Chemistry of Protein-Phenolic Interactions Toward the Microbiota and Microbial Infections
- 1Department of Biotechnology, Faculty of Science, Bartin University, Bartin, Turkey
- 2Division of Food and Nutrition Science, Chalmers University of Technology, Gothenburg, Sweden
- 3Hafik Kamer Ornek MYO, Sivas Cumhuriyet University, Sivas, Turkey
- 4Department of Food Engineering, Faculty of Engineering and Natural Sciences, Istanbul Okan University, Istanbul, Turkey
- 5Department of Food Engineering, Faculty of Chemical and Metallurgical Engineering, Istanbul Technical University, Istanbul, Turkey
Along with health concerns, interest in plants as food and bioactive phytochemical sources has been increased in the last few decades. Phytochemicals as secondary plant metabolites have been the subject of many studies in different fields. Breakthrough for research interest on this topic is re-juvenilized with rising relevance in this global pandemics' era. The recent COVID-19 pandemic attracted the attention of people to viral infections and molecular mechanisms behind these infections. Thus, the core of the present review is the interaction of plant phytochemicals with proteins as these interactions can affect the functions of co-existing proteins, especially focusing on microbial proteins. To the best of our knowledge, there is no work covering the protein-phenolic interactions based on their effects on microbiota and microbial infections. The present review collects and defines the recent data, representing the interactions of phenolic compounds -primarily flavonoids and phenolic acids- with various proteins and explores how these molecular-level interactions account for the human health directly and/or indirectly, such as increased antioxidant properties and antimicrobial capabilities. Furthermore, it provides an insight about the further biological activities of interacted protein-phenolic structure from an antiviral activity perspective. The research on the protein-phenolic interaction mechanisms is of great value for guiding how to take advantage of synergistic effects of proteins and polyphenolics for future medical and nutritive approaches and related technologies.
Introduction
Phenolic compounds are secondary metabolites that are synthesized through the shikimic acid and phenylpropanoid pathways found in most plant tissues, including fruits and vegetables. Even though they are not one of the major nutrients, they provide many bioactive properties including antioxidant, anti-cancer, anti-inflammatory, antimicrobial properties. Their bioactive properties provide many health-protective effects including prevention of cancer, cardiovascular diseases, diabetes, and stroke (1).
Protein-phenolic interactions take place in two different mechanisms, which include either covalent or non-covalent interactions. Covalent bonding between two molecules of proteins and phytochemicals is accomplished by two mechanisms, enzymatic and non-enzymatic reaction mechanisms; on the other hand, non-covalent interactions include hydrogen bonding, hydrophobic interactions, electrostatic interactions, and van der Waals binding forces that are reversible reactions. Additionally, some internal factors including characteristics of proteins, types of phytochemicals, protein/phytochemical ratio and external factors including temperature, pH, ionic strength, additional reagents, and other food components influence these interactions (2).
The reciprocal interactions between phenolic compounds and proteins result in various nutritional, functional, and structural changes in both sides. Among these properties, antioxidant activities are of significant importance in terms of organic chemistry, as well as health and nutrition. Mainly, all antioxidative compounds take the action throughout some particular mechanisms which are scavenging of the free radicals in the media, inhibition of enzymes causing free radical formation, chelating the existing metallic ions, induction of endogenous antioxidant enzymes, preventing the lipid peroxidation, DNA damage, protein modification, and/or sugar degradation (3). All these mechanisms are attributed with many diseases within the living organisms; hence, dietary antioxidants have crucial importance to be a part of the diet (4). Dietary phenolic compounds, due to their antioxidant potentials besides their other benefits have been investigated extensively. In one-step further, reaction of dietary phenolic compounds with protein structures might increase the antioxidative properties of phenolics (5). Particularly, flavonoids and phenolic acids have been subjected to extensive studies in order to clarify the reaction mechanisms with protein structures and the consequences of these interactions in terms of biological activities.
