- 1Department of Fundamental Microbiology, University of Lausanne, Lausanne, Switzerland
- 2Plant Pathology, Institute of Integrative Biology, Swiss Federal Institute of Technology (ETH) Zurich, Zurich, Switzerland
- 3Plant Breeding and Genetic Resources, Institute for Plant Production Sciences, Agroscope, Nyon, Switzerland
- 4Department of Biological Sciences, University of Southern Mississippi, Hattiesburg, MS, USA
Strains of Pseudomonas that produce antimicrobial metabolites and control soilborne plant diseases have often been isolated from soils defined as disease-suppressive, i.e., soils, in which specific plant pathogens are present, but plants show no or reduced disease symptoms. Moreover, it is assumed that pseudomonads producing antimicrobial compounds such as 2,4-diacetylphloroglucinol (DAPG) or phenazines (PHZ) contribute to the specific disease resistance of suppressive soils. However, pseudomonads producing antimicrobial metabolites are also present in soils that are conducive to disease. Currently, it is still unknown whether and to which extent the abundance of antimicrobials-producing pseudomonads is related to the general disease resistance of common agricultural soils. Moreover, virtually nothing is known about the conditions under which pseudomonads express antimicrobial genes in agricultural field soils. We present here results of the first side-by-side comparison of 10 representative Swiss agricultural soils with a cereal-oriented cropping history for (i) the resistance against two soilborne pathogens, (ii) the abundance of Pseudomonas bacteria harboring genes involved in the biosynthesis of the antimicrobials DAPG, PHZ, and pyrrolnitrin on roots of wheat, and (iii) the ability to support the expression of these genes on the roots. Our study revealed that the level of soil disease resistance strongly depends on the type of pathogen, e.g., soils that are highly resistant to Gaeumannomyces tritici often are highly susceptible to Pythium ultimum and vice versa. There was no significant correlation between the disease resistance of the soils, the abundance of Pseudomonas bacteria carrying DAPG, PHZ, and pyrrolnitrin biosynthetic genes, and the ability of the soils to support the expression of the antimicrobial genes. Correlation analyses indicated that certain soil factors such as silt, clay, and some macro- and micronutrients influence both the abundance and the expression of the antimicrobial genes. Taken together, the results of this study suggests that pseudomonads producing DAPG, PHZ, or pyrrolnitrin are present and abundant in Swiss agricultural soils and that the soils support the expression of the respective biosynthetic genes in these bacteria to various degrees. The precise role that these pseudomonads play in the general disease resistance of the investigated agricultural soils remains elusive.
Introduction
The ability of soilborne plant pathogens to attack and damage host plants is influenced by biotic and abiotic soil factors (Weller et al., 2002; Haas and Défago, 2005; Lemanceau et al., 2006; Almario et al., 2014). In some soils, even susceptible crop plants suffer only a little or not at all from specific diseases although soilborne pathogens are present (Weller et al., 2002). In general, two different types of natural pathogen suppression are thought to occur in agricultural soils. First, the general disease suppression, where different soilborne pathogens are controlled to a certain degree depending on the total microbial activity in the soil and/or on abiotic soil factors (Weller et al., 2002; Lemanceau et al., 2006). Second, the specific disease suppression, where the soil restricts the activity of a distinct species of plant pathogen based on its interactions with a specific group of microorganisms (Weller et al., 2002; Haas and Défago, 2005; Lemanceau et al., 2006; Berendsen et al., 2012; Raaijmakers and Mazzola, 2016).
Soils with specific disease suppression have been described worldwide (Cook and Rovira, 1976; Stutz et al., 1986; Weller et al., 2002; Lemanceau et al., 2006; Chng et al., 2015) and for diverse soilborne plant pathogens. They include soils suppressive to Gaeumannomyces graminis var. tritici (recently renamed G. tritici (Hernández-Restrepo et al., 2016)) causing take-all of wheat (Weller et al., 2002), Thielaviopsis basicola causing black root rot of tobacco (Stutz et al., 1986; Almario et al., 2014), Fusarium oxysporum causing wilt on tomatoes (Alabouvette, 1986; Tamietti et al., 1993), Pythium spp. causing seedling damping-off (Martin and Hancock, 1986), and Rhizoctonia solani causing damping-off and root rot on various crop species (Mendes et al., 2011). Such soils are commonly referred-to as suppressive soils. By contrast, conducive soils do not restrict the development of soilborne diseases (Haas and Défago, 2005).
Suppressive soils have been found to host distinct microbial communities that are thought to be responsible for the natural disease control effect (Weller et al., 2002; Haas and Défago, 2005; Mendes et al., 2011, 2013; Klein et al., 2013; Kyselkova et al., 2014; Cha et al., 2016). In particular, bacteria of the Pseudomonas fluorescens group have been isolated from suppressive soils and used as plant or soil inoculants. Several strains proved to be very efficient at colonizing roots, protecting plants from different diseases, and increasing plant productivity (Mercado-Blanco and Bakker, 2007; Lugtenberg and Kamilova, 2009; Höfte and Altier, 2010). Thus, it has been suggested that such pseudomonads contribute to soil suppressiveness (Weller et al., 2002; Haas and Défago, 2005; Garbeva et al., 2006; Lemanceau et al., 2006; Weller, 2007; Mazurier et al., 2009). The capacity of many root-associated pseudomonads to release antimicrobial compounds placed them in the focus of research on the nature of soil disease suppressiveness. Many P. fluorescens group strains produce an array of potent antimicrobials, among which 2,4-diacetylphloroglucinol (DAPG), phenazines (PHZ), pyrrolnitrin (PRN), and hydrogen cyanide (HCN) are most prominent (Blumer and Haas, 2000; Haas and Défago, 2005; Raaijmakers and Mazzola, 2012; Mavrodi et al., 2013). All these antimicrobials were shown, mostly in pot and gnotobiotic assays, to play indeed an important role in the Pseudomonas-mediated protection of plants from soilborne pathogenic fungi and oomycetes (Thomashow and Weller, 1988; Voisard et al., 1989; Keel et al., 1992; Maurhofer et al., 1992; Pierson and Thomashow, 1992; Hwang et al., 2002; Chin-A-Woeng et al., 2003; Weller et al., 2007; Mavrodi et al., 2013).
The role of these antimicrobial compounds in disease-suppressive soils is still not fully understood. They are indeed produced in some field soils, as demonstrated for DAPG in the Quincy take-all decline soil (Raaijmakers et al., 1999) and for PHZ in wheat fields of the Columbia Plateau, USA (Mavrodi D.V. et al., 2012). Several studies performed during the last 15 years aimed at investigating whether disease-suppressive soils are specifically enriched for Pseudomonas genotypes producing antimicrobials compared to conducive soils. In fact, in the Pacific Northwest of the USA, DAPG-producing pseudomonads were found to be more abundant in take-all suppressive soils than in adjacent conducive soils (Raaijmakers et al., 1997). However, DAPG producers were not more abundant in Fusarium suppressive soils of Châteaurenard (France) than in adjacent conducive soils, in contrast to PHZ-producing pseudomonads, which were more abundant in the suppressive soils (Mazurier et al., 2009). In some studies, the total abundance of DAPG-producing pseudomonads was found to be similar in suppressive and conducive soils (Ramette et al., 2006; Almario et al., 2013a), but suppressive soils harbored distinct genotypes of DAPG producers (Frapolli et al., 2010). Moreover, abundances of plant-beneficial pseudomonads with antimicrobial activity were mostly investigated in specific disease-suppressive soils, and very little is known about the occurrence of these bacteria in common agricultural soils and on how soil factors might impact these bacteria. In wheat fields of the Pacific Northwest of the USA, it was found that the abundance of DAPG- and PHZ-producing pseudomonads on wheat roots is influenced by irrigation (Mavrodi O.V. et al., 2012).
Based on such observations, the abundance and genotypic diversity of antimicrobials-producing Pseudomonas bacteria in soil seems not a sufficient argument to explain the disease suppressiveness of some soils. It has been suggested that (i) other bacterial species contribute importantly to the disease suppressiveness (Mendes et al., 2011; Kyselkova et al., 2014), and (ii) somehow the expression of antimicrobial genes in Pseudomonas bacteria is favored in suppressive soils and hampered in conducive soils (Ramette et al., 2006; Almario et al., 2014). Indeed, studies on the abundance of antimicrobial metabolite-producing pseudomonads do not consider the complex interactions in the rhizosphere that ultimately modulate the production of the antimicrobials in the rhizosphere (Rochat et al., 2010; de Werra et al., 2011). To date, little is known about the biotic and abiotic factors affecting the expression of biosynthetic genes for these metabolites in soil. Studies conducted under gnotobiotic conditions indicate that the expression of DAPG, HCN, and PRN biosynthesis genes is influenced by the crop species and variety (Rochat et al., 2010; de Werra et al., 2011; Latz et al., 2015) and for DAPG also by the interaction with other microorganisms and the iron availability in the rhizosphere (Notz et al., 2001, 2002; Maurhofer et al., 2004; Jousset et al., 2010, 2011; Almario et al., 2013b). How biotic and abiotic soil factors affect antimicrobial gene expression under natural conditions in agricultural soils remains, however, unexplored.
There is a clear lack of studies investigating the link between natural disease resistance and abundance and expression of antimicrobial Pseudomonas genes in common agricultural soils. To address this gap, in the present study 10 representative Swiss agricultural soils with a cereal-oriented cropping history and differing in their physical and chemical characteristics were compared for their resistance to two soilborne pathogens of wheat, i.e., G. tritici (Gt) and Pythium ultimum (Pu). In parallel, the 10 soils were planted with wheat and pseudomonads harboring the biosynthetic genes required for the production of the antimicrobial compounds DAPG, PHZ, and PRN were quantified on roots using qPCR. In addition, the expression of these genes and the HCN biosynthetic genes was monitored by flow cytometry using fluorescent reporter strains of the representative model pseudomonads P. protegens CHA0 and P. chlororaphis PCL1391. To our best knowledge, this is the first side-by-side comparison using root-associated pseudomonads as bio-indicators to explore relationships between abundance and expression of antimicrobial genes, soil disease resistance and soil physicochemical characteristics in a range of common agricultural soils.
Materials and Methods
Sampling and Physicochemical Analysis of Field Soils
Soil samples were collected in 10 farmer’s fields across Switzerland (Figure 1) in May 2013. The main characteristics of the 10 field soils are listed in Table 1. Field sites had a history of multi-year cereal-oriented crop rotation and were chosen to represent predominant Swiss agricultural soil types and climate conditions. All fields were cropped with winter wheat in the year of sampling. For sampling, soil cores of 15–20 cm depth were extracted with disinfected soil recovery augers between the rows of wheat plants at 20 random locations in each field in order to obtain a representative sample. Then, extracted soil samples were sieved (mesh size, 10 by 10 mm) in order to remove stones, plant residues or other larger material, pooled and thoroughly mixed. For each site, approximately 120 kg of sieved soil were collected and stored in barrels for 3 months at 15°C before the start of the experiments in order to equilibrate the soils, i.e., to minimize effects of different environmental conditions (e.g., temperature, soil moisture) prevailing at the different sampling sites at the time of sampling. The storage temperature was chosen because it can be considered as the average temperature in Switzerland during the growing season of wheat from April to September (according to long-term monthly temperature averages recorded by the Swiss Federal Office MeteoSwiss1). Soil parameter analyses were carried out by the Labor für Boden- und Umweltanalytik (Eric Schweizer AG, Steffisburg, Switzerland) following standard protocols used in Swiss agriculture (Agroscope, 2006). Concentrations of soluble, readily plant-available macronutrients were determined following H2O extraction, for which soil samples were suspended in distilled water at a ratio of 1:10 (g mL-1). Reserve macronutrients and micronutrients were extracted with ammonium acetate EDTA, for which soil samples dried at 65°C were suspended at a ratio of 1:10 (g mL-1) in a solution consisting of acetic acid (0.5 mol L-1), ammonium acetate and EDTA (0.02 mol L-1) adjusted to a pH of 4.65. Soil suspensions were then vigorously shaken for 1 h and filtered prior to analysis by mass spectrometry. The soil parameter analyses were carried out (i) directly after sampling and (ii) after the end of the experiments. Except for nitrate, the soil parameters did not change significantly between the two analyses (Table 1).
FIGURE 1. Location of the 10 field sites in Switzerland used for soil sampling. Cd, Cadenazzo; Cx, Courtedoux; Cz, Cazis; De, Delley; Es, Eschikon; Gr, Grangeneuve; Ta, Taenikon; Ut, Utzenstorf; Vo, Vouvry; Wi, Witzwil. Empty topographical map of Switzerland courtesy of Federal Office of Topography swisstopo.
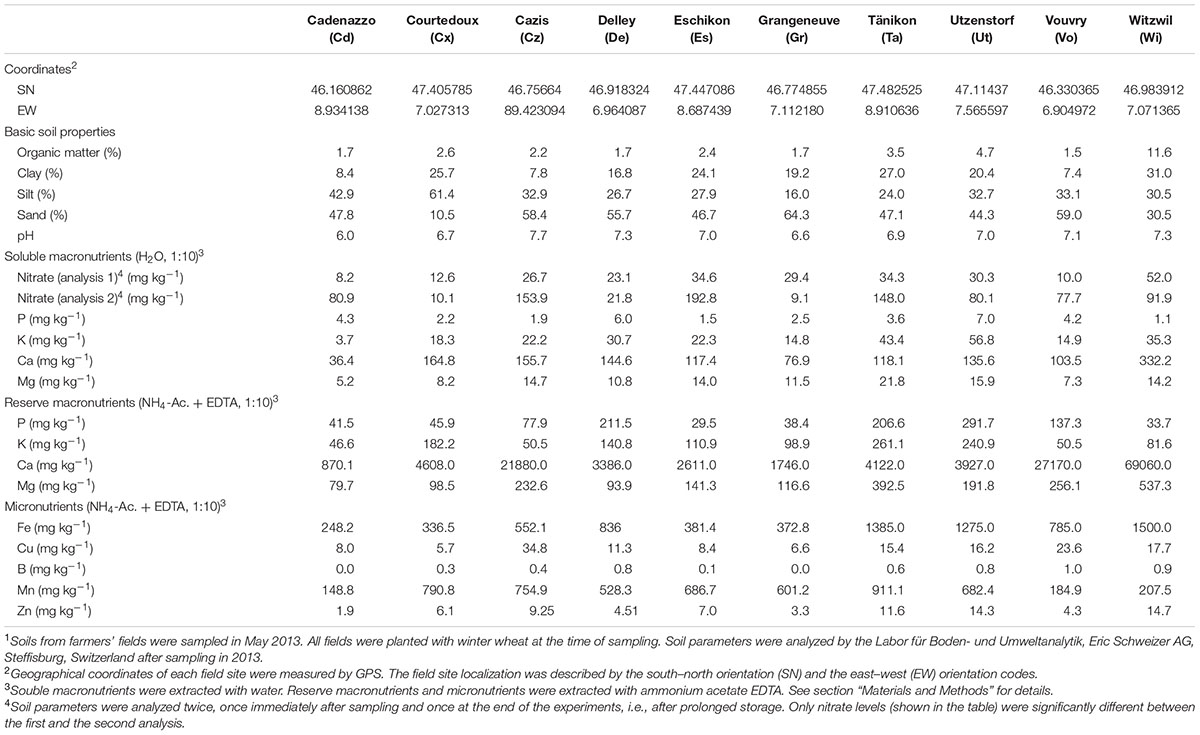
TABLE 1. Characteristics of agricultural soils sampled at different Swiss farmers’ fields cropped with wheat1.
