Purine-Metabolising Enzymes and Apoptosis in Cancer
Abstract
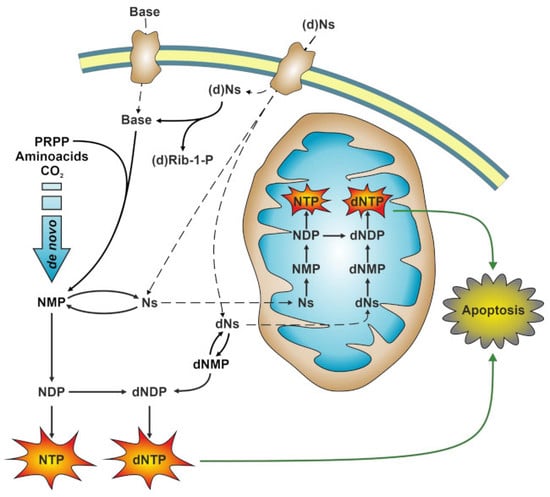
:1. Introduction
2. Ectosolic 5′-Nucleotidase
3. Cytosolic 5′-Nucleotidase II
4. Adenosine Deaminase
5. Purine Nucleoside Phosphorylase
6. Hypoxanthine Guanine Phosphoribosyltransferase
7. Inosine 5′-Monophosphate Dehydrogenase
8. Sterile Alpha Motif and HD Domain-Containing Protein 1
9. Human MutT Homolog 1
10. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ipata, P.L.; Balestri, F.; Camici, M.; Tozzi, M.G. Molecular mechanisms of nucleoside recycling in the brain. Int. J. Biochem. Cell Biol. 2011, 43, 140–145. [Google Scholar] [CrossRef] [PubMed]
- An, S.; Kumar, R.; Sheets, E.D.; Benkovic, S.J. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 2008, 320, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Pedley, A.M.; Benkovic, S.J. A new view into the regulation of purine metabolism: The purinosome. Trends Biochem. Sci. 2017, 42, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Rampazzo, C.; Miazzi, C.; Franzolin, E.; Pontarin, G.; Ferraro, P.; Frangini, M.; Reichard, P.; Bianchi, V. Regulation by degradation, a cellular defense against deoxyribonucleotide pool imbalances. Mutat. Res. 2010, 703, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B. CD73 promotes tumor growth and metastasis. Oncoimmunology 2012, 1, 67–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Z.W.; Dong, K.; Zhang, H.Z. The roles of CD73 in cancer. Biomed. Res. Int. 2014, 2014, 460654. [Google Scholar] [CrossRef] [PubMed]
- Regateiro, F.S.; Cobbold, S.P.; Waldmann, H. CD73 and adenosine generation in the creation of regulatory microenvironments. Clin. Exp. Immunol. 2013, 171, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kazemi, M.H.; Raoofi Mohseni, S.; Hojjat-Farsangi, M.; Anvari, E.; Ghalamfarsa, G.; Mohammadi, H.; Jadidi-Niaragh, F. Adenosine and adenosine receptors in the immunopathogenesis and treatment of cancer. J. Cell Physiol. 2018, 233, 2032–2057. [Google Scholar] [CrossRef] [PubMed]
- Sousa, J.B.; Fresco, P.; Diniz, C.; Goncalves, J. Adenosine receptor ligands on cancer therapy: A review of patent literature. Recent Pat. Anticancer Drug Discov. 2018, 13, 40–69. [Google Scholar] [CrossRef] [PubMed]
- Sadej, R.; Skladanowski, A.C. Dual, enzymatic and non-enzymatic, function of ecto-5’-nucleotidase (eN, CD73) in migration and invasion of A375 melanoma cells. Acta Biochim. Pol. 2012, 59, 647–652. [Google Scholar] [CrossRef]
- Stagg, J.; Divisekera, U.; McLaughlin, N.; Sharkey, J.; Pommey, S.; Denoyer, D.; Dwyer, K.M.; Smyth, M.J. Anti-CD73 antibody therapy inhibits breast tumor growth and metastasis. Proc. Natl. Acad. Sci. USA 2010, 107, 1547–1552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Pei, S.; Wang, H.; Jin, Y.; Yu, F.; Zhou, B.; Zhang, H.; Zhang, D.; Lin, D. Tiamulin inhibits breast cancer growth and pulmonary metastasis by decreasing the activity of CD73. BMC Cancer 2017, 17, 255. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wang, X.; Lu, Q.; Wang, J.; Li, L.; Liao, X.; Zhu, W.; Lv, L.; Zhi, X.; Yu, J.; et al. Extracellular 5’-nucleotidase (CD73) promotes human breast cancer cells growth through AKT/GSK-3beta/beta-catenin/cyclinD1 signaling pathway. Int. J. Cancer 2018, 142, 959–967. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.W.; Wang, H.P.; Lin, F.; Wang, X.; Long, M.; Zhang, H.Z.; Dong, K. CD73 promotes proliferation and migration of human cervical cancer cells independent of its enzyme activity. BMC Cancer 2017, 17, 135. [Google Scholar] [CrossRef] [PubMed]
- Lu, N.; Lin, T.; Wang, L.; Qi, M.; Liu, Z.; Dong, H.; Zhang, X.; Zhai, C.; Wang, Y.; Liu, L.; et al. Association of SOX4 regulated by tumor suppressor miR-30a with poor prognosis in low-grade chondrosarcoma. Tumour Biol. 2015, 36, 3843–3852. [Google Scholar] [CrossRef]
- Cappellari, A.R.; Pillat, M.M.; Souza, H.D.; Dietrich, F.; Oliveira, F.H.; Figueiro, F.; Abujamra, A.L.; Roesler, R.; Lecka, J.; Sevigny, J.; et al. Ecto-5’-Nucleotidase overexpression reduces tumor growth in a xenograph medulloblastoma model. PLoS ONE 2015, 10, e0140996. [Google Scholar] [CrossRef]
- Boyd-Tressler, A.M.; Lane, G.S.; Dubyak, G.R. Up-Regulated ectonucleotidases in fas-associated death domain protein- and receptor-interacting protein kinase 1-deficient jurkat leukemia cells counteract extracellular ATP/AMP accumulation via Pannexin-1 channels during chemotherapeutic drug-induced apoptosis. Mol. Pharm. 2017, 92, 30–47. [Google Scholar] [CrossRef]
- Antonioli, L.; Blandizzi, C.; Pacher, P.; Hasko, G. Immunity, inflammation and cancer: A leading role for adenosine. Nat. Rev. Cancer 2013, 13, 842–857. [Google Scholar] [CrossRef]
- Maj, T.; Wang, W.; Crespo, J.; Zhang, H.; Wang, W.; Wei, S.; Zhao, L.; Vatan, L.; Shao, I.; Szeliga, W.; et al. Oxidative stress controls regulatory T cell apoptosis and suppressor activity and PD-L1-blockade resistance in tumor. Nat. Immunol. 2017, 18, 1332–1341. [Google Scholar] [CrossRef]
- Dumontet, C.; Peyrottes, S.; Rabeson, C.; Cros-Perrial, E.; Geant, P.Y.; Chaloin, L.; Jordheim, L.P. CD73 inhibition by purine cytotoxic nucleoside analogue-based diphosphonates. Eur. J. Med. Chem. 2018, 157, 1051–1055. [Google Scholar] [CrossRef]
- Tozzi, M.G.; Pesi, R.; Allegrini, S. On the physiological role of cytosolic 5’-nucleotidase II (cN-II): Pathological and therapeutical implications. Curr. Med. Chem. 2013, 20, 4285–4291. [Google Scholar] [CrossRef] [PubMed]
- Galmarini, C.M.; Graham, K.; Thomas, X.; Calvo, F.; Rousselot, P.; El Jafaari, A.; Cros, E.; Mackey, J.R.; Dumontet, C. Expression of high Km 5’-nucleotidase in leukemic blasts is an independent prognostic factor in adults with acute myeloid leukemia. Blood 2001, 98, 1922–1926. [Google Scholar] [CrossRef] [PubMed]
- Cividini, F.; Pesi, R.; Chaloin, L.; Allegrini, S.; Camici, M.; Cros-Perrial, E.; Dumontet, C.; Jordheim, L.P.; Tozzi, M.G. The purine analog fludarabine acts as a cytosolic 5’-nucleotidase II inhibitor. Biochem. Pharm. 2015, 94, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Jordheim, L.P.; Chaloin, L. Therapeutic perspectives for cN-II in cancer. Curr. Med. Chem. 2013, 20, 4292–4303. [Google Scholar] [CrossRef] [PubMed]
- Jordheim, L.P.; Puy, J.Y.; Cros-Perrial, E.; Peyrottes, S.; Lefebvre, I.; Perigaud, C.; Dumontet, C. Determination of the enzymatic activity of cytosolic 5’-nucleotidase cN-II in cancer cells: Development of a simple analytical method and related cell line models. Anal. Bioanal. Chem. 2015, 407, 5747–5758. [Google Scholar] [CrossRef] [PubMed]