Oxidative stress induced by viruses is well established. The viral infection interferes with the body's important metabolic processes in addition to participating in the replication of the virus (6). In this sense, not only the antioxidant properties but also the antiviral activities of phenolic compounds have great significance especially during the recent pandemics of viral diseases. Phenolic compounds have potential to inhibit viral replication by regulating viral adsorption, binding to cell receptors, inhibition of virus penetration into the host cell and by competing for pathways of activation of intracellular signals (7). However, the existing studies have focused on the antiviral effects of phenolics alone; thus, the antiviral mechanisms of the protein-phenol conjugates are not fully elucidated. Similarly, it is known that dietary phenolic compounds affect both physiology and gene expressions of gut microorganisms. These effects can result from interactions of phenolic compounds with bacterial proteins.
To the best of our knowledge, there is no recent study framing the chemistry of food protein - phenolic interactions playing roles in their antioxidant and antimicrobial activities. Thus, the present review collects and defines the recent data, representing the interactions of phenolic compounds, mainly flavonoids and phenolic acids with food proteins and explores how these molecular-level interactions account for the human health directly and/or indirectly, such as increased antioxidative properties, altering physiology of gut microorganisms and antimicrobial capabilities. As antimicrobial mechanism of food protein-phenol conjugates is not clear with the existing literature, we focused on the microbial protein– dietary phenol interactions in the present review to better understand the chemistry of protein-phenolic binding.
Chemistry of Protein-Phenolic Interactions
Protein-phenolic interactions take place as covalent and non-covalent interactions (8, 9). Covalent interaction mechanisms are defined as the establishment of irreversible binding between molecules under specific conditions such as availability of phenolic oxidases or alkaline conditions. Non-covalent interactions are defined as the reversible forces including electrostatic interactions, hydrogen bonding, hydrophobic interactions, and ionic bonds (8, 10). These reciprocal actions between proteins and phenolics result in a change in the nutritional, functional, and biological characteristics of both proteins and phenolics.
Covalent bonding between two molecules of proteins and phytochemicals is accomplished by two reaction mechanisms: Enzymatic and non-enzymatic reaction mechanisms. Enzymatic reaction mechanisms mostly use phenolic oxidase such as laccase and tyrosinase. In enzymatic reaction mechanisms, non-enzymatic processes are used to convert phenolics to semiquinone or quinone intermediates. These semiquinone and quinone intermediates are then assailed by nucleophilic side chains and produce C–S or C–N covalent bonds between molecules (10, 11). For non-enzymatic protein-phenolic reaction mechanisms, the free radical grafting and alkaline reaction are two extensively utilized procedures. Phenolic compounds are susceptible to oxidation while in contact with air under alkaline conditions. When they are oxidized, they turn into semiquinones and then to quinones. Covalent cross linkages are formed between proteins-phenolic compounds by reaction of nucleophilic amino acid residues such as Lys, Try, Met and Cys with these extremely reactive products (12, 13). Hydrogen peroxide (H2O2) and ascorbic acid are frequently used as a redox pair in the free-radical grafting method. They produce hydroxyl radicals, and these radicals oxidize amino acids on the side chain of the proteins. Then, they react with phenolic compounds and generate cross-linked conjugates (14, 15).
Non-covalent interactions include reversible reactions and they contain hydrogen bonding, hydrophobic interactions, electrostatic interactions, and van der Waals binding forces (8, 9). Firstly, phenolic compounds are recognized as hydrogen donors that can produce hydrogen bonds with protein carboxyl groups. Hydrogen bonding occur between hydroxyl groups of phenolics and the oxygen or nitrogen, specifically hydroxyl and amino groups of proteins (10, 16). Specifically, associations between compounds non-polar aromatic ring of the phenolic and hydrophobic regions of the protein molecules are the main reason of hydrophobic interactions (11). Furthermore, electrostatic interactions occur between the hydroxyl groups of phenolics and charged groups on the proteins. In general, a non-covalent protein-phenolic complex is the consequence of a combination of some foregoing relationships, with hydrogen bonding and hydrophobic interactions serving as the primary driving forces. According to the literature, most of the interactions between protein and phenolic compounds occur as non-covalent interactions in nature. Despite its instability, the non-covalent protein-phenolic interaction has a significant influence on the establishment and enhancement of associated food systems in the food industry (17).