Assessment of Field Soil Resistance against Root Pathogens
In order to test the natural resistance of the 10 Swiss agricultural soils toward soilborne pathogens, pot experiments were carried out in the greenhouse with cucumber and Pu and wheat and Gt. The Pu inoculum was prepared by inoculating 25 g of autoclaved millet seeds moistened with 10 mL of sterile water with three plugs of a culture of Pu strain ETH-2 (isolated from a Swiss agricultural soil) grown on Oxoid malt agar (Thermo Fisher Scientific, Reinach, Switzerland) for 7 days. Pu millet seed cultures were grown for 7 days at 18°C and then cut into small fragments for soil inoculation. The Gt inoculum was prepared by inoculating 100 g of autoclaved oat seeds without spelts and moistened with 100 mL of sterile, distilled water with 20 plugs of cultures of Gt strain I-17 (Lebreton et al., 2007) grown on Oxoid potato dextrose agar for 14 days. Gt oat cultures were grown for 4 weeks at 24°C in the dark and then dried in a sterile cabinet on sterile filter paper for 3 days. Seeds of cucumber (Cucumis sativus cv. Chinese Snake) and spring wheat (Triticum aestivum cv. Rubli) were surface-sterilized for 30 min in 1.5% (v/v) NaOCl, rinsed with sterile distilled water and pre-germinated on sterile moist filter paper for 2 days in the dark at 24°C. For the plant experiments, 250-mL plastic pots of were filled with a 4:1 mixture (wt/wt) of the respective field soil and quartz sand (grain size of 0.5–2.2 mm diameter). Pathogen inoculum at different concentrations was thoroughly mixed into soil. Pu inoculum was added at 0.125, 0.25, 0.5, or 1.0 g inoculum per pot, whereas Gt inoculum was added at 0.2, 0.6, 2.0, or 6.0 g per pot. Pots in control treatments contained field soil without pathogen addition. Three seedlings of cucumber or wheat, respectively, were then planted per pot. For each treatment, six replicate pots were prepared. Plants were grown in the greenhouse at 70% relative humidity with light (210 μmol m-2 sec-1) for 16 h at 22°C (cucumber) or 18°C (wheat), followed by an 8-h dark period at 18°C (cucumber) or 15°C (wheat). Plants were watered routinely to keep the soil at constant moisture. The position of the pots was changed at random every other day to avoid position effects. After incubation for 10 days (cucumber) or 21 days (wheat), total shoot fresh weights per pot were assessed.
Development of qPCR Methods for Quantification of Pseudomonads Harboring DAPG, PHZ, and PRN Biosynthetic Genes
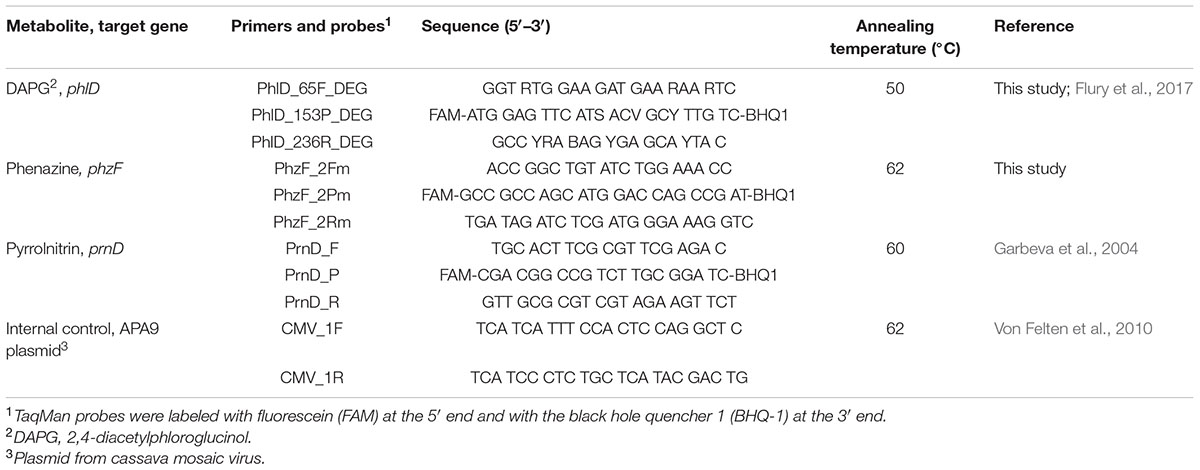
To quantify the abundance of DAPG and PHZ producing bacteria in soil, we developed quantitative real-time polymerase chain reaction (qPCR) assays targeting phlD and phzF genes. These genes encode, respectively, a polyketide synthase involved in the synthesis of phloroglucinols from malonyl-CoA (Bangera and Thomashow, 1996; Achkar et al., 2005) and an isomerase involved in the synthesis of phenazine-1-carboxylic acid (Mavrodi et al., 1998; Blankenfeldt et al., 2004). Alignments were created with publicly available phlD and phzF sequences from GenBank2 and conserved regions were chosen for the design of primers and probes (Table 2), which was carried out with the Primer 3 Plus software (Untergasser et al., 2007). The parameters were amplicon length between 100 and 200 bp, melting temperature (TM) between 50 and 70°C, TM of probe 5°C higher than TM of primers, and the default setting of the program for self-complementarity and 3′-end stability. Partial sequences of phlD (GenBank accession CP003190.1| :6563260-6563937) of strain P. protegens CHA0 (Jousset et al., 2014) and phzF (locus tag, PFLU3_RS28075) of P. synxantha 2–79 (Nesemann et al., 2015) were used for primer design. The specificity of the primers was tested in silico with Primer-Blast (Ye et al., 2012) and in vitro with genomic DNA from 28 DAPG-producing strains and 38 PHZ-producing strains of the P. fluorescens group and nine additional PHZ-producing strains (Supplementary Table S1). Results of these tests revealed that our qPCR assays amplify phlD and phzF genes exclusively from DAPG and PHZ producing species of the P. fluorescens lineage. The PRN biosynthetic genes were quantified on wheat roots by the qPCR method of Garbeva et al. (2004). That assay targets a gene for the class IA oxygenase PrnD that is involved in the final step of PRN biosynthesis (Kirner et al., 1998). In contrast to our phlD and phzF primers, the primers of Garbeva et al. (2004) have broader specificity and, in addition to Pseudomonas, amplify prnD from PRN-producing strains of Burkholderia and Serratia.
The efficiency of phlD and phzF primers at low gene copy numbers was evaluated using in vitro standard curves prepared by serially diluting genomic DNA of P. protegens CHA0 and P. synxantha 2–79. The genomic DNA was prepared by growing both strains in lysogeny broth (LB) (Bertani, 1951) overnight at 24°C on a rotary shaker at 180 rpm and extracting DNA with the Wizard Genomic DNA Purification Kit (Promega AG, Dübendorf, Switzerland). The concentration of purified DNA was quantified by fluorimetry with Qbit (Thermo Fisher Scientific). We also generated an in vivo standard curve for each qPCR assay to quantify the corresponding target genes on wheat roots. To this end, aliquots of 1 g of 21-days-old roots of spring wheat cv. Rubli grown in autoclaved soil were inoculated with decreasing concentrations of a mixture of bacterial cells belonging to different strains carrying the respective target gene. Strains used for in vivo standard curves are listed in Supplementary Table S1. Bacterial cells were harvested from overnight cultures in LB, washed and suspended in sterile 0.9% NaCl solution. Cell suspensions from each strain were set to the same optical density at 600 nm (OD600) and then mixed together at equal proportions. The mixed suspensions were adjusted to an OD600 of 0.125, corresponding to approximately 108 CFU mL-1, serially diluted and inoculated at 101, 102, 103, 104, 105, 106, 107 and 108 CFU g-1 roots for the preparation of the standard curve. For each concentration and for the control without bacteria, three replicates were performed. The inoculated root samples used for standard curves were processed with the same method as the samples from pot experiments with the different soils (see following chapter). In vivo standard curves were prepared as described above for the phlD, phzF and prnD qPCR, using strains listed in Supplementary Table S1. Since all in vivo standard curves were prepared with bacterial cells recovered from wheat roots, the CT values can be directly converted to numbers of bacteria harboring phlD, phzF, or prnD per g root. Our qPCR data also directly reflect the abundance of the antimicrobial biosynthesis genes because phlD, phzF, or prnD are present in single copy in genomes of the P. fluorescens group (Flury et al., 2016). A survey of published bacterial genomes revealed that phzF and prnD are also found as a single copy in other bacterial species such as Burkholderia (phzF and prnD), Pectobacterium (phzF) or Serratia (prnD).
qPCR-Based Quantification of Antimicrobial Genes on Roots of Wheat Grown in Field Soils
To standardize the root material for qPCR quantification of DAPG, PHZ and PRN biosynthetic genes, soil samples from the 10 Swiss field sites were planted with spring wheat cv. Rubli in a greenhouse pot experiment. Plastic pots of 8 cm diameter and 30 cm height were part-filled with field soil and three wheat seedlings, prepared as described above, were planted per pot. Six pots per field soil were prepared. Wheat plants were grown for 2.5 months under the conditions described above for the Gt resistance assays. Root samples were collected, rinsed with tap water, incubated overnight in a sterile 0.9% NaCl solution at 3°C, and then vigorously agitated at 350 rpm for 30 min. Roots were separated from the root-wash suspensions and kept for dry weight assessment. Root-wash suspensions were centrifuged at 3500 rpm for 20 min. The supernatant was discarded and aliquots of 0.5 ml of the resulting root-wash pellet were used for DNA extraction. To each sample, 109 copies of the APA9 plasmid from a cassava mosaic virus were added as an internal standard (Von Felten et al., 2010). DNA extraction was performed with the MPBio soil kit (MP Biomedicals, Illkirch, France) following the protocol of the manufacturer. The concentration of extracted DNA was measured with Qbit. qPCR reactions consisted of 10 μL TaqMan Gene Expression Master Mix (Applied Biosystems, Foster City, CA, USA), 2 μL of the respective forward and reverse primer solutions (10 μM), 2 μL of the respective probe solution (2.5 μM), 0.5 μL of bovine serum albumin solution (20 mg mL-1), and 2 μL of template DNA in a total reaction volume of 20 μL. Primer and probe sequences are indicated in Table 2. Cycling conditions consisted of 2 min at 50°C (to permit uracil-DNA glycosylase activity), an initial denaturation step of 10 min at 95°C, and 40 cycles of 15 s at 95°C, 30 s at annealing temperature (see Table 2) and 30 s at 72°C. In all samples, the added APA9 plasmid was quantified with the primers listed in Table 2, following the method of Von Felten et al. (2010). The results from the APA9 plasmid quantification were used to normalize DNA extraction.
To compare the abundance of phlD and phzF measured with qPCR with the abundance measured using a cultivation-dependent terminal endpoint dilution assay, also called most probable number PCR (MPN-PCR) method (Ramette et al., 2006), a greenhouse experiment was carried out with soil samples from the Cazis and Taenikon field sites and spring wheat cv. Rubli cultivated under the same conditions as described above. After 3 weeks, phzF and phlD qPCR assays were performed on one fraction of each harvested root-wash pellet as described above, while the other fractions were serially diluted into microtiter plate wells (200 μl volume) containing King’s medium B (King et al., 1954) broth amended with 100 mg L-1 cycloheximide, 13 mg L-1 chloramphenicol and 40 mg L-1 ampicillin. The microtiter plates were incubated at 24°C for 3 days and MPN-PCR was performed as described by Ramette et al. (2006), except for adapting annealing temperatures to the primers used in the present study (Table 2).
Quantification of Resident Pu and Gt by qPCR
Pythium ultimum and Gt populations naturally present on roots of wheat were quantified using the qPCR methods developed by Cullen et al. (2007) and Bithell et al. (2012), respectively. The reaction mix was prepared as described above for antimicrobial genes, and cycling conditions were set as described previously (Cullen et al., 2007; Bithell et al., 2012). In vitro standard curves were performed with genomic DNA of Pu isolate ETH-2 (concentration range from 0.1 ng to 200 ag per reaction) and of Gt isolate I-17 (10 ng to 10-4 ng per reaction). Genomic DNA of the two pathogens was extracted with the DNeasy plant mini kit (Qiagen, Hombrechtikon, Switzerland) from lyophilized mycelia prepared from cultures grown in potato dextrose broth (Difco, Becton, Dickinson and Company, Franklin Lakes, USA) for 7 days at 24°C with agitation at 180 rpm.
Construction and Culture of Pseudomonas Reporter Strains
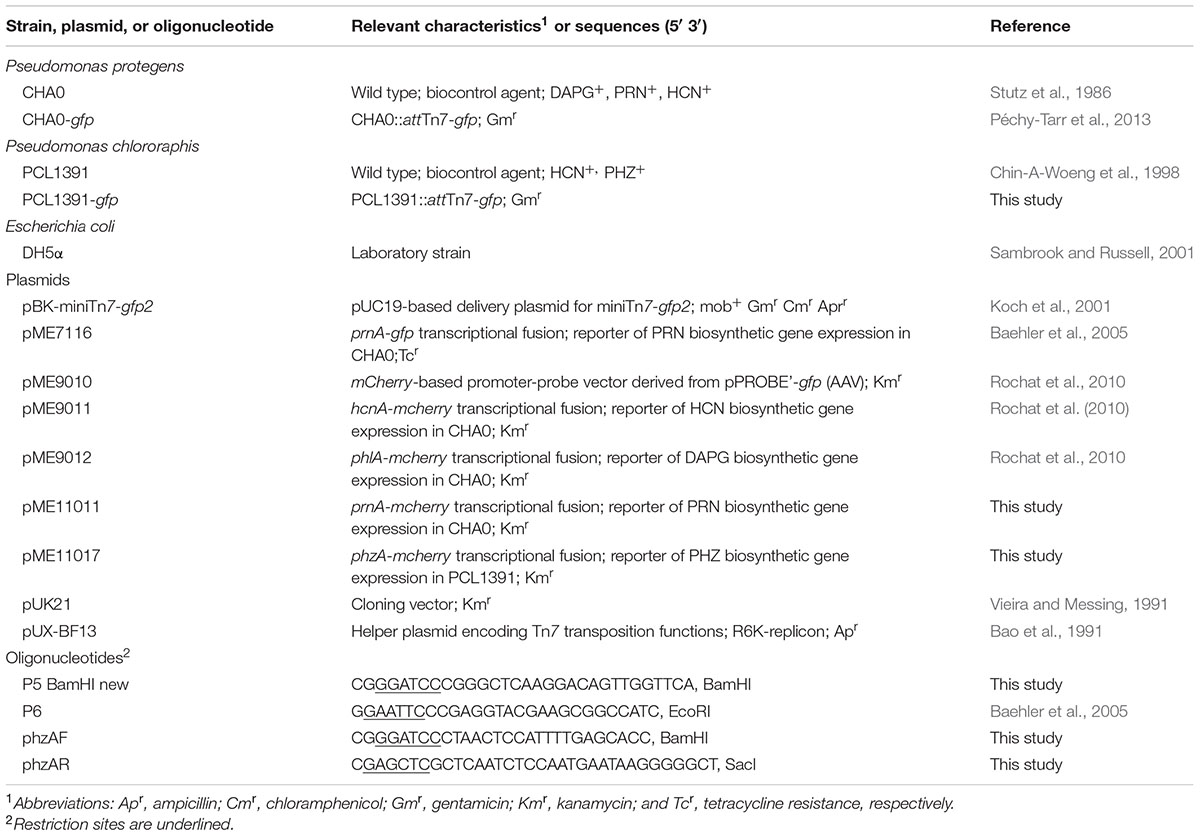
Bacterial strains and plasmids used for generation of Pseudomonas reporter strains for monitoring antimicrobial gene expression are listed in Table 3. Pseudomonas and Escherichia coli strains were routinely cultured at 30 and 37°C, respectively, on nutrient agar plates, in LB and in nutrient yeast broth (Stanisich and Holloway, 1972). When appropriate, selective antibiotics were added to the media at the following concentrations: ampicillin, 100 μg mL-1; chloramphenicol, 50 μg mL-1; gentamicin, 10 μg mL-1; kanamycin, 25 μg mL-1; and tetracycline, 125 μg mL-1. Genomic DNA from P. protegens strain CHA0 and P. chlororaphis strain PCL1391 was isolated as previously described (Schnider-Keel et al., 2000). Plasmids were extracted and purified using the QIAprep Spin Miniprep kit (Qiagen) or the JETStar Plasmid Purification Midi kit (Genomed, Basel, Switzerland). PCRs were done using the PrimeSTAR HS DNA polymerase kit (Takara Bio Inc., Shiga, Japan) as described elsewhere (Péchy-Tarr et al., 2005). All DNA digestion and ligation reactions were done using standard techniques (Sambrook and Russell, 2001; Schnider-Keel et al., 2001). DNA extractions from agarose gels were carried out with the QIAquick Gel Extraction kit (Qiagen). Transformations of electro-competent cells with plasmid or purified ligation products were performed by electroporation (Schnider-Keel et al., 2000). To amplify genomic DNA or to detect the presence of recombinant DNA in E. coli colonies by screening, 100–200 ng of DNA were amplified using the GoTaq DNA polymerase kit (Promega, Dübendorf, Switzerland). All PCR constructs intended for transformation were verified by sequence analysis. DNA sequencing was carried out by GATC Biotech AG (Konstanz, Germany). Sequences were analyzed using the DNASTAR Lasergene software package version 11.0.