- Cividini, F.; Cros-Perrial, E.; Pesi, R.; Machon, C.; Allegrini, S.; Camici, M.; Dumontet, C.; Jordheim, L.P.; Tozzi, M.G. Cell proliferation and drug sensitivity of human glioblastoma cells are altered by the stable modulation of cytosolic 5’-nucleotidase II. Int. J. Biochem. Cell Biol. 2015, 65, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Pesi, R.; Petrotto, E.; Colombaioni, L.; Allegrini, S.; Garcia-Gil, M.; Camici, M.; Jordheim, L.P.; Tozzi, M.G. Cytosolic 5’-Nucleotidase II silencing in a human lung carcinoma cell line opposes cancer phenotype with a concomitant increase in p53 phosphorylation. Int. J. Mol. Sci. 2018, 19, 2115. [Google Scholar] [CrossRef] [PubMed]
- Bricard, G.; Cadassou, O.; Cassagnes, L.E.; Cros-Perrial, E.; Payen-Gay, L.; Puy, J.Y.; Lefebvre-Tournier, I.; Tozzi, M.G.; Dumontet, C.; Jordheim, L.P. The cytosolic 5’-nucleotidase cN-II lowers the adaptability to glucose deprivation in human breast cancer cells. Oncotarget 2017, 8, 67380–67393. [Google Scholar] [CrossRef]
- Careddu, M.G.; Allegrini, S.; Pesi, R.; Camici, M.; Garcia-Gil, M.; Tozzi, M.G. Knockdown of cytosolic 5’-nucleotidase II (cN-II) reveals that its activity is essential for survival in astrocytoma cells. Biochim. Biophys. Acta 2008, 1783, 1529–1535. [Google Scholar] [CrossRef]
- Kulkarni, S.S.; Karlsson, H.K.; Szekeres, F.; Chibalin, A.V.; Krook, A.; Zierath, J.R. Suppression of 5’-nucleotidase enzymes promotes AMP-activated protein kinase (AMPK) phosphorylation and metabolism in human and mouse skeletal muscle. J. Biol. Chem. 2011, 286, 34567–34574. [Google Scholar] [CrossRef]
- Kviklyte, S.; Vertommen, D.; Yerna, X.; Andersen, H.; Xu, X.; Gailly, P.; Bohlooly, Y.M.; Oscarsson, J.; Rider, M.H. Effects of genetic deletion of soluble 5’-nucleotidases NT5C1A and NT5C2 on AMPK activation and nucleotide levels in contracting mouse skeletal muscles. Am. J. Physiol. Endocrinol. Metab. 2017, 313, e48–e62. [Google Scholar] [CrossRef] [PubMed]
- Jordheim, L.P. Expanding the clinical relevance of the 5’-nucleotidase cN-II/NT5C2. Purinergic Signal. 2018, 14, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Cividini, F.; Filoni, D.N.; Pesi, R.; Allegrini, S.; Camici, M.; Tozzi, M.G. IMP-GMP specific cytosolic 5’-nucleotidase regulates nucleotide pool and prodrug metabolism. Biochim. Biophys. Acta 2015, 1850, 1354–1361. [Google Scholar] [CrossRef] [PubMed]
- Allegrini, S.; Filoni, D.N.; Galli, A.; Collavoli, A.; Pesi, R.; Camici, M.; Tozzi, M.G. Expression of bovine cytosolic 5’-Nucleotidase (cN-II) in yeast: Nucleotide pools disturbance and its consequences on growth and homologous recombination. PLoS ONE 2013, 8, e63914. [Google Scholar] [CrossRef] [PubMed]
- Gakis, C. Adenosine deaminase (ADA) isoenzymes ADA1 and ADA2: Diagnostic and biological role. Eur. Respir. J. 1996, 9, 632–633. [Google Scholar] [CrossRef] [PubMed]
- Franco, R.; Casado, V.; Ciruela, F.; Saura, C.; Mallol, J.; Canela, E.I.; Lluis, C. Cell surface adenosine deaminase: Much more than an ectoenzyme. Prog. Neurobiol. 1997, 52, 283–294. [Google Scholar] [CrossRef]
- Moreno, E.; Canet, J.; Gracia, E.; Lluis, C.; Mallol, J.; Canela, E.I.; Cortes, A.; Casado, V. Molecular evidence of adenosine deaminase linking adenosine A2A receptor and CD26 proteins. Front. Pharm. 2018, 9, 106. [Google Scholar] [CrossRef] [PubMed]
- Biri, H.; Ozturk, S.; Kacmaz, M.; Karaca, K.; Tokucoglu, H.; Durak, I. Activities of DNA turnover and free radical metabolizing enzymes in cancerous human prostate tissue. Cancer Investig. 1999, 17, 314–319. [Google Scholar] [CrossRef]
- Namiot, Z.; Stasiewicz, J.; Namiot, A.; Kemona, A.; Kralisz, M.; Gorski, J. Adenosine deaminase activity in patients with the intestinal type of gastric carcinoma. Cancer Lett. 1996, 109, 199–202. [Google Scholar] [CrossRef]
- Specchia, G.; Pavone, V.; Maggio, F.; Lojudice, L.; Iacobazzi, A.; Detullio, L.; Cagnazzo, G.; Liso, V. Adenosine-Deaminase activity in peripheral lymphocytes of patients with gynecologic malignancies. Boll. Inst. Sieroter. Milan. 1985, 64, 404–407. [Google Scholar]
- Sufrin, G.; Tritsch, G.L.; Mittelman, A.; Moore, R.H.; Murphy, G.P. Adenosine-Deaminase activity in patients with renal adenocarcinoma. Cancer 1977, 40, 796–802. [Google Scholar] [CrossRef]
- Dasmahapatra, K.S.; Hill, H.Z.; Dasmahapatra, A.; Suarez, S. Evaluation of adenosine-deaminase activity in patients with head and neck-cancer. J. Surg. Res. 1986, 40, 368–373. [Google Scholar] [CrossRef]
- Kojima, O.; Majima, T.; Uehara, Y.; Yamane, T.; Fujita, Y.; Takahashi, T.; Majima, S. Alteration of adenosine-deaminase levels in peripheral-blood lymphocytes of patients with gastric-cancer. Jpn. J. Surg. 1985, 15, 130–133. [Google Scholar] [CrossRef]
- Russo, M.; Giancane, R.; Apice, G.; Galanti, B. Adenosine-Deaminase and purine nucleoside phosphorylase activities in peripheral lymphocytes from patients with solid tumors. Br. J. Cancer 1981, 43, 196–200. [Google Scholar] [CrossRef]
- Murray, J.L.; Perezsoler, R.; Bywaters, D.; Hersh, E.M. Decreased adenosine-deaminase (Ada) and 5’nucleotidase (5nt) activity in peripheral-blood T-Cells in Hodgkin disease. Am. J. Hematol. 1986, 21, 57–66. [Google Scholar] [CrossRef]
- Camici, M.; Tozzi, M.G.; Allegrini, S.; Delcorso, A.; Sanfilippo, O.; Daidone, M.G.; Demarco, C.; Ipata, P.L. Purine salvage enzyme-activities in normal and neoplastic human tissues. Cancer Biochem. Bioph. 1990, 11, 201–209. [Google Scholar]
- Aghaei, M.; Karami-Tehrani, F.; Salami, S.; Atri, M. Adenosine deaminase activity in the serum and malignant tumors of breast cancer: The assessment of isoenzyme ADA1 and ADA2 activities. Clin. Biochem. 2005, 38, 887–891. [Google Scholar] [CrossRef]
- Mahajan, M.; Tiwari, N.; Sharma, R.; Kaur, S.; Singh, N. Oxidative stress and its relationship with adenosine deaminase activity in various stages of breast cancer. Indian J. Clin. Biochem. 2013, 28, 51–54. [Google Scholar] [CrossRef]
- Durak, I.; Beduk, Y.; Kavutcu, M.; Suzer, O.; Yaman, O.; Ozturk, H.S.; Canbolat, O.; Ulutepe, S. Activity of the enzymes participating in purine metabolism of cancerous and noncancerous human kidney tissues. Cancer Invest. 1997, 15, 212–216. [Google Scholar] [CrossRef]
- Eroglu, A.; Canbolat, O.; Demirci, S.; Kocaoglu, H.; Eryavuz, Y.; Akgul, H. Activities of adenosine deaminase and 5 ‘-nucleotidase in cancerous and noncancerous human colorectal tissues. Med. Oncol. 2000, 17, 319–324. [Google Scholar] [CrossRef]
- Pirincci, N.; Gecit, I.; Gunes, M.; Yuksel, M.B.; Kaba, M.; Tanik, S.; Demir, H.; Aslan, M. Serum adenosine deaminase, catalase and carbonic anhydrase activities in patients with bladder cancer. Clinics 2012, 67, 1443–1446. [Google Scholar] [CrossRef]
- Urunsak, I.F.; Gulec, U.K.; Paydas, S.; Seydaoglu, G.; Guzel, A.B.; Vardar, M.A. Adenosine deaminase activity in patients with ovarian neoplasms. Arch. Gynecol. Obs. 2012, 286, 155–159. [Google Scholar] [CrossRef]
- Sharma, S.D.; Desai, P.B.; Metgudmath, R.B. Evaluation of serum adenosine deaminase and retinol in patients with laryngeal cancer. Indian J. Pharm. Biol. Res. 2013, 1, 5. [Google Scholar] [CrossRef]