In comparison with the non-covalent interactions, covalent interactions are irreversible and more stable in food processes. However, both covalent and non-covalent interactions affect the chemical structure of proteins and phenolic compounds that also change their nutritional, functional, and biological characteristics (11).
Factors Affecting Protein-Phenolic Interactions
The factors affecting protein-phenolic interactions are classified as internal factors (characteristics of proteins, types of phytochemicals, protein/phytochemical ratio) and external factors (temperature, pH, ionic strength, additional reagents, and other food components). The parameters that influence protein-phenolic interactions must be studied in order to investigate relevant applications.
Internal Factors
Characteristics of Proteins
Dietary proteins' propensity to interact with phenolics is influenced by several factors, including their hydrophobicity, molecular weight (MW), conformational configurations, amino acid composition and amino acid sequence (9). According to strong hydrophobic interactions, hydrophobicity supports a strong binding between proteins and phenolics (18). Proteins with more basic amino acids and proline, as well as a bigger, more conformationally open and flexible structure, have a better chance of interacting with polyphenols (9). Furthermore, protein properties such as surface structure, total charges, and secondary structures have been found to have varying degrees of correlation with the non-covalent binding of dietary proteins to specific phenolic substances (9, 10).
Types of Phytochemicals
Occurrence of functional groups such as glycosyl, hydroxyl and methyl groups, as well as molecular weight, hydrophobicity, structural flexibility are the key determinants impacting phenolics' binding capabilities (19). It was reported that high molecular weight polyphenols in tea exhibited a stronger prosperity for interaction with milk proteins attributed to the fact that bigger polyphenols having more binding sites (20). Besides, it was reported in several studies that binding efficiency of tea catechins ((-)-epigallocatechin gallate (EGCG) > epicatechin gallate > epicatechin > catechin) to β-lactoglobulin was related with their molecular size (21, 22). Molecular flexibility of tannins also increase the interactions and binding bonds on proteins (23). In general, it was also reported in several studies that while hydroxylation of flavonoids at ring A and B (Figure 1) increase their interactions to whole milk proteins, methoxylation and methylation decrease the binding affinity and strength (24). Depending on sugar moieties and conjugation sites glycosylation also affect the interaction of phenolics with γ-globulin and hemoglobin. Moreover, hydrogenation of double bond between C2 and C3 also lower the binding affinity of phenolics (19, 24). By the way, hydroxylation of phenolic acids at the 3-position increases their binding affinity to BSA. However, their hydroxylation at the 2-/4-positions decrease their binding affinity. Individually, for increasing the binding strength, the hydroxy groups can be replaced with methoxy groups (4-position) or methyl groups (3-position) (25). Seczyk et al. (26) studied the interactions of pure phenolic compounds (gallic acid, ferulic acid, chlorogenic acid, quercetin, apigenin, and catechin) and phenolics from plant extracts including green tea and green coffee, with protein fractions including albumins and globulins of white bean. They have measured the physicochemical properties of complexes. Their results showed that, in most cases, phenolic type significantly affected the protein-phenolic interactions and measured properties (26).