In order to tag P. protegens CHA0 and P. chlororaphis PCL1391 with green fluorescent protein (GFP), a single copy of a gfp variant gene constitutively expressed from the Ptac promoter, was inserted into the chromosome using the pBK-miniTn7-gfp2 delivery plasmid (Koch et al., 2001) and the Tn7 transposition helper plasmid pUX-BF13 (Bao et al., 1991) as described previously (Péchy-Tarr et al., 2013). For use as reporters to monitor the expression of HCN, DAPG, PRN, and PHZ biosynthetic genes, strain CHA0-gfp was transformed with pME9011 (hcnA-mcherry), pME9012 (phlA-mcherry) or pME11011 (prnA-mcherry), and strain PCL1391-gfp with pME11017 (phzA-mcherry) (Table 3). To construct the prnA-mcherry reporter plasmid pME11011, a 629-bp fragment containing the CHA0 prnA promoter was amplified from pME7116 (Baehler et al., 2005) using primers P5 BamHI new and P6 (Table 3). The obtained fragment was digested with BamHI and EcoRI and ligated into the mcherry-based promoter-probe vector pME9010 (Rochat et al., 2010) opened with the same enzymes. Similarly, for the construction of the phzA-mcherry reporter plasmid pME11017, a 961-bp fragment containing the phzA promoter (Chin-A-Woeng et al., 2001) was amplified from genomic DNA of PCL1391 (Flury et al., 2016) using primers phzAF and phzAR (Table 3). The PCR product was digested with BamHI-SacI and the resulting fragment was first cloned into pUK21 and, from there, into pME9010, both opened with the same restriction enzymes.
Assay to Monitor Antimicrobial Gene Expression in Field Soils
Antimicrobial gene expression and colonization levels of GFP-marked P. protegens CHA0 and P. chlororaphis PCL1391 harboring mCherry-based reporter plasmids were monitored on roots of spring wheat grown in the soils sampled at the 10 different Swiss field sites. Untreated seeds of spring wheat cv. Rubli were surface-sterilized for 12 min in 4% NaClO (vol/vol) and then washed with sterile distilled water. Seeds were germinated on soft agar (Agar Agar Serva at 9 g L-1; Serva, Heidelberg, Germany) for 48 h at room temperature in the dark. The wheat seedlings were then transferred to 200-mL Erlenmeyer flasks (5-cm opening; Simax, Czech Republic) containing 60 g of soil. In each flask, three seedlings were placed into the soil and inoculated with 1 mL of a suspension of washed cells of the respective Pseudomonas reporter strain adjusted within a range from 3.6 107 to 7.8 × 107 cells mL-1. Washed cells were prepared from LB cultures grown without antibiotic addition under the conditions described above to an OD600 of 0.8 to 1.5, depending on the reporter strain used, whereby choosing a growth stage at which no significant expression of the antimicrobial genes occurred. This was done by determining, with a fluorimetry assay (Baehler et al., 2005), the time point at which the relative red fluorescence emitted by the Pseudomonas strains carrying mCherry-based reporter plasmids was not yet significantly different from the red background fluorescence emitted by control strains carrying the empty vector pME9010. Wild-type and GFP-tagged P. protegens CHA0 and P. chlororaphis PCL1391 (with and without pME9010) were included as control treatments for properly setting green and red fluorescence backgrounds for the FACS-based flow cytometry analysis described below. Flasks were sealed with cotton wool plugs and incubated in a growth chamber set to 60% relative humidity for 16 h with light (176 μE m-2 s-1) at 25°C, followed by an 8-h dark period at 20°C. After 5 days of incubation, wheat roots from each flask were removed, washed using distilled water to remove loosely adhering soil particles from roots and pooled in 10 mL of autoclaved, ultrapure water contained in a sterile 50-mL Falcon tube. Tubes were agitated for 20 min at 300 rpm in order to remove the majority of bacteria from the roots. The resulting suspensions were filtered using a 5.0-μm sterile syringe single-use filter (Sartorius Stedim Biotech GmbH, Goettingen, Germany), transferred on ice and immediately analyzed by FACS as described below. Dry weights of wheat roots were recorded and the number of GFP-marked Pseudomonas cells present in the root washes were determined by FACS and recorded as cells g-1 of root.
FACS Analysis
Green fluorescent protein and mCherry expression levels in Pseudomonas reporter cells in natural soils were quantified with a Becton–Dickinson LSRFortessa flow cytometer. Size and granularity of Pseudomonas cells and particles were determined by measuring the forward scatter (FSC-A) and side scatter (SSC-A) signals, respectively. FSC-A signals were collected with a photodiode detector (set to 350 V), in the range of 483 to 493 nm (488/10 BP filter), with a threshold set to 200. SSC-A signals were detected with a photomultiplier tube (PMT) (detector G, set to 300 V) in the range of 483 to 493 nm (488/10 BP filter). Green fluorescence signals were collected with the PMT detector E (set at the voltage of 676 V), in the range of 515 to 545 nm (530/30 BP filter, 505 LP mirror). Red fluorescence signals were detected using the PMT detector C (set to 700 V), between 600 and 620 nm (610/20 BP filter, 600 LP mirror). For FACS analysis, aliquots of 300 μL of filtered root-washes were placed into Nunc MaxiSorp flat-bottom 96-well plates (Sigma–Aldrich, Buchs, Switzerland), and each sample was mixed three times. The analyzed volume was standardized to 200 μL, allowing the detection of 500–10,000 GFP events, depending on the analyzed soil. Gating of GFP-marked bacteria was done by delimiting on the FSC-A/FITC-A density plot particles with green fluorescence values above the background fluorescence noise (i.e., autofluorescence emitted by root and soil particles, cell fragments or bacterial cells not expressing GFP). Control samples obtained from soils amended with pure water, wild-type P. protegens CHA0 and P. chlororaphis PCL1391, and GFP-tagged strains CHA0-gfp and PCL1391-gfp were used to identify the GFP background fluorescence in soil extraction samples. The red fluorescence emitted by the gated GFP-tagged Pseudomonas cells was then analyzed on the PE-Texas Red-A histogram and on the FSC-A/PE-Texas Red-A density plot, allowing to detect and analyze all GFP-marked cells actively expressing their mCherry-based reporter fusion. Control samples including CHA0-gfp and PCL1391-gfp without mCherry-based vector, or carrying the empty pME9010 vector were used to define the mCherry background fluorescence among the GFP-marked Pseudomonas population. The median of red fluorescence emitted by the Pseudomonas cells was calculated using the BD FACSDiva software version 8.0 (Becton–Dickinson). To determine root colonization levels of Pseudomonas reporter strains, the GFP tag was used to count by FACS the number of reporter cells present in the analyzed 200 μL of filtered root-wash and then to calculate their concentration per gram of dry roots.
Data Analysis
Statistical data analysis was carried out with the open source software R version 3.2.3 (R Core Team, 2015). Shoot fresh weights of cucumber and wheat plants obtained from the Pu and Gt infection assays, respectively, were analyzed as a proxy for soil disease resistance. Shoot weights from samples where pathogen inoculum was added were normalized against the shoot weights from non-infested control plants grown in the same soil, to minimize variation due to different nutrient contents in the different soils. Data were checked for normal distribution with the Shapiro–Wilk test and by plotting QQ-Plots. Equality of variance was verified with Bartlett’s test. Analysis of variance was carried out with a non-parametric test (Kruskal–Wallis test, significance level p < 0.05), followed by a post hoc test (kruskalmc, R package ‘Pgirmess’).
The abundance of phlD, phzF, and prnD harboring bacterial cells on wheat roots was calculated with the in vivo standard curves described above. Efficiencies and detection limits of qPCR assays determined by in vivo standard curves are indicated in Supplementary Table S2. Cycle threshold values obtained from the in vivo standard curves and from the samples were normalized for differences in DNA extractions as described by Von Felten et al. (2010). Normalized values were used for further analysis. The average values obtained from the three technical replicates of each qPCR assay were used for statistical analysis, which was performed as described above for shoot weights.
Data on the expression of reporters of antimicrobial genes in the different soils represent the medians of three independent repetitions of the same experiment, with nine replicates per treatment in each experiment. Significant differences between treatments were calculated with a non-parametric Kruskal–Wallis test (significance level p < 0.05), followed by Dunn’s test for post hoc comparisons.
Data used for the heat map showing rankings of pathogen suppression, abundance of antimicrobial genes, and expression of antimicrobial genes by reporter strains in the different soils were normalized using the function ‘scale’ (R package ‘stats’). Correlations between pathogen suppression, gene abundance, gene expression and abiotic soil parameters were inferred with Spearman’s rho rank correlation (significance level p < 0.05). Data were displayed in a heat map with the functions ‘levelplot’ (R package ‘lattice’) or ‘corrplot’ (R package ‘corrplot’).
Results
Resistance of Swiss Agricultural Soils to Soilborne Pathogens
The general resistance of 10 Swiss agricultural soils (Figure 1 and Table 1) to soilborne pathogens was tested in a greenhouse assay in which increasing quantities of Gt or Pu inoculum were added to soil samples planted with spring wheat or cucumber, respectively. Resistance to both pathogens varied between soils. In the Gt-infested soils at 0.6 g inoculum per pot, shoot fresh weights of wheat plants ranged from 31% in Grangeneuve soil to 107% in Taenikon soil of the weights of control plants grown in not artificially infested soils (Figure 2A). The shoot fresh weights of wheat grown in infested Taenikon soil were significantly higher (1.3–3.5-fold) than those of plants grown in infested soils from Cazis, Eschikon, Grangeneuve, Vouvry, and Witzwil. Similar trends were observed at other inoculum quantities (Supplementary Figure S1). Plants grown in Taenikon soil had the highest shoot fresh weights at all Gt concentrations used except the highest (6 g of inoculum per pot), at which the fungal pathogen heavily affected plant growth in all the soils and reduced shoot weights by 60–90% (Supplementary Figure S1D).
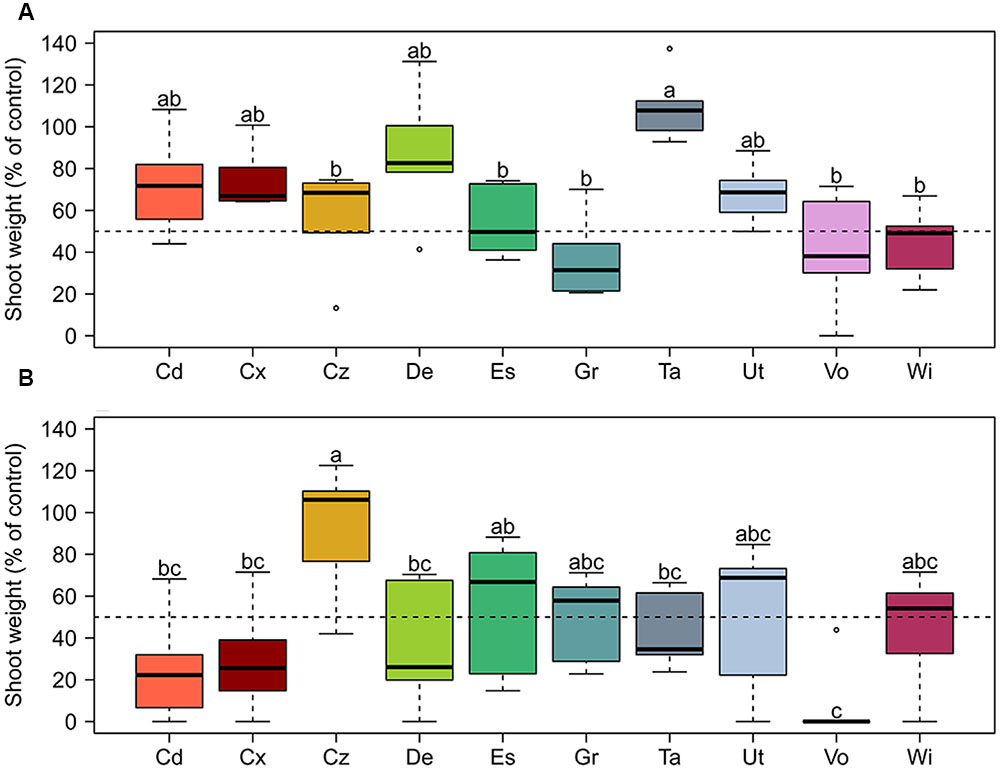
FIGURE 2. Relative resistance of 10 representative Swiss agricultural soils with a cereal-oriented cropping history to the soil-borne pathogens (A) Gaeumannomyces tritici (Gt) and (B) Pythium ultimum (Pu). Increasing concentrations of pathogen inoculum were added to the soil before planting with spring wheat (Gt experiment) or cucumber (Pu experiment) seedlings. Data shown here are for Gt at 0.6 g and Pu at 0.125 g inoculum per pot. Results for the other inoculum concentrations tested are shown in Supplementary Figures S1, S2. Each pathogen concentration and soil was tested in six replicate pots. Soil resistance is shown as fresh shoot weight of plants in artificially pathogen-infested soil compared to fresh shoot weight of control plants grown in non-infested soil. The dotted line indicates 50% of shoot weight compared to the control. Letters indicate significant differences (Kruskal–Wallis test, p < 0.05). Sampling sites: Cd, Cadenazzo; Cx, Courtedoux; Cz, Cazis; De, Delley; Es, Eschikon; Gr, Grangeneuve; Ta, Taenikon; Ut, Utzenstorf; Vo, Vouvry; Wi, Witzwil.
When the soils were artificially infested with Pu inoculum at 0.125 g per pot, median shoot fresh weights of cucumber plants ranged from 0% in Vouvry soil to 106% in Cazis soil of those of control plants grown in non-infested soils (Figure 2B). The shoot fresh weights of cucumber grown in Pu-infested Cazis soil were significantly higher (three–fivefold) than those of plants grown in infested soils from Cadenazzo, Courtedoux, Delley, Taenikon, and Vouvry (Figure 2B) and similar trends were observed for other inoculum densities (Supplementary Figure S2). Plants grown in soil from the Cazis field site had the highest shoot weights at all Pu concentrations, while plants grown in soil from the Vouvry field had the lowest shoot weights at all levels of the pathogenic oomycete except at 0.25 g per pot.
Individual soils did not display equal resistance levels to the two soilborne pathogens. While the soils from the Taenikon and Delley field sites were the most resistant against Gt, they were among the least resistant against Pu (Figure 2 and Supplementary Figures S1, S2). Likewise, the soil from Cazis was the most resistant against Pu, but was only moderately resistant against Gt.
To account for potential effects of resident Gt and Pu populations on the outcome of the soil resistance experiments, a qPCR method targeting the ITS rRNA gene region was used to detect and quantify the two pathogens in the rhizoplane of spring wheat plants grown in the not artificially infested control treatments of all soils. Gt could not be detected in any of the samples. By contrast, Pu was detected on the plant roots in all ten soils, but there was no significant difference in the abundance of the oomycete pathogen among the individual soils (Supplementary Figure S3).
In summary, the 10 agricultural soils strongly varied in their resistance against soilborne pathogens. However, the soil resistance levels observed for the two investigated pathogens in general were different; i.e., some soils displaying high resistance to Pu were highly susceptible to Gt and vice versa, pointing to specificity in the buffering capacity of individual soils toward specific soilborne pathogens.