- Lal, H.; Munjal, S.K.; Wig, U.; Saini, A.S. Serum enzymes in head and neck cancer III. J. Laryngol. Otol. 1987, 101, 1062–1065. [Google Scholar] [CrossRef]
- Mishra, R.; Agarwal, M.K.; Chansuria, J.P. Serum adenosine deaminase levels as an index of tumor growth in head and neck malignancy. Indian J. Otolaryngol. Head Neck Surg. 2000, 52, 360–363. [Google Scholar] [CrossRef]
- Ghaderi, B.; Amini, S.; Maroofi, F.; Jalali, C.; Javanmardi, M.; Roshani, D.; Abdi, M. Adenosine deaminase activity in chronic lymphocytic leukemia and healthy subjects. Iran. J. Cancer Prev. 2016, 9, e5069. [Google Scholar] [CrossRef]
- Whitmore, K.V.; Gaspar, H.B. Adenosine deaminase deficiency—More than just an immunodeficiency. Front. Immunol. 2016, 7, 314. [Google Scholar] [CrossRef]
- Agarwal, R.P. Recovery of 2’-deoxycoformycin-inhibited adenosine deaminase of mouse erythrocytes and leukemia L1210 in vivo. Cancer Res. 1979, 39, 1425–1427. [Google Scholar]
- Dohner, H.; Ho, A.D.; Thaler, J.; Stryckmans, P.; Sonneveld, P.; de Witte, T.; Lechner, K.; Lauria, F.; Bodewadt-Radzun, S.; Suciu, S.; et al. Pentostatin in prolymphocytic leukemia: Phase II trial of the European organization for research and treatment of cancer leukemia cooperative study group. J. Natl. Cancer Inst. 1993, 85, 658–662. [Google Scholar] [CrossRef]
- Willis, C.R.; Goodrich, A.; Park, K.; Waselenko, J.K.; Lucas, M.; Reese, A.; Diehl, L.F.; Grever, M.R.; Byrd, J.C.; Flinn, I.W. A phase I/II study examining pentostatin, chlorambucil, and theophylline in patients with relapsed chronic lymphocytic leukemia and non-Hodgkin’s lymphoma. Ann. Hematol. 2006, 85, 301–307. [Google Scholar] [CrossRef]
- Kay, N.E.; LaPlant, B.R.; Pettinger, A.M.; Call, T.G.; Leis, J.F.; Ding, W.; Parikh, S.A.; Conte, M.J.; Bowen, D.A.; Shanafelt, T.D. Cumulative experience and long term follow-up of pentostatin-based chemoimmunotherapy trials for patients with chronic lymphocytic leukemia. Expert Rev. Hematol. 2018, 11, 337–349. [Google Scholar] [CrossRef]
- Tedeschi, A.; Rossi, D.; Motta, M.; Quaresmini, G.; Rossi, M.; Coscia, M.; Anastasia, A.; Rossini, F.; Cortelezzi, A.; Nador, G.; et al. A phase II multi-center trial of pentostatin plus cyclophosphamide with ofatumumab in older previously untreated chronic lymphocytic leukemia patients. Haematologica 2015, 100, e501–e504. [Google Scholar] [CrossRef] [Green Version]
- Johnston, J.B. Mechanism of action of pentostatin and cladribine in hairy cell leukemia. Leuk. Lymphoma 2011, 52, 43–45. [Google Scholar] [CrossRef]
- Hunt, S.W., 3rd; Hoffee, P.A. Adenosine deaminase from deoxycoformycin-sensitive and -resistant rat hepatoma cells. Purification and characterization. J. Biol. Chem. 1982, 257, 14239–14244. [Google Scholar]
- Camici, M.; Turriani, M.; Tozzi, M.G.; Turchi, G.; Cos, J.; Alemany, C.; Miralles, A.; Noe, V.; Ciudad, C.J. Purine enzyme profile in human colon-carcinoma cell-lines and differential sensitivity to deoxycoformycin and 2’-Deoxyadenosine in combination. Int. J. Cancer 1995, 62, 176–183. [Google Scholar] [CrossRef]
- Bemi, V.; Tazzini, N.; Banditelli, S.; Giorgelli, F.; Pesi, R.; Turchi, C.; Mattana, A.; Sgarrella, F.; Tozzi, M.G.; Camici, M. Deoxyadenosine metabolism in a human colon-carcinoma cell line (LoVo) in relation to its cytotoxic effect in combination with deoxycoformycin. Int. J. Cancer 1998, 75, 713–720. [Google Scholar] [CrossRef]
- Giannecchini, M.; D’Innocenzo, B.; Pesi, R.; Sgarrella, F.; Iorio, M.; Collecchi, P.; Tozzi, M.G.; Camici, M. 2 ‘-deoxyadenosine causes apoptotic cell death in a human colon carcinoma cell line. J. Biochem. Mol. Toxic 2003, 17, 329–337. [Google Scholar] [CrossRef]
- Garcia-Gil, M.; Tozzi, M.G.; Allegrini, S.; Folcarelli, S.; Della Sala, G.; Voccoli, V.; Colombaioni, L.; Camici, M. Novel metabolic aspects related to adenosine deaminase inhibition in a human astrocytoma cell line. Neurochem. Int. 2012, 60, 523–532. [Google Scholar] [CrossRef]
- Garcia-Gil, M.; Tozzi, M.G.; Balestri, F.; Colombaioni, L.; Camici, M. Mitochondrial damage and apoptosis induced by adenosine deaminase inhibition and deoxyadenosine in human neuroblastoma cell lines. J. Cell Biochem. 2016, 117, 1671–1679. [Google Scholar] [CrossRef]
- Garcia-Gil, M.; Tozzi, M.G.; Varani, S.; Della Verde, L.; Petrotto, E.; Balestri, F.; Colombaioni, L.; Camici, M. The combination of adenosine deaminase inhibition and deoxyadenosine induces apoptosis in a human astrocytoma cell line. Neurochem. Int. 2015, 80, 14–22. [Google Scholar] [CrossRef]
- Soleimani, A.; Bahreyni, A.; Roshan, M.K.; Soltani, A.; Ryzhikov, M.; Shafiee, M.; Soukhtanloo, M.; Jaafari, M.R.; Mashkani, B.; Hassanian, S.M. Therapeutic potency of pharmacological adenosine receptors agonist/antagonist on cancer cell apoptosis in tumor microenvironment, current status, and perspectives. J. Cell. Physiol. 2019, 234, 2329–2336. [Google Scholar] [CrossRef]
- Thirupathi, A.; Chang, Y.Z. Role of AMPK and its molecular intermediates in subjugating cancer survival mechanism. Life Sci. 2019, 227, 30–38. [Google Scholar] [CrossRef]
- Saito, M.; Yaguchi, T.; Yasuda, Y.; Nakano, T.; Nishizaki, T. Adenosine suppresses CW2 human colonic cancer growth by inducing apoptosis via A(1) adenosine receptors. Cancer Lett. 2010, 290, 211–215. [Google Scholar] [CrossRef]
- Sai, K.; Yang, D.; Yamamoto, H.; Fujikawa, H.; Yamamoto, S.; Nagata, T.; Saito, M.; Yamamura, T.; Nishizaki, T. A(1) adenosine receptor signal and AMPK involving caspase-9/-3 activation are responsible for adenosine-induced RCR-1 astrocytoma cell death. Neurotoxicology 2006, 27, 458–467. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, S.H.; Hueng, D.Y.; Syu, J.P.; Liao, C.C.; Wu, Y.C. Cordycepin induces apoptosis of C6 glioma cells through the adenosine 2A receptor-p53-caspase-7-PARP pathway. Chem. Biol. Interact. 2014, 216, 17–25. [Google Scholar] [CrossRef]
- Merighi, S.; Mirandola, P.; Milani, D.; Varani, K.; Gessi, S.; Klotz, K.N.; Leung, E.; Baraldi, P.G.; Borea, P.A. Adenosine receptors as mediators of both cell proliferation and cell death of cultured human melanoma cells. J. Invest. Derm. 2002, 119, 923–933. [Google Scholar] [CrossRef]
- Tamura, K.; Kanno, T.; Fujita, Y.; Gotoh, A.; Nakano, T.; Nishizaki, T. A(2a) adenosine receptor mediates HepG2 cell apoptosis by downregulating Bcl-X(L) expression and upregulating Bid expression. J. Cell Biochem. 2012, 113, 1766–1775. [Google Scholar] [CrossRef]
- Yasuda, Y.; Saito, M.; Yamamura, T.; Yaguchi, T.; Nishizaki, T. Extracellular adenosine induces apoptosis in Caco-2 human colonic cancer cells by activating caspase-9/-3 via A(2a) adenosine receptors. J. Gastroenterol. 2009, 44, 56–65. [Google Scholar] [CrossRef]
- Hajiahmadi, S.; Panjehpour, M.; Aghaei, M.; Shabani, M. Activation of A2b adenosine receptor regulates ovarian cancer cell growth: Involvement of Bax/Bcl-2 and caspase-3. Biochem. Cell Biol. 2015, 93, 321–329. [Google Scholar] [CrossRef]
