Value | Adjusted p-Value | Genes |
|---|---|---|---|---|---|
| Aldosterone synthesis and secretion * | 41 | 98 | 1.38 × 10−6 | 3.79 × 10−4 | SCARB1, CAMK2D, ITPR1, ADCY4, ITPR2, ATP1A3, ADCY3, ATP1A2, CACNA1D, ITPR3, CACNA1C, CALML4, CACNA1F, CACNA1H, LIPE, CACNA1I, MC2R, PRKACG, CACNA1S, CAMK2G, PRKCG, KCNJ5, CAMK1D, HSD3B2, PRKCB, HSD3B1, ATP1B4, ATP2B2, PRKCA, AGT, NR4A2, POMC, PLCB3, AGTR1, PRKD2, CAMK1, PRKD1, PLCB1, DAGLB, PLCB2, ATF4 |
| Calcium signaling pathway * | 66 | 188 | 2.48 × 10−6 | 3.79 × 10−4 | RYR1, RYR2, CHRM1, ATP2A3, ATP2A1, CALML4, RYR3, MYLK3, SLC8A1, MYLK, HTR6, GRM5, CCKAR, EDNRB, PRKACG, BDKRB1, PLCE1, NOS1, PDGFRB, PRKCG, PDGFRA, CAMK1D, PRKCB, SPHK1, PRKCA, ITPKB, PLCB3, ADORA2A, LTB4R2, ADORA2B, AGTR1, PLCB1, SLC25A5, PLCB2, CAMK2D, PDE1C, CACNA1B, ITPR1, ADCY4, ITPR2, ADCY3, CACNA1D, ITPR3, CACNA1C, CACNA1F, CACNA1E, EGFR, CACNA1H, CACNA1I, GRIN2A, ERBB4, PTK2B, CACNA1S, PLCG1, CAMK2G, NTSR1, NOS2, NOS3, ATP2B2, PHKB, TPCN2, TPCN1, GRIN2D, CAMK1, PLCD4, MCU |
| Thyroid hormone signaling pathway | 45 | 116 | 5.08 × 10−6 | 5.17 × 10−4 | NOTCH2, NOTCH3, NOTCH1, HDAC1, ITGB3, SLC2A1, ATP1A3, ATP1A2, MED16, HIF1A, ACTB, SLC9A1, MED17, CASP9, MED12, MED14, MED13, MYC, PRKACG, EP300, PLCE1, PLCG1, SLC16A2, RXRG, PRKCG, NCOA2, CREBBP, PRKCB, ATP1B4, TSC2, SLC16A10, PRKCA, ESR1, MTOR, MED13L, BMP4, KAT2A, PLCB3, PIK3CA, CTNNB1, PLCB1, PLCD4, PLCB2, PFKP, MYH6 |
| Ras-proximate-1 (RAP1) signaling pathway * | 67 | 206 | 3.69 × 10−5 | 2.87 × 10−3 | FLT4, ITGB3, CALML4, SIPA1L3, ACTB, IGF1R, FGF9, KDR, RAC2, RAC3, PLCE1, MAP2K3, MAGI1, PDGFRB, PRKCG, PDGFRA, PRKCB, ARAP3, PRKCA, NGF, VAV2, TIAM1, PLCB3, MRAS, ADORA2A, PIK3CA, ADORA2B, PARD3, KIT, RAPGEF2, PFN4, PRKD2, TLN2, PRKD1, PLCB1, PLCB2, MET, RAPGEF6, RGS14, GNAI3, ADCY4, FPR1, ADCY3, LPAR3, RASGRP2, THBS1, EGFR, GRIN2A, KRIT1, PLCG1, FGF23, NGFR, ANGPT4, EGF, VEGFB, GRIN2B, FGF17, EFNA2, FGF19, CTNNB1, TEK, FGFR4, FGFR2, LAT, SIPA1, FGFR1, FGF10 |
| Protein digestion and absorption | 35 | 90 | 5.15 × 10−5 | 3.14 × 10−3 | COL17A1, COL18A1, PRSS1, CPB2, COL14A1, COL11A2, SLC1A1, ATP1A3, ATP1A2, SLC8A1, PRSS3, SLC36A1, CPA2, SLC6A19, SLC15A1, COL27A1, KCNJ13, COL22A1, ATP1B4, SLC16A10, COL1A1, SLC9A3, COL3A1, SLC7A7, COL2A1, COL5A1, COL4A2, COL4A1, XPNPEP2, MEP1A, COL5A3, COL5A2, COL4A3, COL9A1, COL9A2 |