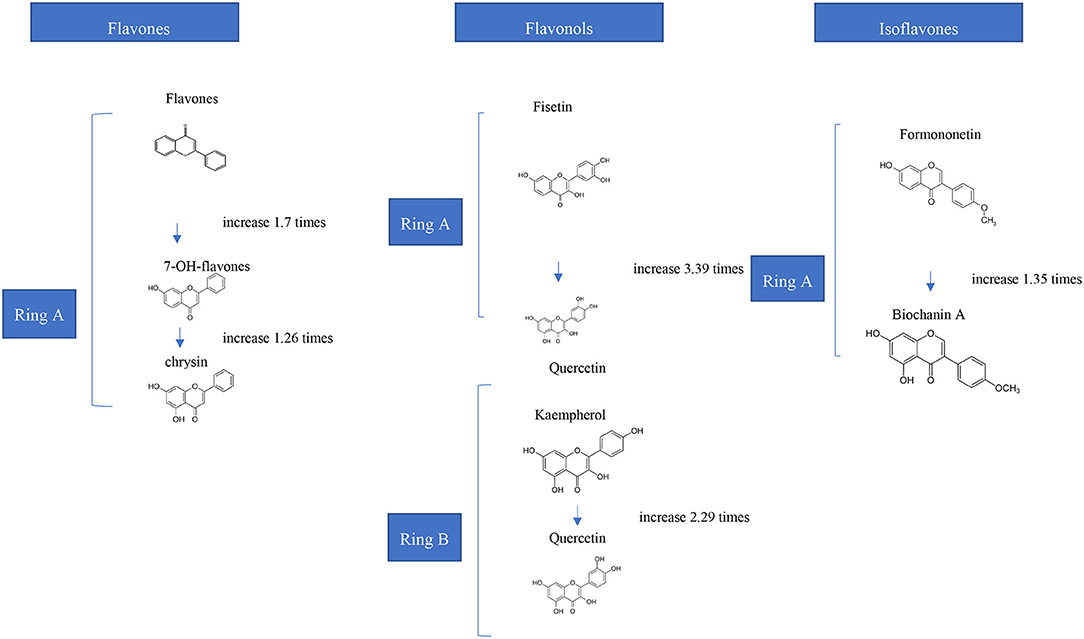
Figure 1. Effects of Hydroxylation of Flavonoids on the Affinities for Bovine Hemoglobin in vitro (24).
Protein/Phenolic Ratio
Protein/phenolic ratio is another important factor that could affect the interactions between phenolic compounds and proteins. Multidentate and monodentate mechanisms can be involved at differing protein/phenolic ratio (27). The multi-dentate process requires a lower phenolic/protein ratio because phenolic chemicals operate as multi-site ligands by binding with several protein molecules or sites. In order to achieve cross linking of proteins to produce dimers or oligomers, phenolics should have large size. In the monodentate process, many phenolics bind with a single protein molecule, necessitating a significantly greater phenolic concentration (11). In complex phenolic combinations, synergistic or antagonistic actions between phenolics toward protein binding might occur, which should be taken into account when working with complex food matrices (28).
External Factors
Temperature
Temperature can alter interactions by changing protein structures, ligand solubility, and the strength of certain non-covalent linkages. Thermal denaturation of proteins has been shown to affect phenolics' binding affinities to proteins on both sides: (1) by exposing previously buried hydrophobic sites by protein unfolding, and on the other hand, (2) by reducing interactions by forming aggregates with limited surface area (29, 30). Thermal modification of protein-phenolic interactions can result in structures due to phenolic oxidation into semiquinone and quinone intermediates (31). The binding affinities of proteins can be measured using fluorescence quenching analysis and It is possible to discern the key driving forces (9). Acknowledging the temperature-dependent relationships of such protein-phenolic combinations might aid in the development of functional food products.
pH
Protein conformational structures, as well as the binding properties of phenolics, may be significantly changed by pH. The tendency of phenolics, such as those found in berries and tea, to precipitate proteins has been shown to be pH-sensitive, especially at the isoelectric points of proteins, indicating that hydrophobic forces are involved in the interactions (32). Due to various reduced binding sites produced by protein secondary structural changes, increasing the pH from acidic to neutral values lowered the affinities of ferulic and chlorogenic acid to BSA (33). Abdollahi et al. (34) studied the interactions between β-lactoglobulin, and ferulic acid at ambient temperature temperature in relation to the dimer, and monomer forms of the protein at pH 7.3 and 2.4, respectively. They have reported that pH affects the binding site and strength of ferulic acid – β-lactoglobulin interaction and binding affinity was found to be higher when β-lactoglobulin was in monomer form (34). Non-covalent binding of dietary polyphenol extracts (tea, coffee, and cocoa) to β-lactoglobulin has been observed to alter with rising pH, probably due to differently charged states of constitutive components at different pH (35). Since the phenolics were auto-oxidized to quinones/semiquinones, which might interact with protein side-chain groups through covalent bonds, further rising the pH to alkaline conditions with oxygen would form covalent protein-phenolic complexes (13, 14, 36).