- Jafari, S.M.; Joshaghani, H.R.; Panjehpour, M.; Aghaei, M. A2B adenosine receptor agonist induces cell cycle arrest and apoptosis in breast cancer stem cells via ERK1/2 phosphorylation. Cell Oncol. 2018, 41, 61–72. [Google Scholar] [CrossRef]
- Abedi, H.; Aghaei, M.; Panjehpour, M.; Hajiahmadi, S. Mitochondrial and caspase pathways are involved in the induction of apoptosis by IB-MECA in ovarian cancer cell lines. Tumour Biol. 2014, 35, 11027–11039. [Google Scholar] [CrossRef]
- Jafari, S.M.; Panjehpour, M.; Aghaei, M.; Joshaghani, H.R.; Enderami, S.E. A3 adenosine receptor agonist inhibited survival of breast cancer stem cells via GLI-1 and ERK1/2 pathway. J. Cell Biochem. 2017, 118, 2909–2920. [Google Scholar] [CrossRef]
- Cohen, S.; Stemmer, S.M.; Zozulya, G.; Ochaion, A.; Patoka, R.; Barer, F.; Bar-Yehuda, S.; Rath-Wolfson, L.; Jacobson, K.A.; Fishman, P. CF102 an A3 adenosine receptor agonist mediates anti-tumor and anti-inflammatory effects in the liver. J. Cell. Physiol. 2011, 226, 2438–2447. [Google Scholar] [CrossRef]
- Kanno, T.; Gotoh, A.; Fujita, Y.; Nakano, T.; Nishizaki, T. A(3) adenosine receptor mediates apoptosis in 5637 human bladder cancer cells by G(q) protein/PKC-dependent AIF upregulation. Cell. Physiol. Biochem. 2012, 30, 1159–1168. [Google Scholar] [CrossRef]
- Kanno, T.; Nakano, T.; Fujita, Y.; Gotoh, A.; Nishizaki, T. Adenosine induces apoptosis in SBC-3 human lung cancer cells through A(3) adenosine receptor-dependent AMID upregulation. Cell. Physiol. Biochem. 2012, 30, 666–677. [Google Scholar] [CrossRef]
- Nagaya, H.; Gotoh, A.; Kanno, T.; Nishizaki, T. A3 adenosine receptor mediates apoptosis in in vitro RCC4-VHL human renal cancer cells by up-regulating AMID expression. J. Urol. 2013, 189, 321–328. [Google Scholar] [CrossRef]
- Aghaei, M.; Panjehpour, M.; Karami-Tehrani, F.; Salami, S. Molecular mechanisms of A3 adenosine receptor-induced G1 cell cycle arrest and apoptosis in androgen-dependent and independent prostate cancer cell lines: Involvement of intrinsic pathway. J. Cancer Res. Clin. Oncol. 2011, 137, 1511–1523. [Google Scholar] [CrossRef]
- Otsuki, T.; Kanno, T.; Fujita, Y.; Tabata, C.; Fukuoka, K.; Nakano, T.; Gotoh, A.; Nishizaki, T. A3 adenosine receptor-mediated p53-dependent apoptosis in Lu-65 human lung cancer cells. Cell. Physiol. Biochem. 2012, 30, 210–220. [Google Scholar] [CrossRef]
- Kim, S.G.; Ravi, G.; Hoffmann, C.; Jung, Y.J.; Kim, M.; Chen, A.; Jacobson, K.A. p53-Independent induction of Fas and apoptosis in leukemic cells by an adenosine derivative, Cl-IB-MECA. Biochem. Pharm. 2002, 63, 871–880. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Tan, H.Y.; Teng, S.; Chan, Y.T.; Wang, D.; Wang, N. The role of AMP-Activated protein kinase as a potential target of treatment of hepatocellular carcinoma. Cancers 2019, 11, 647. [Google Scholar] [CrossRef]
- Tsuchiya, A.; Nishizaki, T. Anticancer effect of adenosine on gastric cancer via diverse signaling pathways. World J. Gastroenterol. 2015, 21, 10931–10935. [Google Scholar] [CrossRef]
- Saitoh, M.; Nagai, K.; Nakagawa, K.; Yamamura, T.; Yamamoto, S.; Nishizaki, T. Adenosine induces apoptosis in the human gastric cancer cells via an intrinsic pathway relevant to activation of AMP-Activated protein kinase. Biochem. Pharmacol. 2004, 67, 2005–2011. [Google Scholar] [CrossRef]
- Nogi, Y.; Kanno, T.; Nakano, T.; Fujita, Y.; Tabata, C.; Fukuoka, K.; Gotoh, A.; Nishizaki, T. AMP converted from intracellularly transported adenosine upregulates p53 expression to induce malignant pleural mesothelioma cell apoptosis. Cell. Physiol. Biochem. 2012, 30, 61–74. [Google Scholar] [CrossRef]
- Zuckerman, V.; Wolyniec, K.; Sionov, R.V.; Haupt, S.; Haupt, Y. Tumour suppression by p53: The importance of apoptosis and cellular senescence. J. Pathol. 2009, 219, 3–15. [Google Scholar] [CrossRef]
- Nakajima, Y.; Kanno, T.; Nagaya, T.; Kuribayashi, K.; Nakano, T.; Gotoh, A.; Nishizaki, T. Adenosine deaminase inhibitor EHNA exhibits a potent anticancer effect against malignant pleural mesothelioma. Cell. Physiol. Biochem. 2015, 35, 51–60. [Google Scholar] [CrossRef]
- Haynes, J.; Killilea, D.W.; Peterson, P.D.; Thompson, W.J. Erythro-9-(2-hydroxy-3-nonyl)adenine inhibits cyclic-3’,5’-guanosine monophosphate-stimulated phosphodiesterase to reverse hypoxic pulmonary vasoconstriction in the perfused rat lung. J. Pharm. Exp. 1996, 276, 752–757. [Google Scholar]
- Tsuchiya, A.; Kanno, T.; Saito, M.; Miyoshi, Y.; Gotoh, A.; Nakano, T.; Nishizaki, T. Intracellularly transported adenosine induces apoptosis in [corrected] MCF-7 human breast cancer cells by accumulating AMID in the nucleus. Cancer Lett. 2012, 321, 65–72. [Google Scholar] [CrossRef]
- Bano, D.; Prehn, J.H.M. Apoptosis-Inducing factor (AIF) in physiology and disease: The tale of a repented natural born killer. EBioMedicine 2018, 30, 29–37. [Google Scholar] [CrossRef]
- Yang, D.; Yaguchi, T.; Nagata, T.; Gotoh, A.; Dovat, S.; Song, C.; Nishizaki, T. AMID mediates adenosine-induced caspase-independent HuH-7 cell apoptosis. Cell Physiol. Biochem. 2011, 27, 37–44. [Google Scholar] [CrossRef]
- Hermes, M.; Osswald, H.; Kloor, D. Role of S-adenosylhomocysteine hydrolase in adenosine-induced apoptosis in HepG2 cells. Exp. Cell Res. 2007, 313, 264–283. [Google Scholar] [CrossRef]
- Kutryb-Zajac, B.; Koszalka, P.; Mierzejewska, P.; Bulinska, A.; Zabielska, M.A.; Brodzik, K.; Skrzypkowska, A.; Zelazek, L.; Pelikant-Malecka, I.; Slominska, E.M.; et al. Adenosine deaminase inhibition suppresses progression of 4T1 murine breast cancer by adenosine receptor-dependent mechanisms. J. Cell Mol. Med. 2018, 22, 5939–5954. [Google Scholar] [CrossRef]
- Erion, M.D.; Takabayashi, K.; Smith, H.B.; Kessi, J.; Wagner, S.; Honger, S.; Shames, S.L.; Ealick, S.E. Purine nucleoside phosphorylase. 1. Structure-function studies. Biochemistry 1997, 36, 11725–11734. [Google Scholar] [CrossRef]
- Bennett, E.M.; Li, C.; Allan, P.W.; Parker, W.B.; Ealick, S.E. Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase. J. Biol. Chem. 2003, 278, 47110–47118. [Google Scholar] [CrossRef]
- Giblett, E.R.; Ammann, A.J.; Wara, D.W.; Sandman, R.; Diamond, L.K. Nucleoside-phosphorylase deficiency in a child with severely defective T-cell immunity and normal B-cell immunity. Lancet 1975, 1, 1010–1013. [Google Scholar] [CrossRef]
- Madrid-Marina, V.; Lestan, B.; Nowak, P.J.; Fox, I.H.; Spychala, J. Altered properties of human T-lymphoblast soluble low Km 5’-nucleotidase: Comparison with B-lymphoblast enzyme. Leuk. Res. 1993, 17, 231–240. [Google Scholar] [CrossRef]
- Huang, M.; Wang, Y.; Gu, J.; Yang, J.; Noel, K.; Mitchell, B.S.; Schramm, V.L.; Graves, L.M. Determinants of sensitivity of human T-cell leukemia CCRF-CEM cells to immucillin-H. Leuk. Res. 2008, 32, 1268–1278. [Google Scholar] [CrossRef] [Green Version]