Abundance of phlD+ Pseudomonads, phzF+ Pseudomonads, and prnD+ Bacteria on Roots of Wheat Grown in Swiss Agricultural Soils
The abundance of bacterial cells harboring phlD, phzF, and prnD required for the biosynthesis of the antimicrobials DAPG, PHZ, and PRN, respectively, was quantified by qPCR on roots of spring wheat grown in the 10 Swiss agricultural soils. As detailed in section “Materials and Methods,” we assume that phlD+ and phzF+ cells quantified in our assays correspond to cell numbers of DAPG and PHZ producing pseudomonads, whereas prnD+ cells correspond to cell numbers of PRN producing bacteria. Since the investigated genes are present as one copy per bacterial cell, we also refer to the abundance of cells harboring an antimicrobial gene as gene abundance.
The abundance of the phlD+ pseudomonads in all studied soils in general was higher than the abundance of the phzF+ and prnD+ bacteria, and in some soils reached 107 cells per gram of root dry weight, whereas the abundance of the latter remained below 106 gene copies per gram of root dry weight (Figure 3). For individual genes, pronounced differences in abundance between soils were observed. The biggest differences were found for phlD+ cells with approximately 104-fold higher numbers on roots of wheat grown in soil from Taenikon compared to those grown in soil from Vouvry (Figure 3A). The abundance of phlD+ pseudomonads was significantly higher in soils from Courtedoux, Delley, Eschikon, Grangeneuve, Taenikon, Utzenstorf, and Witzwil than in soils from Cadenazzo, Cazis, and Vouvry. The abundances of phzF+ pseudomonads and prnD+ bacteria varied at maximum 100-fold between the different soils. The number of pseudomonads harboring the phzF gene was significantly higher on roots samples from Courtedoux and Delley soils compared to samples extracted from Vouvry soil (Figure 3B). The abundance of prnD+ bacteria was significantly higher in soil samples from Cadenazzo, compared to those from Courtedoux, Cazis, and Vouvry (Figure 3C). Taken together, for all three investigated antimicrobial genes pronounced differences in the abundances were found between the individual agricultural soils, indicating that the different soils may sustain to different extents specific populations of pseudomonads producing DAPG, PRN, and/or PHZ.
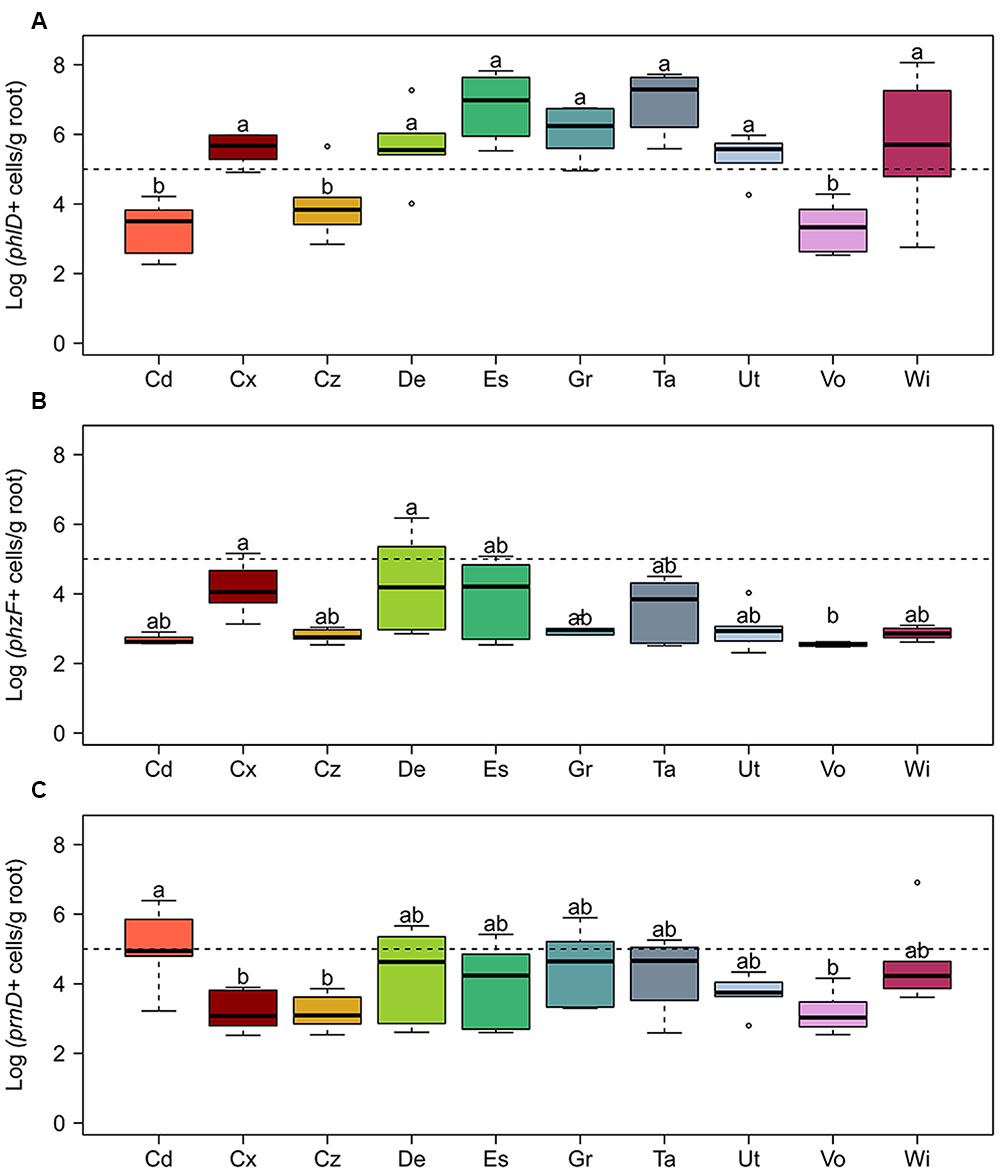
FIGURE 3. Abundance of bacterial cells harboring genes required for the biosynthesis of the antimicrobial compounds (A) 2,4-diacetylphloroglucinol (phlD), (B) phenazine (phzF), and (C) pyrrolnitrin (prnD) on roots of spring wheat in 10 Swiss agricultural soils. Cells harboring the antimicrobial genes were quantified by qPCR. The dotted line indicates 105 cells per gram of root dry weight. For each soil, six replicates were used. Letters indicate significant differences (Kruskal–Wallis test, p < 0.05). Sampling sites: Cd, Cadenazzo; Cx, Courtedoux; Cz, Cazis; De, Delley; Es, Eschikon; Gr, Grangeneuve; Ta, Taenikon; Ut, Utzenstorf; Vo, Vouvry; Wi, Witzwil.
Expression of Antimicrobial Genes in Swiss Agricultural Soils
The relative capacity of the 10 different Swiss agricultural soils to sustain the expression of biosynthetic genes for the antimicrobial compounds DAPG, HCN, PRN, and PHZ on roots was followed using GFP-tagged Pseudomonas strains carrying mCherry-based reporter plasmids inoculated into soil microcosms planted with spring wheat. P. protegens CHA0-gfp carrying plasmid pME9012 (phlA-mcherry), pME9011 (hcnA-mcherry), or pME11011 (prnA-mcherry), or P. chlororaphis PCL1391-gfp carrying pME11017 (phzA-mcherry) served as reporter strains. GFP fluorescence (identifying the tagged reporter strains) and relative mCherry fluorescence intensities (reporting expression levels of respective antimicrobial genes) of cells in root washes extracted from the different soils were recorded with FACS-based flow cytometry. Data presented as total antimicrobial gene expression by all cells of the respective reporter strain per gram dry weight of roots, and they were calculated by multiplying the median gene expression per cell with the total number of reporter cells per gram dry weight of roots (Supplementary Table S3).
The levels of total expression of all investigated antimicrobial genes varied significantly among the 10 field soils, with the highest variations observed for DAPG and HCN biosynthetic genes (Figure 4). The soil from Cadenazzo supported the highest levels of total phlA expression on the roots, which were approximately 16-fold higher than those measured on roots growing in soil from Taenikon, which yielded the lowest expression levels (Figure 4A). The Cadenazzo soil also supported highest levels of total hcnA expression among all 10 soils, and these levels were approximately 20-fold higher than hcnA expression levels recorded in the soil from the Grangeneuve field site, which was the least favorable to the expression of this antimicrobial gene (Figure 4B). Levels of total expression of PRN and PHZ biosynthetic genes appeared to be less variable among the 10 soils. In particular, prnA expression levels were eightfold higher in the soil from Utzenstorf that supported the highest expression compared to the soil from Eschikon that supported the lowest expression (Figure 4C). Levels of overall phzA expression varied only about fivefold between the most contrasting soils from the Cadenazzo and Taenikon field sites, respectively (Figure 4D). Several individual soils sustained total expression levels for all investigated antimicrobial genes to a similar extent. This was evident notably for soils from Cadenazzo and Vouvry in which all four antimicrobial genes attained high total expression levels, while soils from Eschikon, Grangeneuve and Taenikon supported significantly lower overall expression levels of these genes (Figure 4).
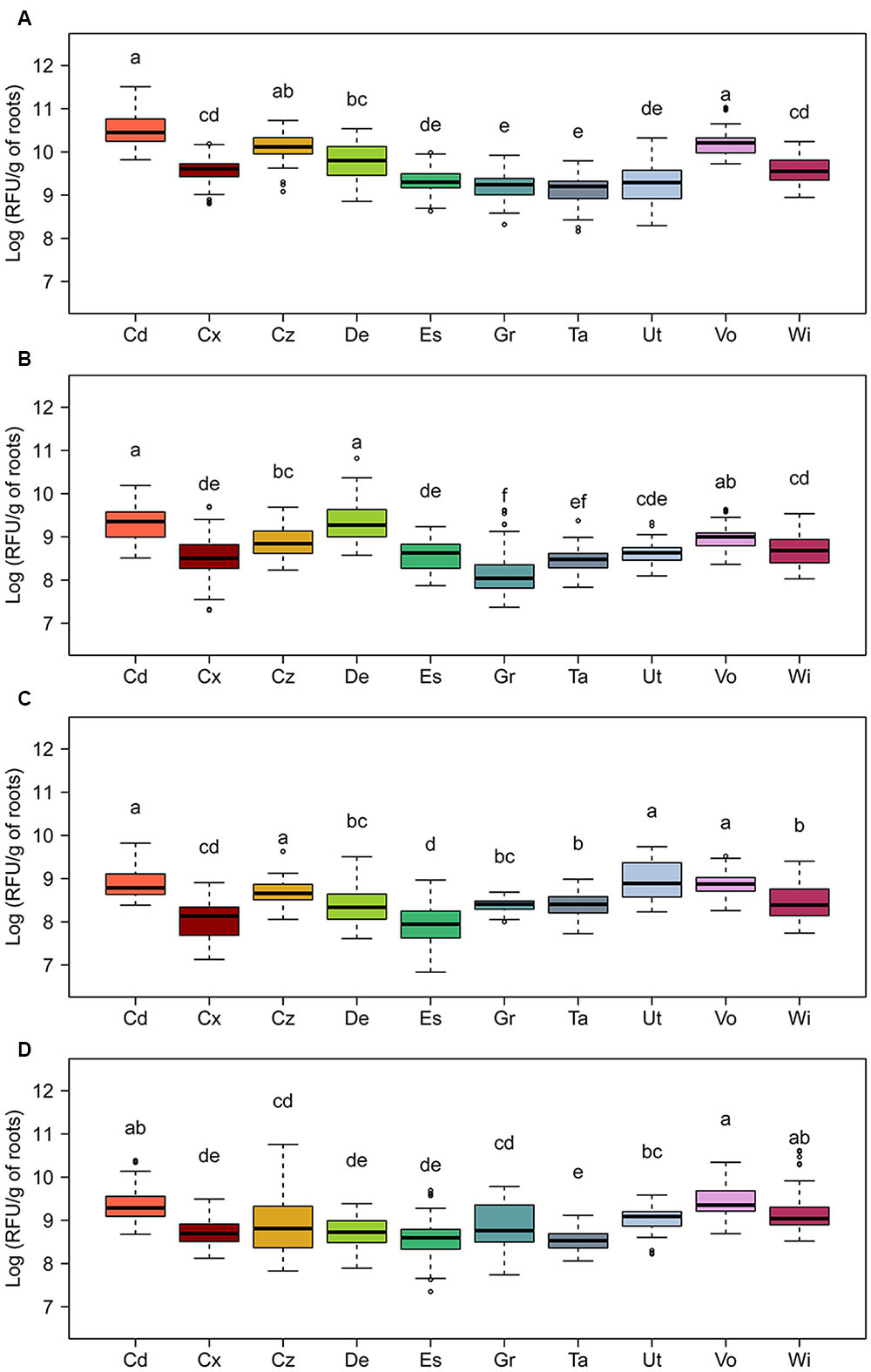
FIGURE 4. Relative expression of genes required for the biosynthesis of the antimicrobial compounds (A) 2,4-diacetylphloroglucinol (phlA), (B) hydrogen cyanide (hcnA), (C) pyrrolnitrin (prnA), and (D) phenazines (phzA) on roots of spring wheat in 10 Swiss agricultural soils. Expression was monitored by fluorescence-activated cell-sorting-based flow cytometry using GFP-tagged strains of Pseudomonas protegens (CHA0-gfp) carrying reporter plasmids pME9012 (phlA-mcherry), pME9011 (hcnA-mcherry), or pME11011 (prnA-mcherry) or of P. chlororaphis (PCL1391-gfp) carrying reporter plasmid pME11017 (phzA-mcherry). Seedlings inoculated with the reporter strains were grown in soil microcosms for 5 days prior to analysis of bacterial cells in root washes. Data are shown as relative fluorescence units (RFU) per gram of root dry weight, and were calculated as the median mCherry expression per GFP-tagged Pseudomonas cell multiplied with the total number of GFP-tagged Pseudomonas cells per gram of root. Results from three independent experiments with nine replicates each are presented. Since Kruskal–Wallis analyses did not reveal significant experiment × treatment interactions, data of the three experiments were pooled for statistical analysis. Letters indicate significant differences (Dunn test, p < 0.05). Sampling sites: Cd, Cadenazzo; Cx, Courtedoux; Cz, Cazis; De, Delley; Es, Eschikon; Gr, Grangeneuve; Ta, Taenikon; Ut, Utzenstorf; Vo, Vouvry; Wi, Witzwil.
The Cadenazzo and Vouvry soils appeared to favor also highest levels of root colonization (1.9 × 107 to 2.4 × 107 cells g-1 of dry root weight) by P. protegens and P. chlororaphis among the 10 soils tested, while the soils from Eschikon, Grangeneuve, and Taenikon were among those supporting lower levels of root colonization (2.9 × 106 to 9.7 × 106 cells g-1 of dry root weight) (Figure 5 and Supplementary Table S3). However, whereas the soil from Cadenazzo was indeed also the field soil supporting highest phlA, hcnA, prnA, and phzA expression levels in individual reporter cells, the median single cell expression of these genes was significantly lower in the soil from Vouvry (Supplementary Table S3). For the Vouvry soil, it thus seems that the high total expression in the reporter population on the roots was mainly due to the higher colonization levels attained in this soil. Likewise, the relatively high single cell expression but low colonization levels in the Eschikon and Taenikon soils (Figure 5 and Supplementary Table S3) resulted in the significantly lower overall expression as compared to the Cadenazzo and Vouvry soils (Figure 5). By contrast, in the soil from Grangeneuve, single cell expression levels of antimicrobial genes as well as colonization levels were consistently relatively low (Supplementary Table S3). Similar overall gene expression levels may therefore reflect contrasting levels of single cell gene expression and colonization in individual soils.