Moreover, Seczyk et al. (26) studied the interactions of pure phenolic compounds (gallic acid, ferulic acid, chlorogenic acid, quercetin, apigenin, and catechin) and phenolics from plant extracts including green tea and green coffee, with protein fractions including albumins and globulins of white bean, also determining the effect of ionic strength and pH on protein-phenolic interactions. It was reported that protein-phenolic interactions were affected by pH according to the results of relative protein solubility. The relative protein solubility was increased gradually as alkalinity increased from pH 5.0 to pH 11.0 (26).
Other Factors (Ionic Strength, Additional Reagents, Other Food Components)
Salt concentration in food products also affects the protein-phenolic interactions as it increases the ionic strength. The binding affinities of quercetin to BSA were reduced when ionic strength was raised by adding NaCl, probably due to a stronger hydration hull of the proteins with dissolved electrostatic connections (33). On the other hand, increased ionic strength in a buffering system improved the interactions between chlorogenic acid and BSA, which were largely mediated by hydrophobic forces (37). Furthermore, binding of EGCG to β-lactoglobulin in the presence of CaCl2 resulted in a bigger network of electrostatic calcium–EGCG bridging, resulting in greater levels of EGCG remaining in the complexes (38). Moreover, the increase of phenolic oxidase or free radicals such as OH radicals affect protein-phenolic interactions through oxidation of phenolics with irreversible covalent bonds. On the other hand, due to the suppression of enzyme activity, reducing substances such as Na2SO3 and ascorbic acid significantly decreased the oxidase-mediated connections (15, 39). Additionally, other macro nutrients including carbohydrates and fats also interact with proteins, however their potential for interfering with dietary protein-phenolic interactions is rarely examined (8).
Implications of Dietary Protein-Phenolic Interactions
In general, it is considered that the reciprocal interactions in between food phenolics and proteins had a negative effect on the desired properties of both compounds such as functional properties and/or bioavailabilities (40), yet the improvements of varying techniques and approaches in recent years led to opposite results -in terms of boosted functional properties of proteins and bioavailability/antioxidant capacities of phenolic compounds- might also be possible and strictly dependent on the types of the compounds (5).
Dietary phenolics and proteins might interact following different mechanisms, which are dependent on the type of protein and phenolic compound, composition of food matrix, processing conditions as well as the digestion (9). Those interactions (Figure 2) were revealed having either covalent bonds as conjugates (chemical bonds) (Figure 2A) or non-covalent bonds as complexes (physical bonds), which have varying effects on techno-functional properties of the compounds but particularly the health promoting properties of both phenolic compounds and the food proteins. As explained in the previous part, food protein-phenolic conjugates occur either with free radical induction or under alkaline conditions while protein-phenolic complexes might comprise even as a result of a forced turbulence such as mixing/vortex action (41). Conjugates-covalent bonds mostly have resistant and irreversible interactions in between the molecules, on the other hand the complexes-non-covalent bonds are comprised of reversible hydrophobic interactions (Figure 2B), hydrogen bonds (Figure 2C), as well as ionic bonds which indicate that both types of food protein- phenolic compounds interaction might easily be generated by food processing and digestion periods. However, very few of research in the literature provided an insight about the type of protein-phenolic interaction mechanism in the matrix and most of the studies rather prefer to focus on the reciprocal consequences of the interactions, instead of the mechanisms. Depending on the prevalence of non-covalent interactions compared with the covalent bond formations during food processing and/or throughout the digestive tract after consumption, all reported protein-phenolic interactions in the food systems are assumed as “complexes,” unless other specified (9, 17). Consequently, the interaction in between the protein-phenolic compounds in food matrices are dynamic processes lasting until the digestion is completed and resulting distinctive indirect health promoting activities such as improved antioxidant capacities, in vivo/in vitro bioavailabilities for phenolics and better digestibility and nutritional levels for proteins. Furthermore, it was critically reviewed in a recent study that multistep interactions in between the ingested phenolic compounds and further derived functional proteins in the body such as enzymes, transporters, receptors and transcription factors are also matter of facts (42). Engineering the phenolic compounds as well as proteins to boost their varying range of health-attributed properties with increased retentions/stabilities are considered as one of the major factors to design and produce novel functional food products (43).