| Cortisol synthesis and secretion | 26 | 65 | 2.69 × 10−4 | 9.92 × 10−3 | SCARB1, ITPR1, ADCY4, ITPR2, ADCY3, CACNA1D, ITPR3, CACNA1C, CACNA1F, CACNA1H, CYP17A1, CACNA1I, MC2R, PRKACG, CACNA1S, PDE8B, PDE8A, HSD3B2, HSD3B1, AGT, POMC, PLCB3, AGTR1, PLCB1, PLCB2, ATF4 |
| ATP-binding cassette ABC transporters | 20 | 45 | 2.59 × 10−4 | 9.92 × 10−3 | ABCA1, ABCA2, ABCC4, ABCD2, ABCC2, ABCB1, ABCB4, ABCC5, ABCA3, ABCA4, ABCC6, ABCA9, TAP2, ABCA7, ABCA12, ABCB11, ABCA13, ABCA10, ABCB10, ABCD1 |
| Circadian entrainment * | 35 | 97 | 2.93 × 10−4 | 9.92 × 10−3 | RYR1, GRIA1, RYR2, CAMK2D, GNAI3, ITPR1, ADCY4, ADCY3, CACNA1D, ITPR3, CACNA1C, CALML4, RYR3, CACNA1H, CACNA1I, GRIN2A, RASD1, PRKACG, NOS1, CAMK2G, PRKG1, PRKCG, KCNJ5, PRKCB, PRKCA, GRIN2B, GRIN2D, PER1, PLCB3, GNB2, NOS1AP, GNB1, GNB3, PLCB1, PLCB2 |
| Cushing syndrome | 51 | 155 | 2.15 × 10−4 | 9.92 × 10−3 | RB1, SCARB1, PRKACG, PDE8B, PDE8A, MEN1, USP8, AXIN1, AIP, PLCB3, AGTR1, PLCB1, PLCB2, ATF4, KMT2D, CAMK2D, KMT2A, TCF7, GNAI3, ITPR1, ADCY4, ITPR2, ADCY3, CACNA1D, ITPR3, CACNA1C, CACNA1F, EGFR, CACNA1H, CYP17A1, CACNA1I, PDE11A, MC2R, RASD1, DVL2, DVL3, CACNA1S, WNT2, CAMK2G, TCF7L2, WNT10A, TCF7L1, FZD2, HSD3B2, FZD5, WNT3A, HSD3B1, AGT, POMC, APC, CTNNB1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piaggio, F.; Tozzo, V.; Bernardi, C.; Croce, M.; Puzone, R.; Viaggi, S.; Patrone, S.; Barla, A.; Coviello, D.; J. Jager, M.; et al. Secondary Somatic Mutations in G-Protein-Related Pathways and Mutation Signatures in Uveal Melanoma. Cancers 2019, 11, 1688. https://doi.org/10.3390/cancers11111688
Piaggio F, Tozzo V, Bernardi C, Croce M, Puzone R, Viaggi S, Patrone S, Barla A, Coviello D, J. Jager M, et al. Secondary Somatic Mutations in G-Protein-Related Pathways and Mutation Signatures in Uveal Melanoma. Cancers. 2019; 11(11):1688. https://doi.org/10.3390/cancers11111688
Chicago/Turabian StylePiaggio, Francesca, Veronica Tozzo, Cinzia Bernardi, Michela Croce, Roberto Puzone, Silvia Viaggi, Serena Patrone, Annalisa Barla, Domenico Coviello, Martine J. Jager, and et al. 2019. "Secondary Somatic Mutations in G-Protein-Related Pathways and Mutation Signatures in Uveal Melanoma" Cancers 11, no. 11: 1688. https://doi.org/10.3390/cancers11111688