- Evans, G.B.; Furneaux, R.H.; Lewandowicz, A.; Schramm, V.L.; Tyler, P.C. Exploring structure-activity relationships of transition state analogues of human purine nucleoside phosphorylase. J. Med. Chem. 2003, 46, 3412–3423. [Google Scholar] [CrossRef]
- Arpaia, E.; Benveniste, P.; Di Cristofano, A.; Gu, Y.; Dalal, I.; Kelly, S.; Hershfield, M.; Pandolfi, P.P.; Roifman, C.M.; Cohen, A. Mitochondrial basis for immune deficiency. Evidence from purine nucleoside phosphorylase-deficient mice. J. Exp. Med. 2000, 191, 2197–2208. [Google Scholar] [CrossRef]
- Zhu, C.; Johansson, M.; Permert, J.; Karlsson, A. Enhanced cytotoxicity of nucleoside analogs by overexpression of mitochondrial deoxyguanosine kinase in cancer cell lines. J. Biol. Chem. 1998, 273, 14707–14711. [Google Scholar] [CrossRef]
- Moll, U.M.; Zaika, A. Nuclear and mitochondrial apoptotic pathways of p53. FEBS Lett. 2001, 493, 65–69. [Google Scholar] [CrossRef] [Green Version]
- Oka, S.; Ohno, M.; Tsuchimoto, D.; Sakumi, K.; Furuichi, M.; Nakabeppu, Y. Two distinct pathways of cell death triggered by oxidative damage to nuclear and mitochondrial DNAs. EMBO J. 2008, 27, 421–432. [Google Scholar] [CrossRef] [Green Version]
- Alonso, R.; Lopez-Guerra, M.; Upshaw, R.; Bantia, S.; Smal, C.; Bontemps, F.; Manz, C.; Mehrling, T.; Villamor, N.; Campo, E.; et al. Forodesine has high antitumor activity in chronic lymphocytic leukemia and activates p53-independent mitochondrial apoptosis by induction of p73 and BIM. Blood 2009, 114, 1563–1575. [Google Scholar] [CrossRef]
- Balakrishnan, K.; Nimmanapalli, R.; Ravandi, F.; Keating, M.J.; Gandhi, V. Forodesine, an inhibitor of purine nucleoside phosphorylase, induces apoptosis in chronic lymphocytic leukemia cells. Blood 2006, 108, 2392–2398. [Google Scholar] [CrossRef] [Green Version]
- Gandhi, V.; Balakrishnan, K. Pharmacology and mechanism of action of forodesine, a T-cell targeted agent. Semin. Oncol. 2007, 34, S8–S12. [Google Scholar] [CrossRef]
- Dohner, H.; Stilgenbauer, S.; Benner, A.; Leupolt, E.; Krober, A.; Bullinger, L.; Dohner, K.; Bentz, M.; Lichter, P. Genomic aberrations and survival in chronic lymphocytic leukemia. N. Engl. J. Med. 2000, 343, 1910–1916. [Google Scholar] [CrossRef]
- Korycka, A.; Blonski, J.Z.; Robak, T. Forodesine (BCX-1777, Immucillin H)—A new purine nucleoside analogue: Mechanism of action and potential clinical application. Mini Rev. Med. Chem. 2007, 7, 976–983. [Google Scholar] [CrossRef]
- Balakrishnan, K.; Ravandi, F.; Bantia, S.; Franklin, A.; Gandhi, V. Preclinical and clinical evaluation of forodesine in pediatric and adult B-cell acute lymphoblastic leukemia. Clin. Lymphoma Myeloma Leuk. 2013, 13, 458–466. [Google Scholar] [CrossRef]
- Ito, Y.; Makita, S.; Tobinai, K. Development of new agents for peripheral T-cell lymphoma. Expert Opin. Biol. 2019, 19, 197–209. [Google Scholar] [CrossRef]
- Zhang, J.; Kale, V.; Chen, M. Gene-directed enzyme prodrug therapy. AAPS J. 2015, 17, 102–110. [Google Scholar] [CrossRef]
- Afshar, S.; Asai, T.; Morrison, S.L. Humanized ADEPT comprised of an engineered human purine nucleoside phosphorylase and a tumor targeting peptide for treatment of cancer. Mol. Cancer 2009, 8, 185–193. [Google Scholar] [CrossRef]
- Parker, W.B.; King, S.A.; Allan, P.W.; Bennett, L.L., Jr.; Secrist, J.A., 3rd; Montgomery, J.A.; Gilbert, K.S.; Waud, W.R.; Wells, A.H.; Gillespie, G.Y.; et al. In vivo gene therapy of cancer with E. coli purine nucleoside phosphorylase. Hum. Gene Ther. 1997, 8, 1637–1644. [Google Scholar] [CrossRef]
- Parker, W.B.; Allan, P.W.; Shaddix, S.C.; Rose, L.M.; Speegle, H.F.; Gillespie, G.Y.; Bennett, L.L., Jr. Metabolism and metabolic actions of 6-methylpurine and 2-fluoroadenine in human cells. Biochem. Pharm. 1998, 55, 1673–1681. [Google Scholar] [CrossRef]
- Parker, W.B.; Sorscher, E.J. Use of E. coli purine nucleoside phosphorylase in the treatment of solid tumors. Curr. Pharm. Des. 2017, 23, 22. [Google Scholar] [CrossRef]
- Martiniello-Wilks, R.; Wang, X.Y.; Voeks, D.J.; Dane, A.; Shaw, J.M.; Mortensen, E.; Both, G.W.; Russell, P.J. Purine nucleoside phosphorylase and fludarabine phosphate gene-directed enzyme prodrug therapy suppresses primary tumour growth and pseudo-metastases in a mouse model of prostate cancer. J. Gene Med. 2004, 6, 1343–1357. [Google Scholar] [CrossRef]
- Arvidsson, Y.; Sumantran, V.; Watt, F.; Uramoto, H.; Funa, K. Neuroblastoma-specific cytotoxicity mediated by the Mash1-promoter and E. coli purine nucleoside phosphorylase. Pediatr. Blood Cancer 2005, 44, 77–84. [Google Scholar] [CrossRef]
- Singh, P.P.; Joshi, S.; Russell, P.J.; Nair, S.; Khatri, A. Purine nucleoside phosphorylase mediated molecular chemotherapy and conventional chemotherapy: A tangible union against chemoresistant cancer. BMC Cancer 2011, 11, 368. [Google Scholar] [CrossRef]
- Singh, P.P.; Joshi, S.; Russell, P.J.; Verma, N.D.; Wang, X.; Khatri, A. Molecular chemotherapy and chemotherapy: A new front against late-stage hormone-refractory prostate cancer. Clin. Cancer Res. 2011, 17, 4006–4018. [Google Scholar] [CrossRef]
- Krohne, T.U.; Shankara, S.; Geissler, M.; Roberts, B.L.; Wands, J.R.; Blum, H.E.; Mohr, L. Mechanisms of cell death induced by suicide genes encoding purine nucleoside phosphorylase and thymidine kinase in human hepatocellular carcinoma cells in vitro. Hepatology 2001, 34, 511–518. [Google Scholar] [CrossRef]
- Arnold, W.J.; Kelley, W.N. Human hypoxanthine-guanine phosphoribosyltransferase. Purification and subunit structure. J. Biol. Chem. 1971, 246, 7398–7404. [Google Scholar]
- Wilson, J.M.; O’Toole, T.E.; Argos, P.; Shewach, D.S.; Daddona, P.E.; Kelley, W.N. Human adenine phosphoribosyltransferase. Complete amino acid sequence of the erythrocyte enzyme. J. Biol. Chem. 1986, 261, 13677–13683. [Google Scholar]
- Sahasranaman, S.; Howard, D.; Roy, S. Clinical pharmacology and pharmacogenetics of thiopurines. Eur. J. Clin. Pharm. 2008, 64, 753–767. [Google Scholar] [CrossRef]
- Moon, W.; Loftus, E.V., Jr. Review article: Recent advances in pharmacogenetics and pharmacokinetics for safe and effective thiopurine therapy in inflammatory bowel disease. Aliment. Pharm. 2016, 43, 863–883. [Google Scholar] [CrossRef]
- Swann, P.F.; Waters, T.R.; Moulton, D.C.; Xu, Y.Z.; Zheng, Q.; Edwards, M.; Mace, R. Role of postreplicative DNA mismatch repair in the cytotoxic action of thioguanine. Science 1996, 273, 1109–1111. [Google Scholar] [CrossRef]
- Yan, T.; Berry, S.E.; Desai, A.B.; Kinsella, T.J. DNA mismatch repair (MMR) mediates 6-thioguanine genotoxicity by introducing single-strand breaks to signal a G2-M arrest in MMR-proficient RKO cells. Clin. Cancer Res. 2003, 9, 2327–2334. [Google Scholar]