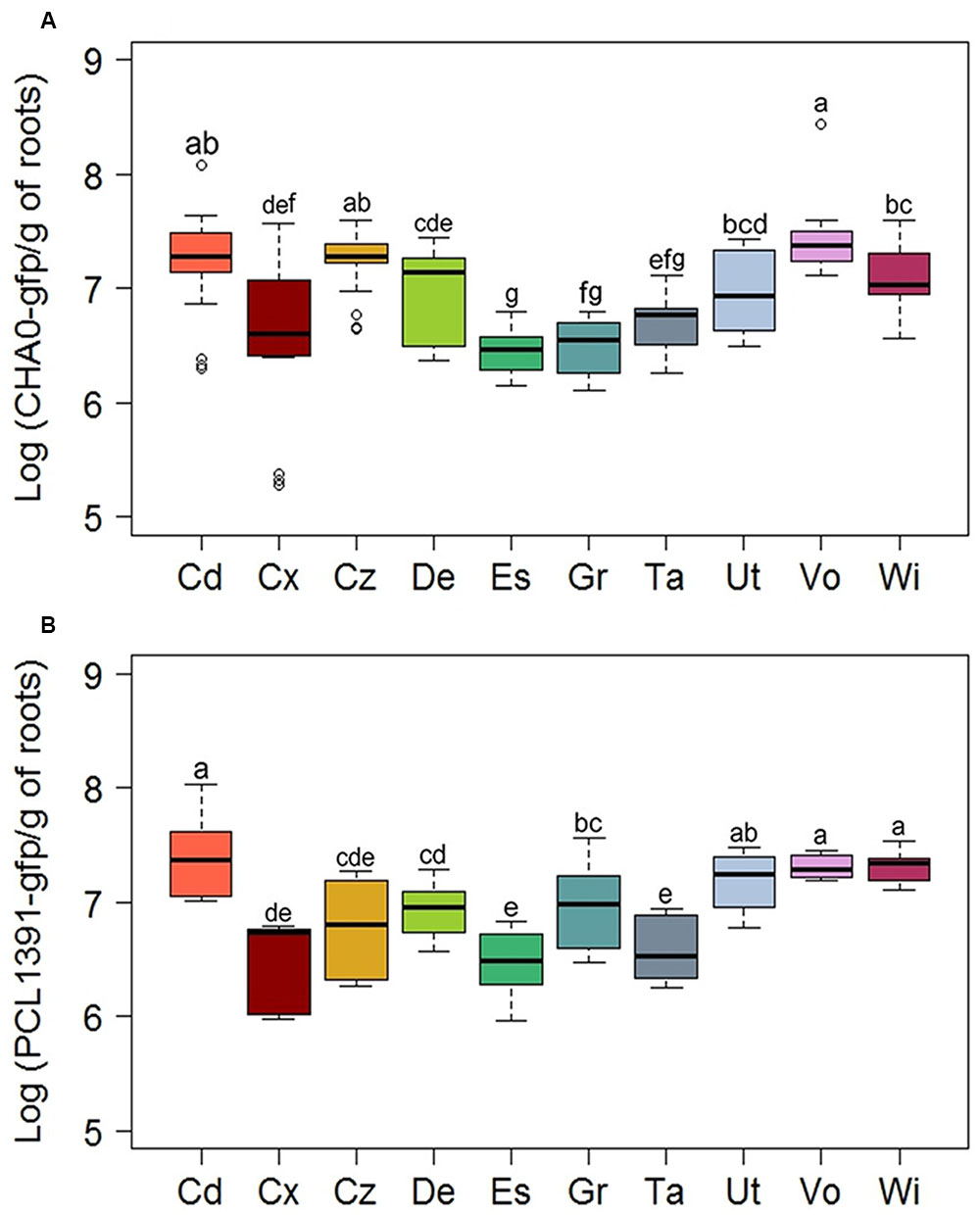
FIGURE 5. Root colonization levels of Pseudomonas reporter strains in 10 Swiss agricultural soils planted with spring wheat. Cells of GFP-tagged strains of (A) P. protegens (CHA0-gfp) or (B) P. chlororaphis (PCL1391-gfp) were monitored by fluorescence-activated cell-sorting-based flow cytometry. Data show numbers of GFP-tagged cells per gram of root dry weight. Results from three independent experiments with nine replicates each are presented. Since Kruskal–Wallis analyses did not reveal significant experiment × treatment interactions, data of the three experiments were pooled for statistical analysis. Letters indicate significant differences (Dunn test, p < 0.05). Sampling sites: Cd, Cadenazzo; Cx, Courtedoux; Cz, Cazis; De, Delley; Es, Eschikon; Gr, Grangeneuve; Ta, Taenikon; Ut, Utzenstorf; Vo, Vouvry; Wi, Witzwil.
In summary, the expression of the investigated antimicrobial genes strongly varied between the studied agricultural soils. Some soils seem to favor higher levels of overall antimicrobial gene expression whereas others apparently support the expression of these genes much less. The findings suggest that the expression of different antimicrobial genes may be induced by the same soil factors (or similar combinations thereof).
Relationships between Pathogen Resistance and Abundance and Expression of Antimicrobial Genes in Swiss Agricultural Soils
Results obtained from experiments on pathogen resistance of soils, abundance and expression of antimicrobial genes were displayed in a gradient map (Figure 6). Soils were grouped in three clusters. The first cluster consisted of soils supporting high abundances but low expression levels of antimicrobial genes (i.e., soils from Eschikon and Taenikon field sites). The second cluster consisted of soils supporting high antimicrobial gene expression levels but low abundances of bacteria harboring antimicrobial genes (i.e., soils from Cadenazzo and Vouvry field sites). The third cluster consisted of soils ranging in between the two other clusters. Two soils displayed high resistance against soilborne plant pathogens, i.e., the soil from Taenikon against Gt and the soil from Cazis against Pu. Although the soil from Taenikon was the one supporting the highest abundance of pseudomonads harboring the DAPG biosynthetic gene phlD, resistance to the soilborne pathogens did not generally cluster with high abundances or expression levels of antimicrobial genes (Figure 6).
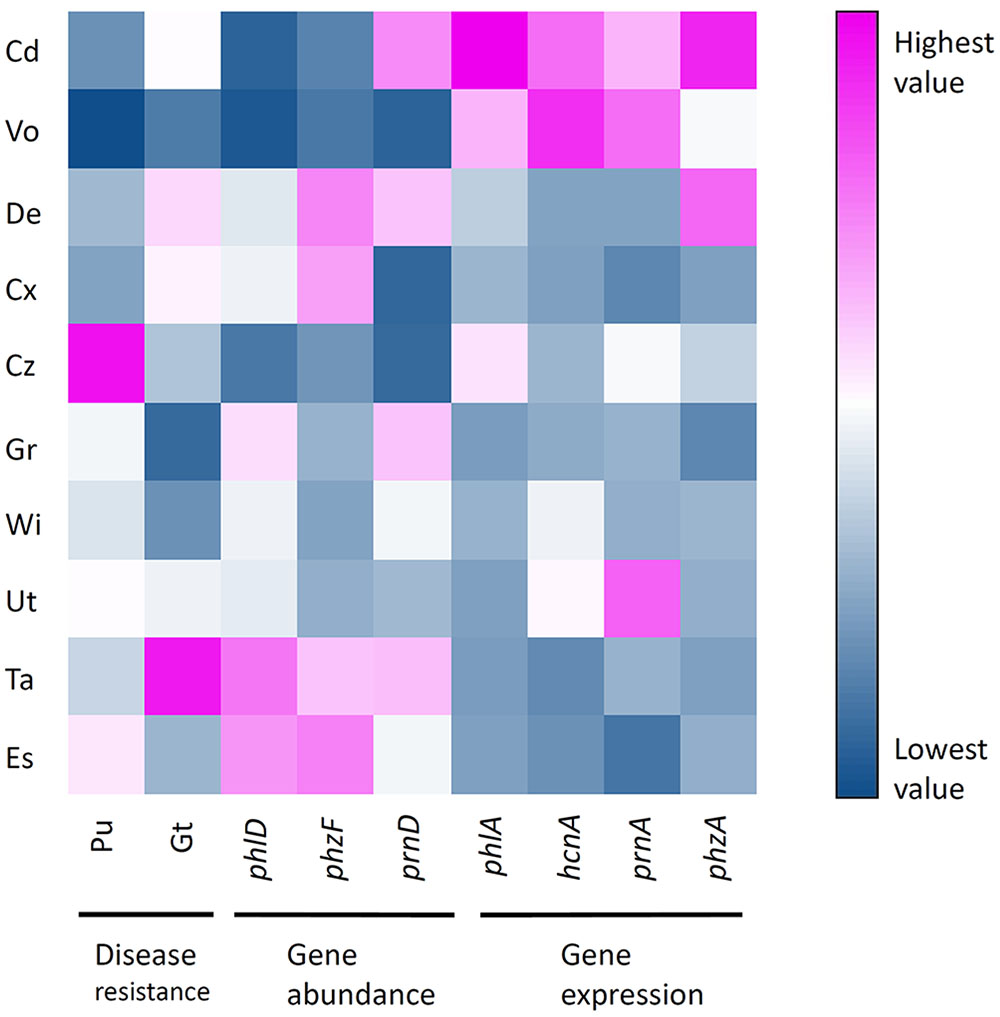
FIGURE 6. Heatmap showing normalized-values of disease resistance, antimicrobial gene abundance and antimicrobial gene expression measured in 10 representative Swiss agricultural soils with a cereal-oriented cropping history. The color scale depicts highest (fuchsia) via intermediate (white) to lowest (blue) values for each variable. Sampling sites: Cd, Cadenazzo; Cx, Courtedoux; Cz, Cazis; De, Delley; Es, Eschikon; Gr, Grangeneuve; Ta, Taenikon; Ut, Utzenstorf; Vo, Vouvry; Wi, Witzwil.
Correlation analysis revealed that the expression of the four studied antimicrobial genes was positively correlated (Figure 7B). A similar trend could be observed for the abundance of the antimicrobial genes (Figure 7A). Resistance to soilborne pathogens was not significantly correlated to the abundance of antimicrobial genes, although a weak positive correlation could be observed between the abundance of bacteria harboring individual antimicrobial genes and resistance to pathogens (Figure 7A). These results suggest that the expression of the investigated antimicrobial genes is similarly influenced by biotic and abiotic factors prevailing in the respective soils. The same could be true for the abundance of bacteria harboring these antimicrobial genes. Our results further indicate that the abundance of such bacteria probably plays only a limited role in the resistance of the investigated cereal crop-oriented agricultural soils to the soilborne pathogens Pu and Gt.
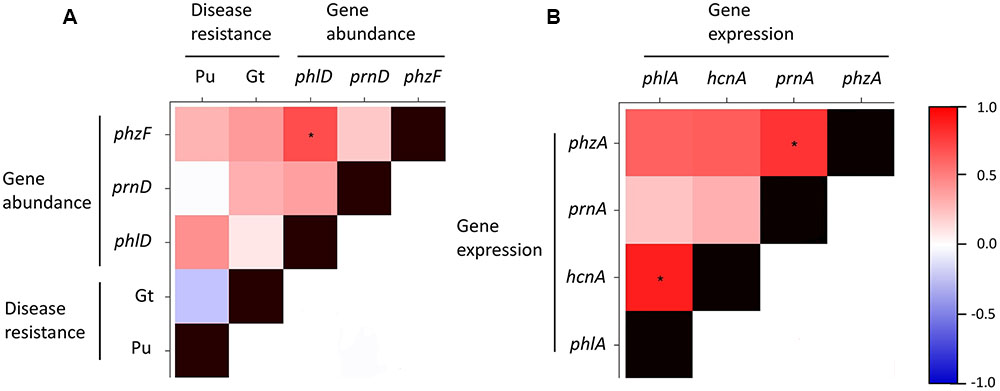
FIGURE 7. Heatmap showing Spearman’s rank correlations for disease resistance and abundance of antimicrobial genes (A) and for expression levels of antimicrobial genes (B) in 10 representative Swiss agricultural soils with a cereal-oriented cropping history. The abundance of antimicrobial genes is defined as the number of bacterial cells harboring the indicated gene. Significant correlations (p < 0.05) are highlighted with asterisks. The color scale to the right of the matrix indicates rho correlation coefficients.
Relationships between Soil Parameters, Pathogen Resistance, Abundance and Expression of Antimicrobial Genes
The physical and chemical properties of the 10 Swiss agricultural soils investigated in this study were analyzed (Figure 1 and Table 1) and correlated with their resistance to pathogens, the abundance of antimicrobial genes and the expression of antimicrobial genes (Figure 8). Macronutrients were extracted from soils with the water or ammonium acetate-EDTA (AA-EDTA) soil extraction procedures routinely used in Swiss agriculture to account for, respectively, soluble, i.e., readily plant-available macronutrients and bound, reserve macronutrients that become available to plants at mid or long term (Agroscope, 2006; Stünzi, 2007). Plant micronutrients were extracted with AA-EDTA.
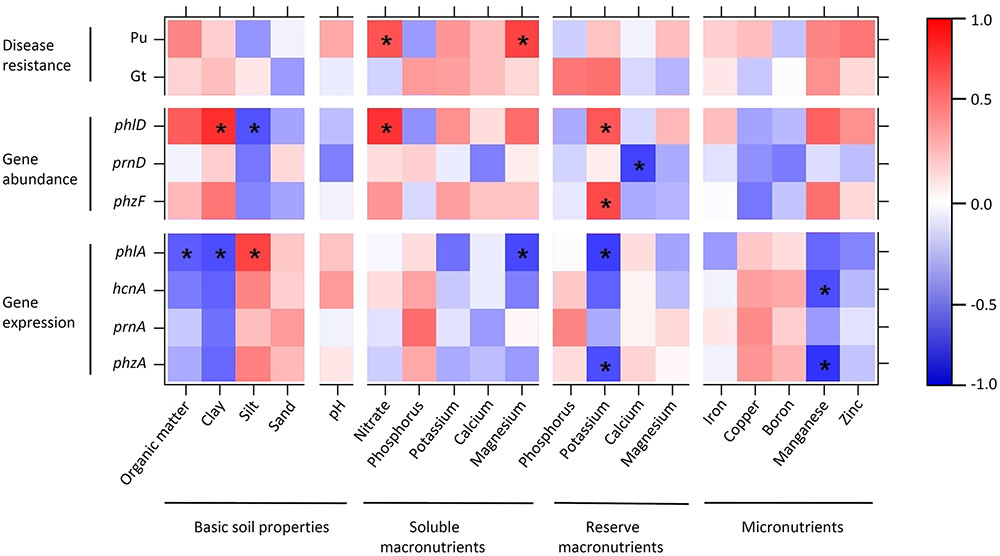
FIGURE 8. Heatmap showing Spearman’s rank correlations between soil parameters, disease resistance, and abundance and expression of antimicrobial genes in 10 representative Swiss agricultural soils with a cereal-oriented cropping history. The abundance of antimicrobial genes is defined as the number of bacterial cells harboring the indicated gene. Significant correlations (p < 0.05) are highlighted with asterisks. The color scale to the right of the matrix indicates rho correlation coefficients.
Organic carbon and clay, and silt and sand inversely influenced antimicrobial gene abundance and expression. Abundance of DAPG biosynthesis genes (recorded for phlD) was significantly positively correlated with clay and significantly negatively correlated with silt, while expression of these genes (recorded for phlA) was significantly negatively correlated with organic carbon and clay and significantly positively correlated with silt (Figure 8). Similar trends were also observed for the pyrrolnitrin and phenazine biosynthetic genes prnA and phzF, respectively. No clear positive or negative correlation was found between pH and antimicrobial gene abundance or expression (Figure 8). Nitrate concentration in soil was positively correlated with the abundance of bacteria harboring the investigated antimicrobial genes; in particular, it was significantly positively correlated with the abundance of phlD+ pseudomonads (Figure 8). Antimicrobial gene expression was neither clearly positively nor clearly negatively correlated with nitrate. Of the other macronutrients, potassium inversely influenced abundance and expression of antimicrobial genes. Reserve potassium extracted with AA-EDTA was significantly positively correlated with the abundance of phlD+ and phzF+ pseudomonads and significantly negatively with phlA and phzA expression (Figure 8). A similar trend was also observed for water-extracted, i.e., readily plant available magnesium. Among the measured micronutrients, a significant effect was only observed for manganese, which was significantly negatively correlated with phzA and hcnA expression (Figure 8).
Resistance to soilborne pathogens was not significantly correlated to any soil factor in the case of Gt, and significantly positively correlated to nitrate and magnesium extracted with water in the case of Pu (Figure 8). The resistance to both pathogens was positively, but not significantly correlated to organic carbon, clay, potassium, manganese, and zinc.
Taken together, these analyses indicate that soil physical and chemical properties have contrasting and subtle effects on the abundance and expression of antimicrobial genes. No pronounced correlations between soil properties and general disease resistance of soils could be observed for the 10 agricultural field sites investigated. Remarkably, all soil factors that were positively correlated with the abundance of phlD+ and phzF+ pseudomonads were also positively correlated with Pu resistance.