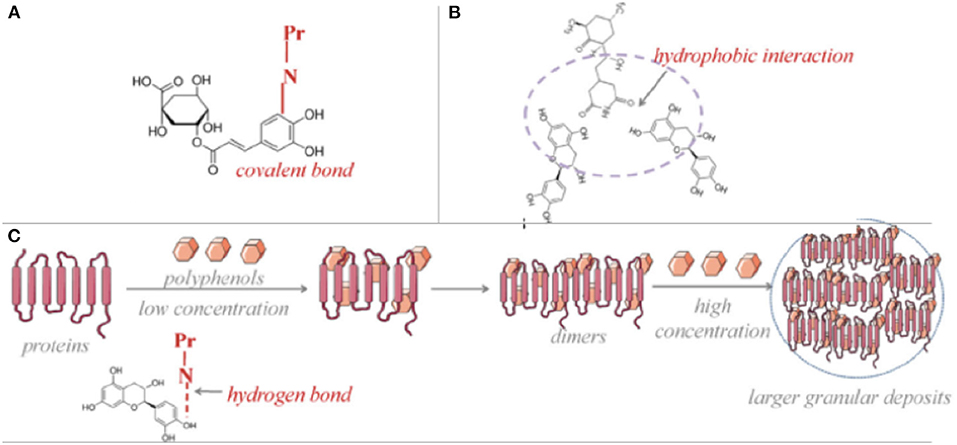
Figure 2. Mechanisms of protein-phenolic compounds interactions as conjugation via covalent bond (A), complexation via hydrophilic interactions (B) and hydrogen bonds (C) [Adapted from (113)].
Since the stability and bioaccessibility of phenolic compounds were induced with protein interactions, phenolic compounds protect their antioxidative properties for longer times, with being less affected by external processing/environmental conditions and/or digestion periods (44). This protection effect was considered to increase the antioxidative potential of phenolic compounds indirectly throughout the whole digestion process. In this section, the latest studies covering the flavonoid and phenolic acid interactions with food proteins are reviewed for a positive health effect point of view, but the focus is mainly on the improved antioxidant activities. Only very recent papers (in the last 2 years) are covered to present the most novel outcomes.
Flavonoid-Protein Interactions
Covalently structured soy protein isolates and black rice anthocyanins were exposed to in vitro gastrointestinal digestion, and it was indicated that the conjugated protein-anthocyanin structure had a higher degree of hydrolysis by around 20%, increased antioxidant capacity by 30 nmol/mg by DPPH assay and 1.5 nmol/mg by ABTS+. assay (Trolox equivalent) compared to the control sample, however a decrease by 13% was reported for transepithelial transport of peptides across Caco-2 cell monolayer (45). Complex and conjugate forms of soybean protein isolate (5 mg/ml) and EGCG (0.2 mM) were investigated for the differences of protein digestibility as well as the antioxidant capacities following the in vitro digestion. It was observed that the protein digestibility of non-covalent complexes decreased to 59.14-65.71% compared with the control which was also higher than that of the covalent conjugates. Likewise, antioxidant activities by both ABTS+. and FRAP assays were greater than the control sample in varying ranges for all stages of digestions for both interactions however, ABTS+. trend was consistent for non-covalent structure with a promising scavenging activity increase from 29.02 to 360.51 μg Trolox/g protein (46). In another study with a similar approach, soybean protein isolate-EGCG complexes were used to fabricate alginate hydrogel beads in order to obtain higher antioxidant activities from the core material during the digestion process. It was indicated that 2:1 soy protein:EGCG covalent compounds enabled to produce 20% more stable and highly bioaccessible alginate beads by 20%, while non-covalent complexes enabled this ratio as around 10% (47). On the other hand, lentil protein isolates conjugated with quercetin were indicated in another study as to increase the radical scavenging activity with DPPH and FRAP assays by 66 and 46%, compared to the phenolic alone, respectively (48).