- Chaabane, W.; Appell, M.L. Interconnections between apoptotic and autophagic pathways during thiopurine-induced toxicity in cancer cells: The role of reactive oxygen species. Oncotarget 2016, 7, 75616–75634. [Google Scholar] [CrossRef]
- Isakovic, A.M.; Dulovic, M.; Markovic, I.; Kravic-Stevovic, T.; Bumbasirevic, V.; Trajkovic, V.; Isakovic, A. Autophagy suppression sensitizes glioma cells to IMP dehydrogenase inhibition-induced apoptotic death. Exp. Cell Res. 2017, 350, 32–40. [Google Scholar] [CrossRef]
- Kim, H.R.; Roe, J.S.; Lee, J.E.; Hwang, I.Y.; Cho, E.J.; Youn, H.D. A p53-inducible microRNA-34a downregulates Ras signaling by targeting IMPDH. Biochem. Biophys. Res. Commun. 2012, 418, 682–688. [Google Scholar] [CrossRef]
- Natsumeda, Y.; Ohno, S.; Kawasaki, H.; Konno, Y.; Weber, G.; Suzuki, K. Two distinct cDNAs for human IMP dehydrogenase. J. Biol. Chem. 1990, 265, 5292–5295. [Google Scholar]
- Shah, C.P.; Kharkar, P.S. Newer human inosine 5’-monophosphate dehydrogenase 2 (hIMPDH2) inhibitors as potential anticancer agents. J. Enzym. Inhib. Med. Chem. 2018, 33, 972–977. [Google Scholar] [CrossRef]
- Nair, V.; Shu, Q. Inosine monophosphate dehydrogenase as a probe in antiviral drug discovery. Antivir. Chem. Chemother. 2007, 18, 245–258. [Google Scholar] [CrossRef]
- Bentley, R. Mycophenolic acid: A one hundred year odyssey from antibiotic to immunosuppressant. Chem. Rev. 2000, 100, 3801–3826. [Google Scholar] [CrossRef]
- Knight, S.R.; Morris, P.J. Does the evidence support the use of mycophenolate mofetil therapeutic drug monitoring in clinical practice? A systematic review. Transplantation 2008, 85, 1675–1685. [Google Scholar] [CrossRef]
- Shah, C.P.; Kharkar, P.S. Inosine 5’-monophosphate dehydrogenase inhibitors as antimicrobial agents: Recent progress and future perspectives. Future Med. Chem. 2015, 7, 1415–1429. [Google Scholar] [CrossRef]
- Shu, Q.; Nair, V. Inosine monophosphate dehydrogenase (IMPDH) as a target in drug discovery. Med. Res. Rev. 2008, 28, 219–232. [Google Scholar] [CrossRef]
- Pankiewicz, K.W.; Petrelli, R.; Singh, R.; Felczak, K. Nicotinamide adenine dinucleotide based therapeutics, update. Curr. Med. Chem. 2015, 22, 3991–4028. [Google Scholar] [CrossRef]
- Cuny, G.D.; Suebsuwong, C.; Ray, S.S. Inosine-5’-monophosphate dehydrogenase (IMPDH) inhibitors: A patent and scientific literature review (2002–2016). Expert Opin. Pat. 2017, 27, 677–690. [Google Scholar] [CrossRef]
- Valvezan, A.J.; Turner, M.; Belaid, A.; Lam, H.C.; Miller, S.K.; McNamara, M.C.; Baglini, C.; Housden, B.E.; Perrimon, N.; Kwiatkowski, D.J.; et al. mTORC1 couples nucleotide synthesis to nucleotide demand resulting in a targetable metabolic vulnerability. Cancer Cell 2017, 32, 624–638. [Google Scholar] [CrossRef]
- Huang, F.; Ni, M.; Chalishazar, M.D.; Huffman, K.E.; Kim, J.; Cai, L.; Shi, X.; Cai, F.; Zacharias, L.G.; Ireland, A.S.; et al. Inosine monophosphate dehydrogenase dependence in a subset of small cell lung cancers. Cell Metab. 2018, 28, 369–382. [Google Scholar] [CrossRef]
- Kofuji, S.; Hirayama, A.; Eberhardt, A.O.; Kawaguchi, R.; Sugiura, Y.; Sampetrean, O.; Ikeda, Y.; Warren, M.; Sakamoto, N.; Kitahara, S.; et al. IMP dehydrogenase-2 drives aberrant nucleolar activity and promotes tumorigenesis in glioblastoma. Nat. Cell Biol. 2019, 21, 1003–1014. [Google Scholar] [CrossRef]
- McLean, J.E.; Hamaguchi, N.; Belenky, P.; Mortimer, S.E.; Stanton, M.; Hedstrom, L. Inosine 5’-monophosphate dehydrogenase binds nucleic acids in vitro and in vivo. Biochem. J. 2004, 379, 243–251. [Google Scholar] [CrossRef]
- Mortimer, S.E.; Xu, D.; McGrew, D.; Hamaguchi, N.; Lim, H.C.; Bowne, S.J.; Daiger, S.P.; Hedstrom, L. IMP dehydrogenase type 1 associates with polyribosomes translating rhodopsin mRNA. J. Biol. Chem. 2008, 283, 36354–36360. [Google Scholar] [CrossRef]
- Park, J.H.; Ahn, S.H. IMP dehydrogenase is recruited to the transcription complex through serine 2 phosphorylation of RNA polymerase II. Biochem. Biophys. Res. Commun. 2010, 392, 588–592. [Google Scholar] [CrossRef]
- Kozhevnikova, E.N.; van der Knaap, J.A.; Pindyurin, A.V.; Ozgur, Z.; van Ijcken, W.F.; Moshkin, Y.M.; Verrijzer, C.P. Metabolic enzyme IMPDH is also a transcription factor regulated by cellular state. Mol. Cell 2012, 47, 133–139. [Google Scholar] [CrossRef]
- Tricot, G.J.; Jayaram, H.N.; Lapis, E.; Natsumeda, Y.; Nichols, C.R.; Kneebone, P.; Heerema, N.; Weber, G.; Hoffman, R. Biochemically directed therapy of leukemia with tiazofurin, a selective blocker of inosine 5’-phosphate dehydrogenase activity. Cancer Res. 1989, 49, 3696–3701. [Google Scholar]
- Laliberte, J.; Yee, A.; Xiong, Y.; Mitchell, B.S. Effects of guanine nucleotide depletion on cell cycle progression in human T lymphocytes. Blood 1998, 91, 2896–2904. [Google Scholar]
- Kiguchi, K.; Collart, F.R.; Henning-Chubb, C.; Huberman, E. Cell differentiation and altered IMP dehydrogenase expression induced in human T-lymphoblastoid leukemia cells by mycophenolic acid and tiazofurin. Exp. Cell Res. 1990, 187, 47–53. [Google Scholar] [CrossRef]
- Kiguchi, K.; Collart, F.R.; Henning-Chubb, C.; Huberman, E. Induction of cell differentiation in melanoma cells by inhibitors of IMP dehydrogenase: Altered patterns of IMP dehydrogenase expression and activity. Cell Growth Differ. 1990, 1, 259–270. [Google Scholar]
- Hunakova, L.; Bies, J.; Sedlak, J.; Duraj, J.; Jakubikova, J.; Takacsova, X.; Novotny, L.; Chorvath, B. Differential sensitivity of ovarian carcinoma cell lines to apoptosis induced by the IMPDH inhibitor benzamide riboside. Neoplasma 2000, 47, 274–279. [Google Scholar]
- Moosavi, M.A.; Yazdanparast, R.; Sanati, M.H.; Nejad, A.S. 3-Hydrogenkwadaphnin targets inosine 5’-monophosphate dehydrogenase and triggers post-G1 arrest apoptosis in human leukemia cell lines. Int. J. Biochem. Cell Biol. 2005, 37, 2366–2379. [Google Scholar] [CrossRef]
- Pan, H.; Hu, Q.; Wang, J.; Liu, Z.; Wu, D.; Lu, W.; Huang, J. Myricetin is a novel inhibitor of human inosine 5’-monophosphate dehydrogenase with anti-leukemia activity. Biochem. Biophys. Res. Commun. 2016, 477, 915–922. [Google Scholar] [CrossRef]
- Yang, H.; Fang, Z.; Wei, Y.; Bohannan, Z.S.; Ganan-Gomez, I.; Pierola, A.A.; Paradiso, L.J.; Iwamura, H.; Garcia-Manero, G. Preclinical activity of FF-10501-01, a novel inosine-5’-monophosphate dehydrogenase inhibitor, in acute myeloid leukemia. Leuk. Res. 2017, 59, 85–92. [Google Scholar] [CrossRef]
- Khanna, N.; Jayaram, H.N.; Singh, N. Benzamide riboside induced mitochondrial mediated apoptosis in human lung cancer H520 cells. Life Sci. 2004, 75, 179–190. [Google Scholar] [CrossRef]
- Takebe, N.; Cheng, X.; Fandy, T.E.; Srivastava, R.K.; Wu, S.; Shankar, S.; Bauer, K.; Shaughnessy, J.; Tricot, G. IMP dehydrogenase inhibitor mycophenolate mofetil induces caspase-dependent apoptosis and cell cycle inhibition in multiple myeloma cells. Mol. Cancer 2006, 5, 457–466. [Google Scholar] [CrossRef]