Discussion
Resistance to soilborne diseases and beneficial microbial populations involved have been studied extensively in soils specifically suppressive to one particular pathogen species (Weller et al., 2002; Mendes et al., 2011; Raaijmakers and Mazzola, 2016). However, virtually nothing is known about interactions between soilborne pathogens and beneficial microorganisms in common agricultural soils, i.e., in soils that lack specific disease suppressiveness. Similarly, it is not clear whether a particular soil may simultaneously exhibit suppressiveness toward multiple soilborne pathogens. To our best knowledge, the present study is the first to compare side-by-side a range of common agricultural soils for their resistance toward two soilborne pathogens, P. ultimum (Pu) and G. tritici (Gt), as well as for the abundance and expression of biosynthetic genes required for the production of antimicrobial compounds by plant-beneficial pseudomonads.
First, we investigated the capacity of different Swiss agricultural soils to buffer the attack of two soilborne pathogens by testing the growth of crop plants in these soils after amendment with increasing quantities of pathogens up to very high concentrations. Individual soils differed markedly in their respective resistance to the two pathogens. Several soils that showed comparatively high suppressiveness toward Pu were poorly or only moderately resistant to Gt and vice versa. These results indicate that conventional agricultural soils do not necessarily exhibit a general level of resistance toward a range of soilborne pathogens, but rather display variable resistance levels toward specific pathogens, which are likely modulated by different microbial and abiotic soil factors. Nevertheless, soils suppressive to more than one pathogen have been reported, notably the Fusarium wilt of pea suppressive soils in Mt. Vernon, WA, USA (Landa et al., 2002; Weller et al., 2007), which are also suppressive to take-all of wheat (Allende Molar, 2006; Allende-Molar and Weller, personal communication).
Soil resistance to soilborne pathogens has often been linked to the abundance of pseudomonads producing antimicrobial compounds (Stutz et al., 1986; Raaijmakers et al., 1997; Weller et al., 2002, 2007; Haas and Défago, 2005; Raaijmakers et al., 2008; Mazurier et al., 2009; Almario et al., 2014). For this reason, we have used a qPCR approach targeting biosynthetic genes for DAPG, PHZ, and PRN in bacterial cells present on roots of wheat grown in the agricultural soils that we tested for disease resistance. While the qPCR assays used in this study to quantify phlD+ and phzF+ bacteria are specific for fluorescent pseudomonads (see Supplementary Table S1), the assay used to quantify prnD+ cells additionally detects Burkholderia and Serratia (Garbeva et al., 2004). However, 16S rRNA amplicon sequencing performed on root samples from wheat grown in the same soils (Dennert et al., unpublished results) showed that the relative abundance of Pseudomonas (4.4–25.2%) was markedly higher than the relative abundances of Burkholderia (0.003–0.53%) and Serratia (0.003–1.21%). Therefore, we assume that the prnD genes detected in the present study predominantly derive from pseudomonads. Most studies that quantified pseudomonads harboring antimicrobial genes so far were carried out using cultivation-dependent approaches, e.g., colony plating/colony hybridization assays or endpoint dilution assays followed by PCR (Raaijmakers et al., 1997; Meyer et al., 2010; Mavrodi D.V. et al., 2012; Mavrodi O.V. et al., 2012). An examination of genome sequences published by Flury et al. (2016) and other investigators indicated that fluorescent pseudomonads harbor only one copy of phlD, phzF, or prnD per cell. Accordingly, we did not find any significant difference between the abundances of phlD and phzF quantified by qPCR or a cultivation-dependent endpoint dilution assay in samples from Taenikon (Supplementary Figure S4). Still, qPCR assays can potentially detect viable but non-culturable or even dead cells, thus caution is required when comparing our findings with results of cultivation-dependent experiments.
Studies simultaneously investigating the abundance of several antimicrobial genes in soil are rare. Raaijmakers et al. (1997) used a colony-hybridization assay to quantify pseudomonads harboring DAPG or PHZ genes in different take-all suppressive and conducive US soils. They found pseudomonads harboring DAPG genes to be enriched in suppressive soils, but much less abundant or below the detection limit in conducive soils and they did not detect pseudomonads harboring PHZ genes in any of the tested soils. In our study, we found phzF+ pseudomonads to be present in all investigated agricultural soils. However, in general their abundance was quite low, ranging from 2.5 to 5.3 log cells/g root, compared to another US study on soils of the Columbia Plateau of the Pacific Northwest, in which the abundance of PHZ-producing pseudomonads detected by endpoint-dilution assays coupled with phzF-specific PCR was up to 100-fold higher in certain soils (Mavrodi D.V. et al., 2012; Mavrodi O.V. et al., 2012). It has been suggested that PHZ producing pseudomonads are more abundant in dryland fields without irrigation, where yearly rainfall ranges from 150 to 300 mm, compared to irrigated fields (Mavrodi O.V. et al., 2012). At the sampling sites of the present study, average annual rainfall was high, ranging from 610 mm per year to 1780 mm per year, which could be a possible reason for the rather low abundance of phzF+ pseudomonads detected here.
The abundance of DAPG producing pseudomonads in the investigated soils varied strongly, from 2.2 to 8.0 log cells/g root. We detected higher but also lower numbers compared to previous studies, which may be explained by the very different types of soils we investigated (Table 1). Two studies using terminal endpoint dilution assays followed by phlD-specific PCR, one by Meyer et al. (2010) on two Swiss agricultural soils and one by Mavrodi O.V. et al. (2012) on irrigated fields of the Pacific Northwest, detected phlD+ pseudomonads at levels ranging from 4.5 to 6.5 log CFU/g on roots of wheat. The population levels of pseudomonads on roots of wheat grown in the Delley soil reported by Meyer et al. (2010) were similar to the numbers of phlD+ pseudomonads detected by qPCR in our study. Bacteria harboring prnD were detected in the 10 Swiss soils investigated here at abundances comparable to those found in different types of agricultural soils in a previous study (Garbeva et al., 2004).
The abundance of antimicrobial genes, respectively, of the bacteria harboring these genes, does only reflect the bacterial population that potentially could produce a particular antimicrobial compound but is not useful to identify the conditions that favor the proliferation and the consequent niche domination for a given bacterial species, as well as the production of specific antimicrobials in situ. The present study is the first to monitor side-by-side the expression of several biosynthetic genes for antimicrobial compounds in different agricultural soils, i.e., in particular phlA, hcnA, prnA, and phzA, reflecting the biosynthesis of DAPG, HCN, pyrrolnitrin, and phenazines (Baehler et al., 2005; Chin-A-Woeng et al., 2005; Rochat et al., 2010). This was done by FACS-based monitoring of dual-labeled Pseudomonas reporter strains that carry a GFP cell tag and a mCherry-based reporter allowing to record the relative expression of a specific antimicrobial gene in individual cells of a Pseudomonas population. A similar technique was used by de Werra et al. (2008) and Rochat et al. (2010) for measuring the expression of phlA, hcnA, and prnA genes by Pseudomonas reporter strains on roots of different plant varieties in soilless systems. However, this is the first time that the combination of FACS and fluorescent reporter strains was applied to measure antimicrobial gene expression on plant roots grown in different natural soils. Moreover, to our best knowledge, there were no reports so far on phenazine gene expression in soil or on plant roots. To date, only a handful of studies attempted to determine the expression of antimicrobial genes of pseudomonads in soil (DeCoste et al., 2011; Novinscak and Filion, 2011; Almario et al., 2013b), mainly because of practical challenges. In our study, our initial attempts of tracking fluorescence of reporter strains extracted from roots and soil with microscopy revealed impracticable because of the strong autofluorescence of the investigated soil and root material. To address this problem, we modified the FACS approach of Rochat et al. (2010) and used GFP as a constitutively expressed cell marker and mCherry-based reporters for monitoring antimicrobial gene expression. This approach allowed to specifically track the Pseudomonas reporter cells and their expression of select antimicrobial genes in natural soil. However, due to the lack of suitable fluorescent proteins allowing reliable visualization of bacteria cells and gene expression in natural soil, a major limitation of this method is that it is impossible to monitor the expression of two or more antimicrobial genes in the same bacterial cell simultaneously. An alternative method of measuring gene expression in soils involves the extraction of total RNA followed by quantitative reverse transcription PCR of bacterial transcripts of interest. This method was used to quantify the expression of DAPG and HCN biosynthetic genes in natural and artificial soils (DeCoste et al., 2011; Novinscak and Filion, 2011; Almario et al., 2013b). However, the main problem with this approach is the inefficiency of RNA extraction from a complex material such as soil along with the limited abundance of transcripts for antimicrobial genes.
We observed an interesting trend where in soils from Cadenazzo and Vouvry the expression of the four studied genes was markedly enhanced, while lower levels of gene expression occurred in soils from Eschikon, Grangeneuve, and Taenikon. The observation that a specific group of soils is capable of promoting or hampering the expression of four distinct antimicrobial genes in pseudomonads suggests that something in the abiotic or biotic composition of these soils can modulate the production of corresponding plant-beneficial compounds in the P. protegens and P. chlororaphis reporter strains. Furthermore, we observed that the soil type also influenced the capacity of both pseudomonads to colonize wheat, e.g., plants grown in soils from Cadenazzo and Vouvry supported higher populations than those grown in the Eschikon soil (Figure 5). However, the differences in root colonization by the two pseudomonads do not explain entirely the difference in gene expression observed in the different soils (Figures 4, 5 and Supplementary Table S3).
The relative importance of the abundance of Pseudomonas spp. for suppression of soilborne pathogens was questioned recently (Kyselkova et al., 2014). Indeed, DAPG and HCN producing pseudomonads could be isolated not only from suppressive but also from conducive soils, leading to the hypothesis that differential environmental factors prevailing in the two types of soils may shape the expression of relevant biocontrol genes in Pseudomonas bacteria and thus disease suppression (Ramette et al., 2006). Moreover, the abundance of the DAPG biosynthesis gene phlD was not indicative of the resistance of soils to black root rot of tobacco (Almario et al., 2013b; Kyselkova et al., 2014). Our results support the hypothesis that the abundance of pseudomonads producing antimicrobial metabolites may, in fact, be less important for disease suppression in the rhizosphere than previously hypothesized, at least for certain soils. We failed to detect any significant positive correlation between the resistances to the two soilborne pathogens Pu or Gt and the abundance of Pseudomonas harboring antimicrobial genes in the investigated agricultural soils (Figure 7). The lack of a significant positive correlation between disease resistance and abundance of antimicrobial pseudomonads could also be because only the phlD+ bacteria were detected at more than 105 cells/g root, which is close to the threshold considered to be relevant for biocontrol activity (Raaijmakers et al., 1999; Haas and Défago, 2005) in many soils. By analogy, the populations of PHZ and PRN producing pseudomonads might therefore also be too low to contribute to efficient pathogen suppression in the soils of our study. Although correlations between gene abundances and disease resistance were never significant, it is still worth to note that for both pathogens they were mostly positive (Figure 7A). The expression of antimicrobial genes by reporter strains cannot be directly correlated to pathogen suppression data in the present study. Nevertheless, our findings suggest that high expression levels of antimicrobial genes in a particular soil may not necessarily be indicative of high levels of pathogen suppression (Figure 6). Furthermore, soils supporting the high expression of antimicrobial genes mostly harbored lower numbers of antimicrobials-producing pseudomonads (Figure 6). We thus speculate that the specific biotic and abiotic factors operating in the distinct agricultural soils might differently influence the abundance and expression of antimicrobial genes. At the present stage, it remains therefore elusive to which extent and how exactly pseudomonads producing DAPG, PHZ, and PRN contribute to the disease resistance of the investigated agricultural soils. Moreover, only two soilborne pathogens were investigated here and individual soils strongly differed in their response to Pu and Gt. It is likely that different relations to gene abundances and expression would be found for other pathogen and plant species.
Soil nutrients are known to be important factors influencing resistance to pathogens (Martin and Hancock, 1986; Löbmann et al., 2016) and the abundance of pseudomonads producing antimicrobial compounds (Meyer et al., 2010; Kim et al., 2013). Recently, Löbmann et al. (2016) found that soils can influence the resistance to Pu in two ways: through abiotic effects that inhibit pathogen growth, which was the case in soils with a high pH, high calcium, and high clay content; or through balanced nutrient contents, which were hypothesized to stimulate the proliferation of plant-beneficial microorganisms. Löbmann et al. (2016) postulated this effect in soils with high P, K, Mg, sand and organic matter contents, although the involved plant-beneficial microorganisms were not identified in their study. In our study, we found significant positive correlations between Pu resistance and the contents of nitrate and Mg in soil. However, the same two nutrients were also positively correlated with the abundance of the DAPG biosynthesis gene phlD, so we cannot conclude on how exactly these nutrients have a positive impact on soil resistance. They could have a negative effect on the pathogen, have a positive effect on the growth of the plants and increase their pathogen resistance, or have an indirect positive effect on soil resistance by promoting beneficial bacteria.
We found no significant correlation between pH and abundance of Pseudomonas harboring antimicrobial genes, probably because the soils of this study all had a pH close to neutral and ranged from 6.0 (Cadenazzo) to 7.7 (Cazis) (Table 1). The influence of pH of agricultural soils on the population sizes of pseudomonads producing antimicrobial compounds is poorly understood. Previously, indigenous pseudomonads were found to be more abundant in soils with a neutral pH than in soils with an acidic pH (Kim et al., 2013). Likewise, P. protegens CHA0 inoculated into soils with an acidic pH reached lower population sizes compared to when it was inoculated into soils with neutral or basic pH (Mascher et al., 2013).
We obtained contrasting results for clay and silt, where clay was positively and silt negatively correlated with gene abundance (Figure 8). For gene expression, the opposite was the case. The precise relation between Pseudomonas abundance and clay content is not known. However, high clay content in soil was previously shown to reduce the expression of the HCN biosynthetic gene hcnC (Novinscak and Filion, 2011) and the biocontrol activity of phenazine-producing strains (Ownley et al., 2003). The pronounced effect of clay on biocontrol activity of pseudomonads was also observed in studies carried out in artificial soils (Keel et al., 1989; Almario et al., 2013b, 2014). In particular, vermiculitic clay supported a higher level of biocontrol activity and HCN production than illitic clay (Keel et al., 1989), and the expression of phlA was greater in the presence of vermiculite than in the presence of illite (Almario et al., 2013b).
The plant macronutrients nitrate, potassium and magnesium also inversely influenced abundance and expression of antimicrobial genes. To the best of our knowledge, it has not been investigated if high macronutrient contents in the soil directly stimulate the growth of antimicrobial pseudomonads. We hypothesize that the positive correlation between abundance of antimicrobial pseudomonads and certain macronutrients such as nitrate could also be due to an indirect effect, e.g., via stimulation of plant growth and increased root exudation. Support for this hypothesis comes from a recent study on maize roots, where nitrogen concentration in soil was found to be positively correlated with plant root exudation and abundance of rhizosphere bacteria (Zhu et al., 2016). For the micronutrients, a significant, though negative, correlation was found only for manganese and the expression of the HCN and PHZ biosynthetic genes (Figure 8). However, in previous studies, several micronutrients were described as factors that affect abundance, gene expression, metabolite production and biocontrol activity in pseudomonads. For instance, copper negatively influenced the abundance of Pseudomonas spp. in agricultural soils (Brandt et al., 2006), while the bioavailability of iron affected the expression of phlA and the production of HCN in artificial soils (Keel et al., 1989; Almario et al., 2013b). The biocontrol activity of phenazine-producing strains was positively correlated to zinc and negatively to iron and manganese levels in soil (Ownley et al., 2003). Moreover, the production of several antimicrobial metabolites, notably DAPG, pyoluteorin and PRN by strain P. protegens CHA0 was stimulated by zinc, but these experiments were performed in culture media (Duffy and Défago, 1997, 1999). The lack of significant correlations between micronutrient contents in soil and abundance and expression of antimicrobial genes in our study could be due to the relatively high micronutrient content in the sampled soils. In fact, all studied soils had sufficient amounts of micronutrients according to the classification for farmers approved by the Swiss government (Flisch et al., 2009). Boron concentration was an exception, as three out of 10 sampled soils tested as poor (Table 1).