In a study mainly focusing on the polyphenol stability during a storage period (12 days at 37°C), strawberry polyphenol extract (dominating by flavonols and phenolic acids) was used to make non-covalent complexes with canola protein extract. Degradation rate of the compounds such as pelargonidin-3-O-glucoside, cyanidin-3-O-glucoside, pelargonidin-3-O-rutinoside, pelargonidin-3-O-malonylglucoside, kaempferol-3-O-malonylglucoside were found to decrease significantly in a range of 17–44%. However, no significant difference was observed for total antioxidant activity of defined phenolic compounds after complexation with canola protein extract (49).
Bioaccessibility of coffee beverage phenolics was observed to increase by applying different protein sources as skimmed milk or soy proteins. Total bioaccessibility of coffee phenolics increased with skimmed milk and soy protein application by 37.01–64.21% and 24.74–47.32%, respectively while individual phenolics bioaccessibility in vitro increased by 4.40–27.29% and 12.02–28.61% under heat treatment (25, 90, 121°C). An antioxidant activity analysis was not conducted however, the study was observed to have the potential to increase the antioxidative properties (50).
Besides the antioxidative property improvement potentials of the phenolic interactions with food proteins, their capability to decrease the allergen effects of particular food proteins is another research interest of protein-phenolic interaction, with a growing attention. The potential of covalent and non-covalent interactions between the major milk proteins (α-casein and β-lactoglobulin) and cyanidin-3-O-glucoside were investigated aiming to reduce/mask the allergenic activity of milk proteins. Covalent conjugated forms significantly reduced the digestibility of milk proteins (a decrease range 5–11% with varying phenolic:protein ratios as 20, 30, 50) for both proteins, while showing lower IgE binding properties compared with the non-covalent complex forms as a decrease by 54 and 88% for α-casein and β-lactoglobulin conjugates (when the phenolic:protein ratio is 50), respectively (16). Similar promising results were presented by Pu et al. (51) who intended to examine the allergen reduction potentials of six different flavonoids such as EGCG, naringenin, quercetin, kaempferol, myricetin, and phloretin non-covalent complexations with β-lactoglobulin. Molar ratios of protein and phenolics were applied as 1:1–1:5. All phenolic complexations were indicated to perform antigenic inhibition for β-lactoglobulin in a descending order; 72.6, 68.4, 59.7, 52.3, 51.4, and 40.8% for EGCG, phloretin, naringenin, myricetin, kaempferol, and quercetin (51). Due to the promising results of existing antigenic factor inhibition studies for varying food proteins with distinctive types of phenolic compounds, the research interest on this area has been growing (52).
Phenolic Acid-Protein Interactions
According to the literature, it is a very well know issue that the phenolic acid interactions with varying food proteins have a significant potential to increase their antioxidant activities as well as improvements in many other reciprocal techno-functional properties. For instance, the non-covalent interactions of whey protein isolate and casein with chlorogenic acid (240 μmol/g protein) were evaluated in terms of the interaction effect on the protein digestibility. It was observed that at the end of 2 h, degree of hydrolysis was increased by 7.7 and 5.3% for casein and whey protein isolate, respectively compared with the samples without phenolic interactions. The same samples also yielded a significant antioxidant capacity increase by 460 and 400% for casein and whey protein isolate, respectively based on the ABTS+. assay (53). A different study focused on the type of complexations in β-lactoglobulin (150 μM) and chlorogenic acid (1.5 mM) under different thermal conditions as well as the alteration of antioxidant capacities. It was indicated that, with the temperatures below 60°C, the interactions were observed as non-covalent however, above that temperature covalent interactions started to be established. Both interactions retarded the loss of antioxidant capacities based on ABTS and FRAP assays. Non-covalent complexations had no significant increase on antioxidant capacity by ABTS assay, however covalent conjugates showed increased capacities (the highest increase as 56.18% at 121°C). Based on FRAP assay, both interactions increased the antioxidant capacities but the highest one (23.36%) was observed for 85°C conjugates (54).