- Meli, M.; Tolomeo, M.; Grifantini, M.; Franchetti, P.; Cappellacci, L.; Simoni, D.; Invidiata, F.P.; Aiello, S.; Dusonchet, L. The synergistic apoptotic effects of thiophenfurin, an inosine monophosphate dehydrogenase inhibitor, in combination with retinoids in HL60 cells. Oncol. Rep. 2007, 17, 185–192. [Google Scholar] [CrossRef]
- Ishitsuka, K.; Hideshima, T.; Hamasaki, M.; Raje, N.; Kumar, S.; Podar, K.; Le Gouill, S.; Shiraishi, N.; Yasui, H.; Roccaro, A.M.; et al. Novel inosine monophosphate dehydrogenase inhibitor VX-944 induces apoptosis in multiple myeloma cells primarily via caspase-independent AIF/Endo G pathway. Oncogene 2005, 24, 5888–5896. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.E.; Ha, T.K.; Yoon, J.H.; Lee, J.S. Myricetin induces cell death of human colon cancer cells via BAX/BCL2-dependent pathway. Anticancer Res. 2014, 34, 701–706. [Google Scholar]
- Floryk, D.; Thompson, T.C. Antiproliferative effects of AVN944, a novel inosine 5-monophosphate dehydrogenase inhibitor, in prostate cancer cells. Int. J. Cancer 2008, 123, 2294–2302. [Google Scholar] [CrossRef] [Green Version]
- Messina, E.; Gazzaniga, P.; Micheli, V.; Guaglianone, M.R.; Barbato, S.; Morrone, S.; Frati, L.; Agliano, A.M.; Giacomello, A. Guanine nucleotide depletion triggers cell cycle arrest and apoptosis in human neuroblastoma cell lines. Int. J. Cancer 2004, 108, 812–817. [Google Scholar] [CrossRef]
- Li, M.; Su, B.S.; Chang, L.H.; Gao, Q.; Chen, K.L.; An, P.; Huang, C.; Yang, J.; Li, Z.F. Oxymatrine induces apoptosis in human cervical cancer cells through guanine nucleotide depletion. Anticancer Drugs 2014, 25, 161–173. [Google Scholar] [CrossRef]
- Gu, J.J.; Gathy, K.; Santiago, L.; Chen, E.; Huang, M.; Graves, L.M.; Mitchell, B.S. Induction of apoptosis in IL-3-dependent hematopoietic cell lines by guanine nucleotide depletion. Blood 2003, 101, 4958–4965. [Google Scholar] [CrossRef]
- Goldstone, D.C.; Ennis-Adeniran, V.; Hedden, J.J.; Groom, H.C.; Rice, G.I.; Christodoulou, E.; Walker, P.A.; Kelly, G.; Haire, L.F.; Yap, M.W.; et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 2011, 480, 379–382. [Google Scholar] [CrossRef]
- Powell, R.D.; Holland, P.J.; Hollis, T.; Perrino, F.W. Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 2011, 286, 43596–43600. [Google Scholar] [CrossRef]
- Baldauf, H.M.; Pan, X.; Erikson, E.; Schmidt, S.; Daddacha, W.; Burggraf, M.; Schenkova, K.; Ambiel, I.; Wabnitz, G.; Gramberg, T.; et al. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat. Med. 2012, 18, 1682–1687. [Google Scholar] [CrossRef]
- Rice, G.I.; Bond, J.; Asipu, A.; Brunette, R.L.; Manfield, I.W.; Carr, I.M.; Fuller, J.C.; Jackson, R.M.; Lamb, T.; Briggs, T.A.; et al. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 2009, 41, 829–832. [Google Scholar] [CrossRef]
- Leshinsky-Silver, E.; Malinger, G.; Ben-Sira, L.; Kidron, D.; Cohen, S.; Inbar, S.; Bezaleli, T.; Levine, A.; Vinkler, C.; Lev, D.; et al. A large homozygous deletion in the SAMHD1 gene causes atypical Aicardi-Goutieres syndrome associated with mtDNA deletions. Eur. J. Hum. Genet. 2011, 19, 287–292. [Google Scholar] [CrossRef]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.M.; Gallia, G.L.; et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Lee, W.; Jiang, Z.; Chen, Z.; Jhunjhunwala, S.; Haverty, P.M.; Gnad, F.; Guan, Y.; Gilbert, H.N.; Stinson, J.; et al. Genome and transcriptome sequencing of lung cancers reveal diverse mutational and splicing events. Genome Res. 2012, 22, 2315–2327. [Google Scholar] [CrossRef] [Green Version]
- Jones, S.; Zhang, X.; Parsons, D.W.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Kamiyama, H.; Jimeno, A.; et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008, 321, 1801–1806. [Google Scholar] [CrossRef]
- Clifford, R.; Louis, T.; Robbe, P.; Ackroyd, S.; Burns, A.; Timbs, A.T.; Wright Colopy, G.; Dreau, H.; Sigaux, F.; Judde, J.G.; et al. SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage. Blood 2014, 123, 1021–1031. [Google Scholar] [CrossRef]
- Walker, B.A.; Wardell, C.P.; Melchor, L.; Hulkki, S.; Potter, N.E.; Johnson, D.C.; Fenwick, K.; Kozarewa, I.; Gonzalez, D.; Lord, C.J.; et al. Intraclonal heterogeneity and distinct molecular mechanisms characterize the development of t(4; 14) and t(11; 14) myeloma. Blood 2012, 120, 1077–1086. [Google Scholar] [CrossRef]
- Sjoblom, T.; Jones, S.; Wood, L.D.; Parsons, D.W.; Lin, J.; Barber, T.D.; Mandelker, D.; Leary, R.J.; Ptak, J.; Silliman, N.; et al. The consensus coding sequences of human breast and colorectal cancers. Science 2006, 314, 268–274. [Google Scholar] [CrossRef]
- Wang, J.L.; Lu, F.Z.; Shen, X.Y.; Wu, Y.; Zhao, L.T. SAMHD1 is down regulated in lung cancer by methylation and inhibits tumor cell proliferation. Biochem. Biophys. Res. Commun. 2014, 455, 229–233. [Google Scholar] [CrossRef]
- Zhu, C.; Gao, W.; Zhao, K.; Qin, X.; Zhang, Y.; Peng, X.; Zhang, L.; Dong, Y.; Zhang, W.; Li, P.; et al. Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase. Nat. Commun. 2013, 4, 2722. [Google Scholar] [CrossRef] [Green Version]
- Miazzi, C.; Ferraro, P.; Pontarin, G.; Rampazzo, C.; Reichard, P.; Bianchi, V. Allosteric regulation of the human and mouse deoxyribonucleotide triphosphohydrolase sterile alpha-motif/histidine-aspartate domain-containing protein 1 (SAMHD1). J. Biol. Chem. 2014, 289, 18339–18346. [Google Scholar] [CrossRef]
- Amie, S.M.; Bambara, R.A.; Kim, B. GTP is the primary activator of the anti-HIV restriction factor SAMHD1. J. Biol. Chem. 2013, 288, 25001–25006. [Google Scholar] [CrossRef]
- Beloglazova, N.; Flick, R.; Tchigvintsev, A.; Brown, G.; Popovic, A.; Nocek, B.; Yakunin, A.F. Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J. Biol. Chem. 2013, 288, 8101–8110. [Google Scholar] [CrossRef]
- Seamon, K.J.; Sun, Z.; Shlyakhtenko, L.S.; Lyubchenko, Y.L.; Stivers, J.T. SAMHD1 is a single-stranded nucleic acid binding protein with no active site-associated nuclease activity. Nucleic Acids Res. 2015, 43, 6486–6499. [Google Scholar] [CrossRef]
- Kohnken, R.; Kodigepalli, K.M.; Wu, L. Regulation of deoxynucleotide metabolism in cancer: Novel mechanisms and therapeutic implications. Mol. Cancer 2015, 14, 176. [Google Scholar] [CrossRef]
- Kodigepalli, K.M.; Li, M.; Liu, S.L.; Wu, L. Exogenous expression of SAMHD1 inhibits proliferation and induces apoptosis in cutaneous T-cell lymphoma-derived HuT78 cells. Cell Cycle 2017, 16, 179–188. [Google Scholar] [CrossRef]
- Kodigepalli, K.M.; Bonifati, S.; Tirumuru, N.; Wu, L. SAMHD1 modulates in vitro proliferation of acute myeloid leukemia-derived THP-1 cells through the PI3K-Akt-p27 axis. Cell Cycle 2018, 17, 1124–1137. [Google Scholar] [CrossRef]