Conclusion
Results of this study suggest that resistance of soils to pathogens, and abundance and expression of antimicrobial genes are not generally positively or negatively correlated in a wide range of diverse agricultural soils. Complex interactions depending on the host–pathogen system and the soil composition determine pathogen resistance of soils and the abundance and expression of antimicrobial genes. The abundance of antimicrobial metabolites producing pseudomonads in the investigated agricultural soils and the expression of biosynthetic genes for these compounds as studied here using reporter strains seem to be differentially shaped by multiple soil factors. This could explain, at least in part, why soils that sustain high numbers of these bacteria, often support only low levels of antimicrobial gene expression and vice versa. Therefore, to better understand the links between soil characteristics and abundance and expression of antimicrobial genes in pseudomonads, future studies should include extreme soils (i.e., highly acidic or alkaline soils or soils enriched in or depleted of specific nutrients). Pseudomonads are probably only one among many microbial groups determining the natural pathogen tolerance or resistance of agricultural soils (Mendes et al., 2011; Kyselkova et al., 2014; Cha et al., 2016; Raaijmakers and Mazzola, 2016). In future work, the potential of Pseudomonas bacteria as bio-indicators of soil resistance has to be re-evaluated with care. It will probably be difficult to identify specific groups of microorganisms as general indicators of soil health and disease resistance, since natural disease suppression likely requires individual compositions of the beneficial microbiota depending on the soil type, the crop species, the soilborne disease and maybe even the cropping system.
Author Contributions
NI, FD, MM, and CK designed the research. MM and CK supervised the study. MF and FM organized the soil sampling and soil analysis. FD performed pathogen resistance greenhouse experiments. FD, DM, and OM developed the qPCR assays. FD performed the DNA extractions and qPCR assays. NI developed the reporter strains and performed the gene expression experiments. FD and NI analyzed the data. TL, JS, MW, and CV assisted with experiments and/or data evaluation. FD, NI, MM, and CK wrote the manuscript. All authors critically revised the manuscript and approved the final version.
Funding
This study was funded by the National Research Program 68 ‘Sustainable use of soil as a resource’ of the Swiss National Science Foundation (Grant no. NRP68 406840_143141 awarded to MM and CK and Grant no. NRP68 406840 143065 awarded to Ted Turlings and FM) and a travel grant for doctoral students of the Walter Hochstrasser Foundation, ETH Zürich, awarded to FD.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Michele Gusberti and Karent P. Kupferschmied for technical assistance with fieldwork and greenhouse assays. We thank Linda S. Thomashow and David M. Weller (USDA-ARS Wheat Health, Genetics and Quality Research Unit, Washington State University, Pullman, WA, USA) for kindly providing access to the PHZ+ strain collection and for information about Gaeumannomyces tritici inoculum preparation. We thank Alain Sarniguet (INRA, Le Rheu, France) for kindly providing G. tritici strain I-17. qPCR data were obtained at the Genetic Diversity Centre (GDC), ETH Zürich. We thank farmers and research station managers for access to their fields: Mario Bertossa (Cadenazzo), Padruot Salzgeber (Cazis), Michel Petitat (Courtedoux), Karl Camp (Delley), Hanspeter Renfer (Eschikon), André Chassot (Grangeneuve), Thomas Anken (Taenikon), Jürg Hiltbrunner (Utzenstorf), and Quentin Lassueur (Vouvry).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00427/full#supplementary-material
Footnotes
- ^http://www.meteoswiss.admin.ch/home/climate/past/homogenous-monthly-data.html
- ^https://www.ncbi.nlm.nih.gov/genbank/
References
Achkar, J., Xian, M., Zhao, H., and Frost, J. (2005). Biosynthesis of phloroglucinol. J. Am. Chem. Soc. 127, 5332–5333. doi: 10.1021/ja042340g
Agroscope (2006). Schweizerische Referenzmethoden der Eidgenössischen landwirtschaftlichen Forschungsanstalten, Vol. 1, Bern-Liebefeld.
Alabouvette, C. (1986). Fusarium-wilt suppressive soils from the Châteaurenard region: review of a 10-year study. Agronomie 6, 273–284. doi: 10.1051/agro:19860307
Allende Molar, R. (2006). Role of 2,4-Diacetylphloroglucinol-Producing Pseudomonas fluorescens in the Suppression of Take-All and Pythium Root Rot of Wheat. Ph.D. thesis, Washington State University, Pullman, WA.
Almario, J., Moënne-Loccoz, Y., and Muller, D. (2013a). Monitoring of the relation between 2,4-diacetylphloroglucinol-producing Pseudomonas and Thielaviopsis basicola populations by real-time PCR in tobacco black root-rot suppressive and conducive soils. Soil Biol. Biochem. 57, 144–155. doi: 10.1016/j.soilbio.2012.09.003
Almario, J., Muller, D., Défago, G., and Moënne-Loccoz, Y. (2014). Rhizosphere ecology and phytoprotection in soils naturally suppressive to Thielaviopsis black root rot of tobacco. Environ. Microbiol. 16, 1949–1960. doi: 10.1111/1462-2920.12459
Almario, J., Prigent-Combaret, C., Muller, D., and Moënne-Loccoz, Y. (2013b). Effect of clay mineralogy on iron bioavailability and rhizosphere transcription of 2,4-diacetylphloroglucinol biosynthetic genes in biocontrol Pseudomonas protegens. Mol. Plant Microbe Interact. 26, 566–574. doi: 10.1094/MPMI-11-12-0274-R
Baehler, E., Bottiglieri, M., Péchy-Tarr, M., Maurhofer, M., and Keel, C. (2005). Use of green fluorescent protein-based reporters to monitor balanced production of antifungal compounds in the biocontrol agent Pseudomonas fluorescens CHA0. J. Appl. Microbiol. 99, 24–38. doi: 10.1111/j.1365-2672.2005.02597.x
Bangera, M. G., and Thomashow, L. S. (1996). Characterization of a genomic locus required for synthesis of the antibiotic 2,4-diacetylphloroglucinol by the biological control agent Pseudomonas fluorescens Q2-87. Mol. Plant Microbe Interact. 9, 83–90. doi: 10.1094/MPMI-9-0083
Bao, Y., Lies, D. P., Fu, H., and Roberts, G. P. (1991). An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109, 167–168. doi: 10.1016/0378-1119(91)90604-A
Berendsen, R. L., Pieterse, C. M., and Bakker, P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Bertani, G. (1951). Studies on lysogenesis I.: the mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62, 293–300.
Bithell, S. L., McKay, A., Butler, R. C., Herdina, Ophel-Keller, K., Hartley, D., et al. (2012). Predicting take-all severity in second-year wheat using soil DNA concentrations of Gaeumannomyces graminis var. tritici determined with qPCR. Plant Dis. 96, 443–451. doi: 10.1094/pdis-05-11-0445
Blankenfeldt, W., Kuzin, A. P., Skarina, T., Korniyenko, Y., Tong, L., Bayer, P., et al. (2004). Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens. Proc. Natl. Acad. Sci. U.S.A. 101, 16431–16436. doi: 10.1073/pnas.0407371101
Blumer, C., and Haas, D. (2000). Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis. Arch. Microbiol. 173, 170–177. doi: 10.1007/s002039900127
Brandt, K. K., Petersen, A., Holm, P. E., and Nybroe, O. (2006). Decreased abundance and diversity of culturable Pseudomonas spp. populations with increasing copper exposure in the sugar beet rhizosphere. FEMS Microbiol. Ecol. 56, 281–291. doi: 10.1111/j.1574-6941.2006.00081.x
Cha, J. Y., Han, S., Hong, H. J., Cho, H., Kim, D., Kwon, Y., et al. (2016). Microbial and biochemical basis of a Fusarium wilt-suppressive soil. ISME J. 10, 119–129. doi: 10.1038/ismej.2015.95
Chin-A-Woeng, T. E. C., Bloemberg, G. V., and Lugtenberg, B. J. J. (2003). Phenazines and their role in biocontrol by Pseudomonas bacteria. New Phytol. 157, 503–523. doi: 10.1046/j.1469-8137.2003.00686.x
Chin-A-Woeng, T. F., Bloemberg, G. V., van der Bij, A. J., van der Drift, K. M., Schripsema, J., Kroon, B., et al. (1998). Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. Mol. Plant Microbe Interact. 11, 1069–1077. doi: 10.1094/MPMI.1998.11.11.1069
Chin-A-Woeng, T. E. C., Thomas-Oates, J. E., Lugtenberg, B. J. J., and Bloemberg, G. V. (2001). Introduction of the phzH gene of Pseudomonas chlororaphis PCL1391 extends the range of biocontrol ability of phenazine-1-carboxylic acid producing Pseudomonas spp. strains. Mol. Plant Microbe Interact. 14, 1006–1015. doi: 10.1094/MPMI.2001.14.8.1006
Chin-A-Woeng, T. F., van den Broek, D., Lugtenberg, B. J., and Bloemberg, G. V. (2005). The Pseudomonas chlororaphis PCL1391 sigma regulator psrA represses the production of the antifungal metabolite phenazine-1-carboxamide. Mol. Plant Microbe Interact. 18, 244–253. doi: 10.1094/MPMI-18-0244
Chng, S., Cromey, M. G., Dodd, S. L., Stewart, A., Butler, R. C., and Jaspers, M. V. (2015). Take-all decline in New Zealand wheat soils and the microorganisms associated with the potential mechanisms of disease suppression. Plant Soil 397, 239–259. doi: 10.1007/s11104-015-2620-4
Cook, J. R., and Rovira, A. D. (1976). The role of bacteria in the biological control of Gaeumannomyces graminis by suppressive soils. Soil Biol. Biochem. 8, 269–273. doi: 10.1016/0038-0717(76)90056-0
Cullen, D. W., Toth, I. K., Boonham, N., Walsh, K., Barker, I., and Lees, A. K. (2007). Development and validation of conventional and quantitative polymerase chain reaction assays for the detection of storage rot potato pathogens, Phytophthora erythroseptica, Pythium ultimum and Phoma foveata. J. Phytopathol. 155, 309–315. doi: 10.1111/j.1439-0434.2007.01233.x
de Werra, P., Baehler, E., Huser, A., Keel, C., and Maurhofer, M. (2008). Detection of plant-modulated alterations in antifungal gene expression in Pseudomonas fluorescens CHA0 on roots by flow cytometry. Appl. Environ. Microbiol. 74, 1339–1349. doi: 10.1128/AEM.02126-07
de Werra, P., Huser, A., Tabacchi, R., Keel, C., and Maurhofer, M. (2011). Plant- and microbe-derived compounds affect the expression of genes encoding antifungal compounds in a pseudomonad with biocontrol activity. Appl. Environ. Microbiol. 77, 2807–2812. doi: 10.1128/AEM.01760-10
DeCoste, N. J., Gadkar, V. J., and Filion, M. (2011). Relative and absolute quantitative real-time PCR-based quantifications of hcnC and phlD gene transcripts in natural soil spiked with Pseudomonas sp. strain LBUM300. Appl. Environ. Microbiol. 77, 41–47. doi: 10.1128/AEM.01387-10
Duffy, B. K., and Défago, G. (1997). Zinc improves biocontrol of Fusarium crown and root rot of tomato by Pseudomonas fluorescens and represses the production of pathogen metabolites inhibitory to bacterial antibiotic biosynthesis. Phytopathology 87, 1250–1257. doi: 10.1094/PHYTO.1997.87.12.1250
Duffy, B. K., and Défago, G. (1999). Environmental factors modulating antibiotic and siderophore biosynthesis by Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 65, 2429–2438.
Flisch, R., Sinaj, S., Charles, R., and Richner, W. (2009). GRUDAF 2009. Principles for fertilisation in arable and fodder production (in German). Agrarforschung 16, 1–100.
Flury, P., Aellen, N., Ruffner, B., Péchy-Tarr, M., Fataar, S., Metla, Z., et al. (2016). Insect pathogenicity in plant-beneficial pseudomonads: phylogenetic distribution and comparative genomics. ISME J. 10, 2527–2542. doi: 10.1038/ismej.2016.5
Flury, P., Vesga, P., Péchy-Tarr, M., Aellen, N., Dennert, F., Hofer, N., et al. (2017). Antimicrobial and insecticidal: cyclic lipopeptides and hydrogen cyanide produced by plant-beneficial Pseudomonas strains CHA0, CMR12a and PCL1391 contribute to insect killing. Front. Microbiol. 8:100. doi: 10.3389/fmicb.2017.00100
Frapolli, M., Défago, G., and Moënne-Loccoz, Y. (2010). Denaturing gradient gel electrophoretic analysis of dominant 2,4-diacetylphloroglucinol biosynthetic phlD alleles in fluorescent Pseudomonas from soils suppressive or conducive to black root rot of tobacco. Soil Biol. Biochem. 42, 649–656. doi: 10.1016/j.soilbio.2010.01.005
Garbeva, P., Postma, J., Van Veen, J., and Van Elsas, J. (2006). Effect of above-ground plant species on soil microbial community structure and its impact on suppression of Rhizoctonia solani AG3. Environ. Microbiol. 8, 233–246. doi: 10.1111/j.1462-2920.2005.00888.x
Garbeva, P., Voesenek, K., and Elsas, J. D. (2004). Quantitative detection and diversity of the pyrrolnitrin biosynthetic locus in soil under different treatments. Soil Biol. Biochem. 36, 1453–1463. doi: 10.1016/j.soilbio.2004.03.009
Haas, D., and Défago, G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3, 307–319. doi: 10.1038/nrmicro1129
Hernández-Restrepo, M., Groenewald, J. Z., Elliott, M. L., Canning, G., McMillan, V. E., and Crous, P. W. (2016). Take-all or nothing. Stud. Mycol. 83, 19–48. doi: 10.1016/j.simyco.2016.06.002
Höfte, M., and Altier, N. (2010). Fluorescent pseudomonads as biocontrol agents for sustainable agricultural systems. Res. Microbiol. 161, 464–471. doi: 10.1016/j.resmic.2010.04.007
Hwang, J., Chilton, W., and Benson, D. (2002). Pyrrolnitrin production by Burkholderia cepacia and biocontrol of Rhizoctonia stem rot of poinsettia. Biol. Control 25, 56–63. doi: 10.1016/S1049-9644(02)00044-0
Jousset, A., Rochat, L., Lanoue, A., Bonkowski, M., Keel, C., and Scheu, S. (2011). Plants respond to pathogen infection by enhancing the antifungal gene expression of root-associated bacteria. Mol. Plant Microbe Interact. 24, 352–358. doi: 10.1094/MPMI-09-10-0208
Jousset, A., Rochat, L., Scheu, S., Bonkowski, M., and Keel, C. (2010). Predator-prey chemical warfare determines the expression of biocontrol genes by rhizosphere-associated Pseudomonas fluorescens. Appl. Environ. Microbiol. 76, 5263–5268. doi: 10.1128/AEM.02941-09
Jousset, A., Schuldes, J., Keel, C., Maurhofer, M., Daniel, R., Scheu, S., et al. (2014). Full-genome sequence of the plant growth-promoting bacterium Pseudomonas protegens CHA0. Genome Announc. 2:e322-14. doi: 10.1128/genomeA.00322-14
Keel, C., Schnider, U., Maurhofer, M., Voisard, C., Laville, J., Burger, U., et al. (1992). Suppression of root diseases by Pseudomonas fluorescens CHA0: importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol. Mol. Plant Microbe Interact. 5, 4–13. doi: 10.1094/MPMI-5-004
Keel, C., Voisard, C., Berling, C., Kahr, G., and Défago, G. (1989). Iron sufficiency, a prerequisite for the suppression of tobacco black root rot by Pseudomonas fluorescens strain CHA0 under gnotobiotic conditions. Phytopathology 79, 584–589. doi: 10.1094/Phyto-79-584
Kim, J.-S., Mele, P. M., and Crowley, D. E. (2013). Application of PCR primer sets for detection of Pseudomonas sp. functional genes in the plant rhizosphere. J. Agric. Chem. Environ. 2, 8–15. doi: 10.4236/jacen.2013.21002
King, E. O., Ward, M. K., and Raney, D. E. (1954). Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44, 301–307.
Kirner, S., Hammer, P. E., Hill, D. S., Altmann, A., Fischer, I., Weislo, L. J., et al. (1998). Functions encoded by pyrrolnitrin biosynthetic genes from Pseudomonas fluorescens. J. Bacteriol. 180, 1939–1943.
Klein, E., Ofek, M., Katan, J., Minz, D., and Gamliel, A. (2013). Soil suppressiveness to fusarium disease: shifts in root microbiome associated with reduction of pathogen root colonization. Phytopathology 103, 23–33. doi: 10.1094/PHYTO-12-11-0349
Koch, B., Jensen, L. E., and Nybroe, O. (2001). A panel of Tn7-based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into Gram-negative bacteria at a neutral chromosomal site. J. Microbiol. Methods 45, 187–195. doi: 10.1016/S0167-7012(01)00246-9
Kyselkova, M., Almario, J., Kopecky, J., Sagova-Mareckova, M., Haurat, J., Muller, D., et al. (2014). Evaluation of rhizobacterial indicators of tobacco black root rot suppressiveness in farmers’ fields. Environ. Microbiol. Rep. 6, 346–353. doi: 10.1111/1758-2229.12131
Landa, B. B., Mavrodi, O. V., Raaijmakers, J. M., McSpadden Gardener, B. B., Thomashow, L. S., and Weller, D. M. (2002). Differential ability of genotypes of 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens strains to colonize the roots of pea plants. Appl. Environ. Microbiol. 68, 3226–3237. doi: 10.1128/AEM.68.7.3226-3237.2002
Latz, E., Eisenhauer, N., Scheu, S., and Jousset, A. (2015). Plant identity drives the expression of biocontrol factors in a rhizosphere bacterium across a plant diversity gradient. Funct. Ecol. 29, 1225–1234. doi: 10.1111/1365-2435.12417
Lebreton, L., Gosme, M., Lucas, P., Guillerm-Erckelboudt, A. Y., and Sarniguet, A. (2007). Linear relationship between Gaeumannomyces graminis var. tritici (Ggt) genotypic frequencies and disease severity on wheat roots in the field. Environ. Microbiol. 9, 492–499. doi: 10.1111/j.1462-2920.2006.01166.x
Lemanceau, P., Maurhofer, M., and Défago, G. (2006). “Contribution of studies on suppressive soils to the identification of bacterial biocontrol agents and to the knowledge of their modes of action,” in Plant-Associated Bacteria, ed. S. S. Gnanamanickam (Dordrecht: Springer), 231–267. doi: 10.1007/978-1-4020-4538-7_7
Löbmann, M. T., Vetukuri, R. R., de Zinger, L., Alsanius, B. W., Grenville-Briggs, L. J., and Walter, A. J. (2016). The occurrence of pathogen suppressive soils in Sweden in relation to soil biota, soil properties, and farming practices. Appl. Soil Ecol. 107, 57–65. doi: 10.1016/j.apsoil.2016.05.011
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Martin, F. N., and Hancock, J. G. (1986). Association of chemical and biological factors in soils suppressive to Pythium ultimum. Phytopathology 76, 1221–1231. doi: 10.1094/Phyto-76-1221
Mascher, F., Hase, C., Bouffaud, M. L., Défago, G., and Moënne-Loccoz, Y. (2013). Cell culturability of Pseudomonas protegens CHA0 depends on soil pH. FEMS Microbiol. Ecol. 87, 441–450. doi: 10.1111/1574-6941
Maurhofer, M., Baehler, E., Notz, R., Martinez, V., and Keel, C. (2004). Cross talk between 2,4-diacetylphloroglucinol-producing biocontrol pseudomonads on wheat roots. Appl. Environ. Microbiol. 70, 1990–1998. doi: 10.1128/AEM.70.4.1990-1998.2004
Maurhofer, M., Keel, C., Schnider, U., Voisard, C., Haas, D., and Défago, G. (1992). Influence of enhanced antibiotic production in Pseudomonas fluorescens strain CHA0 on its disease suppressive capacity. Phytopathology 82, 190–195. doi: 10.1094/Phyto-82-190
Mavrodi, D. V., Ksenzenko, V. N., Bonsall, R. F., Cook, R. J., Boronin, A. M., and Thomashow, L. S. (1998). A seven-gene locus for synthesis of phenazine-1-carboxylic acid by Pseudomonas fluorescens 2-79. J. Bacteriol. 180, 2541–2548.
Mavrodi, D. V., Mavrodi, O. V., Parejko, J. A., Bonsall, R. F., Kwak, Y.-S., Paulitz, T. C., et al. (2012). Accumulation of the antibiotic phenazine-1-carboxylic acid in the rhizosphere of dryland cereals. Appl. Environ. Microbiol. 78, 804–812. doi: 10.1128/AEM.06784-11
Mavrodi, D. V., Parejko, J. A., Mavrodi, O. V., Kwak, Y. S., Weller, D. M., Blankenfeldt, W., et al. (2013). Recent insights into the diversity, frequency and ecological roles of phenazines in fluorescent Pseudomonas spp. Environ. Microbiol. 15, 675–686. doi: 10.1111/j.1462-2920.2012.02846.x
Mavrodi, O. V., Mavrodi, D. V., Parejko, J. A., Thomashow, L. S., and Weller, D. M. (2012). Irrigation differentially impacts populations of indigenous antibiotic-producing Pseudomonas spp. in the rhizosphere of wheat. Appl. Environ. Microbiol. 78, 3214–3220. doi: 10.1128/AEM.07968-11
Mazurier, S., Corberand, T., Lemanceau, P., and Raaijmakers, J. M. (2009). Phenazine antibiotics produced by fluorescent pseudomonads contribute to natural soil suppressiveness to Fusarium wilt. ISME J. 3, 977–991. doi: 10.1038/ismej.2009.33
Mendes, R., Garbeva, P., and Raaijmakers, J. M. (2013). The rhizosphere microbiome: significance of plant-beneficial, plant-pathogenic and human-pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Mendes, R., Kruijt, M., de Bruijn, I., Dekkers, E., van der Voort, M., Schneider, J. H., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100. doi: 10.1126/science.1203980
Mercado-Blanco, J., and Bakker, P. A. H. M. (2007). Interactions between plants and beneficial Pseudomonas spp.: exploiting bacterial traits for crop protection. Antonie van Leeuwenhoek 92, 367–389. doi: 10.1007/s10482-007-9167-1
Meyer, J. B., Lutz, M. P., Frapolli, M., Pechy-Tarr, M., Rochat, L., Keel, C., et al. (2010). Interplay between wheat cultivars, biocontrol pseudomonads, and soil. Appl. Environ. Microbiol. 76, 6196–6204. doi: 10.1128/AEM.00752-10
Nesemann, K., Braus-Stromeyer, S. A., Thuermer, A., Daniel, R., Mavrodi, D. V., Thomashow, L. S., et al. (2015). Draft genome sequence of the phenazine-producing Pseudomonas fluorescens strain 2-79. Genome Announc. 3:e130-15. doi: 10.1128/genomeA.00130-15
Notz, R., Maurhofer, M., Dubach, H., Haas, D., and Défago, G. (2002). Fusaric acid-producing strains of Fusarium oxysporum alter 2,4-diacetylphloroglucinol biosynthetic gene expression in Pseudomonas fluorescens CHA0 in vitro and in the rhizosphere of wheat. Appl. Environ. Microbiol. 68, 2229–2235. doi: 10.1128/AEM.68.5.2229-2235.2002
Notz, R., Maurhofer, M., Schnider-Keel, U., Duffy, B., Haas, D., and Défago, G. (2001). Biotic factors affecting expression of the 2,4-diacetylphloroglucinol biosynthesis gene phlA in Pseudomonas fluorescens biocontrol strain CHA0 in the rhizosphere. Phytopathology 91, 873–881. doi: 10.1094/PHYTO.2001.91.9.873
Novinscak, A., and Filion, M. (2011). Effect of soil clay content on RNA isolation and on detection and quantification of bacterial gene transcripts in soil by quantitative reverse transcription-PCR. Appl. Environ. Microbiol. 77, 6249–6252. doi: 10.1128/AEM.00055-11
Ownley, B. H., Duffy, B. K., and Weller, D. M. (2003). Identification and manipulation of soil properties to improve the biological control performance of phenazine-producing Pseudomonas fluorescens. Appl. Environ. Microbiol. 69, 3333–3343. doi: 10.1128/aem.69.6.3333-3343.2003
Péchy-Tarr, M., Bottiglieri, M., Mathys, S., Lejbølle, K. B., Schnider-Keel, U., Maurhofer, M., et al. (2005). RpoN (σ54) controls production of antifungal compounds and biocontrol activity in Pseudomonas fluorescens CHA0. Mol. Plant Microbe Interact. 18, 260–272. doi: 10.1094/MPMI-18-0260
Péchy-Tarr, M., Borel, N., Kupferschmied, P., Turner, V., Binggeli, O., Radovanovic, D., et al. (2013). Control and host-dependent activation of insect toxin expression in a root-associated biocontrol pseudomonad. Environ. Microbiol. 15, 736–750. doi: 10.1111/1462-2920.12050
Pierson, L. S., and Thomashow, L. S. (1992). Cloning and heterologous expression of the phenazine biosynthetic locus from Pseudomonas aureofaciens 30-84. Mol. Plant Microbe Interact. 5, 330–339. doi: 10.1094/MPMI-5-330
R Core Team (2015). R: A Language for Statistical Computing [Online]. Vienna: R Foundation for Statistical Computing.
Raaijmakers, J. M., Bonsall, R. F., and Weller, D. (1999). Effect of population density of Pseudomonas fluorescens on production of 2,4-diacetylphloroglucinol in the rhizosphere of wheat. Phytopathology 89, 470–475. doi: 10.1094/PHYTO.1999.89.6.470
Raaijmakers, J. M., and Mazzola, M. (2012). Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu. Rev. Phytopathol. 50, 403–424. doi: 10.1146/annurev-phyto-081211-172908
Raaijmakers, J. M., and Mazzola, M. (2016). Soil immune responses. Science 352, 1392–1393. doi: 10.1126/science.aaf3252
Raaijmakers, J. M., Paulitz, T. C., Steinberg, C., Alabouvette, C., and Moënne-Loccoz, Y. (2008). The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321, 341–361. doi: 10.1007/s11104-008-9568-6
Raaijmakers, J. M., Weller, D. M., and Thomashow, L. S. (1997). Frequency of antibiotic-producing Pseudomonas spp. in natural environments. Appl. Environ. Microbiol. 63, 881–887.
Ramette, A., Moenne-Loccoz, Y., and Défago, G. (2006). Genetic diversity and biocontrol potential of fluorescent pseudomonads producing phloroglucinols and hydrogen cyanide from Swiss soils naturally suppressive or conducive to Thielaviopsis basicola-mediated black root rot of tobacco. FEMS Microbiol. Ecol. 55, 369–381. doi: 10.1111/j.1574-6941.2005.00052.x
Rochat, L., Péchy-Tarr, M., Baehler, E., Maurhofer, M., and Keel, C. (2010). Combination of fluorescent reporters for simultaneous monitoring of root colonization and antifungal gene expression by a biocontrol pseudomonad on cereals with flow cytometry. Mol. Plant Microbe Interact. 23, 949–961. doi: 10.1094/MPMI-23-7-0949
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press.
Schnider-Keel, U., Lejbølle, K. B., Baehler, E., Haas, D., and Keel, C. (2001). The sigma factor AlgU (AlgT) controls exopolysaccharide production and tolerance towards desiccation and osmotic stress in the biocontrol agent Pseudomonas fluorescens CHA0. Appl. Environ. Microbiol. 67, 5683–5693. doi: 10.1128/AEM.67.12.5683-5693.2001
Schnider-Keel, U., Seematter, A., Maurhofer, M., Blumer, C., Duffy, B., Gigot-Bonnefoy, C., et al. (2000). Autoinduction of 2, 4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J. Bacteriol. 182, 1215–1225. doi: 10.1128/JB.182.5.1215-1225.2000
Stanisich, V., and Holloway, B. (1972). A mutant sex factor of Pseudomonas aeruginosa. Genet. Res. 19, 91–108. doi: 10.1017/S0016672300014294
Stünzi, H. (2007). Bodenuntersuchungsmethoden für K, Mg und Ca im Vergleich (Comparison of soil analyis methods for K, Mg and Ca). Agrarforschung 14, 358–363.
Stutz, E. W., Défago, G., and Kern, H. (1986). Naturally occurring fluorescent pseudomonads involved in suppression of black root rot of tobacco. Phytopathology 76, 181–185. doi: 10.1094/Phyto-76-181
Tamietti, G., Ferraris, L., Matta, A., and Abbatista Gentile, I. (1993). Physiological responses of tomato plants grown in Fusarium suppressive soil. J. Phytopathol. 138, 66–76. doi: 10.1111/j.1439-0434.1993.tb01361.x
Thomashow, L. S., and Weller, D. M. (1988). Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J. Bacteriol. 170, 3499–3508. doi: 10.1128/jb.170.8.3499-3508.1988
Untergasser, A., Nijveen, H., Rao, X., Bisseling, T., Geurts, R., and Leunissen, J. A. M. (2007). Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35, W71–W74. doi: 10.1093/nar/gkm306
Vieira, J., and Messing, J. (1991). New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100, 189–194. doi: 10.1016/0378-1119(91)90365-I
Voisard, C., Keel, C., Haas, D., and Défago, G. (1989). Cyanide production by Pseudomonas fluorescens helps suppress black root rot of tobacco under gnotobiotic conditions. EMBO J. 8, 351–358.
Von Felten, A., Défago, G., and Maurhofer, M. (2010). Quantification of Pseudomonas fluorescens strains F113, CHA0 and Pf153 in the rhizosphere of maize by strain-specific real-time PCR unaffected by the variability of DNA extraction efficiency. J. Microbiol. Methods 81, 108–115. doi: 10.1016/j.mimet.2010.02.003
Weller, D. M. (2007). Pseudomonas biocontrol agents of soilborne pathogens: looking back over 30 years. Phytopathology 97, 250–256. doi: 10.1094/PHYTO-97-2-0250
Weller, D. M., Landa, B. B., Mavrodi, O. V., Schroeder, K. L., De La Fuente, L., Blouin Bankhead, S., et al. (2007). Role of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. in the defense of plant roots. Plant Biol. 9, 4–20. doi: 10.1055/s-2006-924473
Weller, D. M., Raaijmakers, J. M., Gardener, B. B., and Thomashow, L. S. (2002). Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40, 309–348. doi: 10.1146/annurev.phyto.40.030402.110010
Ye, J., Coulouris, G., Zaretskaya, I., Cutcutache, I., Rozen, S., and Madden, T. L. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. doi: 10.1186/1471-2105-13-134
Keywords: Pseudomonas, PGPR, plant-beneficial activity, antimicrobial metabolites, Pythium ultimum, Gaeumannomyces tritici, soil, disease suppressiveness
Citation: Imperiali N, Dennert F, Schneider J, Laessle T, Velatta C, Fesselet M, Wyler M, Mascher F, Mavrodi O, Mavrodi D, Maurhofer M and Keel C (2017) Relationships between Root Pathogen Resistance, Abundance and Expression of Pseudomonas Antimicrobial Genes, and Soil Properties in Representative Swiss Agricultural Soils. Front. Plant Sci. 8:427. doi: 10.3389/fpls.2017.00427
Received: 16 December 2016; Accepted: 13 March 2017;
Published: 29 March 2017.
Edited by:
Jesús Mercado-Blanco, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Francisco X. Nascimento, Universidade Federal de Santa Catarina, BrazilBrigitte Mauch-Mani, University of Neuchâtel, Switzerland
Copyright © 2017 Imperiali, Dennert, Schneider, Laessle, Velatta, Fesselet, Wyler, Mascher, Mavrodi, Mavrodi, Maurhofer and Keel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christoph Keel, christoph.keel@unil.ch Monika Maurhofer, monika.maurhofer@usys.ethz.ch
†These authors have contributed equally to this work.
 Nicola Imperiali
Nicola Imperiali Francesca Dennert
Francesca Dennert Jana Schneider2
Jana Schneider2 Michele Wyler
Michele Wyler Olga Mavrodi
Olga Mavrodi Dmitri Mavrodi
Dmitri Mavrodi Monika Maurhofer
Monika Maurhofer Christoph Keel
Christoph Keel