- Fujikawa, K.; Kamiya, H.; Yakushiji, H.; Nakabeppu, Y.; Kasai, H. Human MTH1 protein hydrolyzes the oxidized ribonucleotide, 2-hydroxy-ATP. Nucleic Acids Res. 2001, 29, 449–454. [Google Scholar] [CrossRef] [Green Version]
- Oda, H.; Nakabeppu, Y.; Furuichi, M.; Sekiguchi, M. Regulation of expression of the human MTH1 gene encoding 8-oxo-dGTPase. Alternative splicing of transcription products. J. Biol. Chem. 1997, 272, 17843–17850. [Google Scholar] [CrossRef]
- Rai, P.; Sobol, R.W. Mechanisms of MTH1 inhibition-induced DNA strand breaks: The slippery slope from the oxidized nucleotide pool to genotoxic damage. DNA Repair 2019, 77, 18–26. [Google Scholar] [CrossRef]
- Trachootham, D.; Zhou, Y.; Zhang, H.; Demizu, Y.; Chen, Z.; Pelicano, H.; Chiao, P.J.; Achanta, G.; Arlinghaus, R.B.; Liu, J.; et al. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer Cell 2006, 10, 241–252. [Google Scholar] [CrossRef]
- Storz, P. Reactive oxygen species in tumor progression. Front. Biosci. J. Virtual Libr. 2005, 10, 1881–1896. [Google Scholar] [CrossRef]
- Ishikawa, K.; Takenaga, K.; Akimoto, M.; Koshikawa, N.; Yamaguchi, A.; Imanishi, H.; Nakada, K.; Honma, Y.; Hayashi, J. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science 2008, 320, 661–664. [Google Scholar] [CrossRef]
- Ames, B.N. Dietary carcinogens and anticarcinogens. Oxygen radicals and degenerative diseases. Science 1983, 221, 1256–1264. [Google Scholar] [CrossRef]
- Rai, P.; Onder, T.T.; Young, J.J.; McFaline, J.L.; Pang, B.; Dedon, P.C.; Weinberg, R.A. Continuous elimination of oxidized nucleotides is necessary to prevent rapid onset of cellular senescence. Proc. Natl. Acad. Sci. USA 2009, 106, 169–174. [Google Scholar] [CrossRef] [Green Version]
- Gad, H.; Koolmeister, T.; Jemth, A.S.; Eshtad, S.; Jacques, S.A.; Strom, C.E.; Svensson, L.M.; Schultz, N.; Lundback, T.; Einarsdottir, B.O.; et al. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature 2014, 508, 215–221. [Google Scholar] [CrossRef]
- Youn, C.K.; Jun, J.Y.; Hyun, J.W.; Hwang, G.; Lee, B.R.; Chung, M.H.; Chang, I.Y.; You, H.J. hMTH1 depletion promotes oxidative-stress-induced apoptosis through a Noxa- and caspase-3/7-mediated signaling pathway. DNA Repair 2008, 7, 1809–1823. [Google Scholar] [CrossRef]
- Russo, M.T.; Blasi, M.F.; Chiera, F.; Fortini, P.; Degan, P.; Macpherson, P.; Furuichi, M.; Nakabeppu, Y.; Karran, P.; Aquilina, G.; et al. The oxidized deoxynucleoside triphosphate pool is a significant contributor to genetic instability in mismatch repair-deficient cells. Mol. Cell Biol. 2004, 24, 465–474. [Google Scholar] [CrossRef]
- Huber, K.V.; Salah, E.; Radic, B.; Gridling, M.; Elkins, J.M.; Stukalov, A.; Jemth, A.S.; Gokturk, C.; Sanjiv, K.; Stromberg, K.; et al. Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature 2014, 508, 222–227. [Google Scholar] [CrossRef] [Green Version]
- Nakabeppu, Y.; Oka, S.; Sheng, Z.; Tsuchimoto, D.; Sakumi, K. Programmed cell death triggered by nucleotide pool damage and its prevention by MutT homolog-1 (MTH1) with oxidized purine nucleoside triphosphatase. Mutat. Res. 2010, 703, 51–58. [Google Scholar] [CrossRef]
- Rampazzo, C.; Tozzi, M.G.; Dumontet, C.; Jordheim, L.P. The druggability of intracellular nucleotide-degrading enzymes. Cancer Chemother. Pharm. 2016, 77, 883–893. [Google Scholar] [CrossRef]
- Samaranayake, G.J.; Huynh, M.; Rai, P. MTH1 as a chemotherapeutic target: The elephant in the room. Cancers 2017, 9, 47. [Google Scholar] [CrossRef]
- Svilar, D.; Goellner, E.M.; Almeida, K.H.; Sobol, R.W. Base excision repair and lesion-dependent subpathways for repair of oxidative DNA damage. Antioxid. Redox Signal. 2011, 14, 2491–2507. [Google Scholar] [CrossRef]
- Zhou, J.; Ahn, J.; Wilson, S.H.; Prives, C. A role for p53 in base excision repair. EMBO J. 2001, 20, 914–923. [Google Scholar] [CrossRef] [Green Version]
- Achanta, G.; Huang, P. Role of p53 in sensing oxidative DNA damage in response to reactive oxygen species-generating agents. Cancer Res. 2004, 64, 6233–6239. [Google Scholar] [CrossRef]
- Chatterjee, A.; Mambo, E.; Osada, M.; Upadhyay, S.; Sidransky, D. The effect of p53-RNAi and p53 knockout on human 8-oxoguanine DNA glycosylase (hOgg1) activity. FASEB J. 2006, 20, 112–114. [Google Scholar] [CrossRef]
- Wang, R.; Li, C.; Qiao, P.; Xue, Y.; Zheng, X.; Chen, H.; Zeng, X.; Liu, W.; Boldogh, I.; Ba, X. OGG1-initiated base excision repair exacerbates oxidative stress-induced parthanatos. Cell Death Dis. 2018, 9, 628. [Google Scholar] [CrossRef]
- Patel, A.; Burton, D.G.; Halvorsen, K.; Balkan, W.; Reiner, T.; Perez-Stable, C.; Cohen, A.; Munoz, A.; Giribaldi, M.G.; Singh, S.; et al. MutT Homolog 1 (MTH1) maintains multiple KRAS-driven pro-malignant pathways. Oncogene 2015, 34, 2586–2596. [Google Scholar] [CrossRef]
- Abbas, H.H.K.; Alhamoudi, K.M.H.; Evans, M.D.; Jones, G.D.D.; Foster, S.S. MTH1 deficiency selectively increases non-cytotoxic oxidative DNA damage in lung cancer cells: More bad news than good? BMC Cancer 2018, 18, 423. [Google Scholar] [CrossRef]
- Fouquerel, E.; Lormand, J.; Bose, A.; Lee, H.T.; Kim, G.S.; Li, J.; Sobol, R.W.; Freudenthal, B.D.; Myong, S.; Opresko, P.L. Oxidative guanine base damage regulates human telomerase activity. Nat. Struct. Mol. Biol. 2016, 23, 1092–1100. [Google Scholar] [CrossRef] [Green Version]
- Mathews, C.K. DNA precursor metabolism and genomic stability. FASEB J. 2006, 20, 1300–1314. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, I.; Yoshida, Y.; Suda, M.; Minamino, T. DNA damage response and metabolic disease. Cell Metab. 2014, 20, 967–977. [Google Scholar] [CrossRef]
- Kohalmi, S.E.; Glattke, M.; McIntosh, E.M.; Kunz, B.A. Mutational specificity of DNA precursor pool imbalances in yeast arising from deoxycytidylate deaminase deficiency or treatment with thymidylate. J. Mol. Biol. 1991, 220, 933–946. [Google Scholar] [CrossRef]
- Pai, C.C.; Kearsey, S.E. A critical balance: dNTPs and the maintenance of genome stability. Genes 2017, 8, 57. [Google Scholar] [CrossRef]
- Wyant, G.A.; Abu-Remaileh, M.; Frenkel, E.M.; Laqtom, N.N.; Dharamdasani, V.; Lewis, C.A.; Chan, S.H.; Heinze, I.; Ori, A.; Sabatini, D.M. NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science 2018, 360, 751–758. [Google Scholar] [CrossRef] [Green Version]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Camici, M.; Garcia-Gil, M.; Pesi, R.; Allegrini, S.; Tozzi, M.G. Purine-Metabolising Enzymes and Apoptosis in Cancer. Cancers 2019, 11, 1354. https://doi.org/10.3390/cancers11091354
Camici M, Garcia-Gil M, Pesi R, Allegrini S, Tozzi MG. Purine-Metabolising Enzymes and Apoptosis in Cancer. Cancers. 2019; 11(9):1354. https://doi.org/10.3390/cancers11091354
Chicago/Turabian StyleCamici, Marcella, Mercedes Garcia-Gil, Rossana Pesi, Simone Allegrini, and Maria Grazia Tozzi. 2019. "Purine-Metabolising Enzymes and Apoptosis in Cancer" Cancers 11, no. 9: 1354. https://doi.org/10.3390/cancers11091354