In another study, covalent crosslinking effect of soy protein isolate (12%, w/v) on the antioxidant capacity of tannic acid (29, 58, 88, 117, 146 μmol/g protein) was investigated with DPPH, FRAP, and ABTS+. assays. It was indicated that 146 μmol/g protein tannic acids yielded the highest antioxidant capacity in terms of all three assays in a pH (9.0–11.0) dependent manner (55). Soy protein isolate was covalently interacted with chlorogenic acid (20, 40, 60, 80, 100 μmol/g protein) and based on the ABTS and DPPH assays, an antioxidant capacity increase by 10.87% and 33.08 μmol Trolox/g protein was observed for the chlorogenic acid concentration of 100 μmol/g protein (56). Another plant-based material, pea protein isolate was studied in case of non-covalent complexations with chlorogenic acid (50 μmol/g protein). The main interaction in between the phenolic acid and protein sample was declared as electrostatic interactions. Degree of hydrolysis as an indicator for in vitro digestion for pea protein was found to increase by around 5% (57).
Consequently, protein-phenolic interactions are of significant techno-functional reactions ending up with crucial biological activities such as significant antioxidative properties, varying dependent on the types of the compounds, processing conditions and physical media. However, further in-depth research is still required for better tailoring and designing for novel functional foods with boosted health promoting factors.
Implications of Dietary Polyphenols-Viral Protein Interactions
In recent years, the pandemic of viral diseases caused serious economic losses and community associated infections, which forced the scientific community to investigate less toxic antiviral phytomolecules instead of using nucleic acid analogs, protease inhibitors or other toxic synthetic molecules as antiviral therapeutics. Although the use of natural antimicrobial agents for food preservation is a trend that is followed by both consumers and food manufacturers, the studies on the antimicrobial activity of phenolic–protein conjugates are very limited (58). On the other hand, there are few studies on the antibacterial activity of protein-phenolic conjugates (59, 60), and the antiviral mechanisms of the conjugates are still unclear until now. The existing studies have focused on the antiviral effect of phytochemicals alone (7). Most of the studied plant-derived secondary metabolites were polyphenolic compounds. They have three main modes of action in treating/preventing viral diseases.
(1) They can bind to the various non-structural proteins (enzymes) to replicate the virus itself, thus inhibit early and late phase of viral replication.
(2) They can bind to structural protein (spike protein) of virus itself, thus inhibit the virus from binding and penetration to the host cells.
(3) They can bind to cell receptor proteins and inhibit virus penetration into the cell.
Since antiviral mechanism of food protein-phenolic conjugates is not clear with the existing literature, we focused on the viral protein-phenolic interactions in this review to better understand the chemistry of protein-phenolic binding.
In the literature there are many natural product-derived phytochemicals investigated as potential agents against viruses. The majority have been classified as polyphenols among the most promising small molecules identified as virus inhibitors. Most studies interpret whether phytochemicals have an antiviral effect based on changes in virus numbers, but they do not show that this antiviral effect is related to the binding between phytochemical and virus protein. We have not included the details on such studies investigating the antiviral activity of numerous phytochemicals in this review. Rather, our aim is to explain the antiviral effect of polyphenols by focusing on their binding properties with viral proteins through the most studied viruses in the literature.
Recently, molecular docking studies have become popular and provided detailed information about ligand-protein binding properties. According to the results, protein-phenolic interactions have direct effect on viral inhibition. These interactions generally occur between phytochemical and non-structural proteins of the viruses, but binding with structural viral protein was also available though.
Corona Viruses
Novel corona-virus (SARS-CoV-2 or COVID-19) has structural and non-structural proteins, which are the key viral molecules, involved in attachment, replication and reproduction of viral particle in the human host cells. SARS-CoV spike glycoprotein (S protein) is the surface protein (structural protein) that is mainly responsible for the initial attachment with the host cells receptor, Angiotensin-converting enzyme (ACE2) (61). SARS-CoV-2 chymotrypsin-like main protease (3CLpro or Mpro, corresponding to Nsp5), papain like protease (PLpro, a domain within Nsp3) are non-structural viral proteins which facilitates viral assembly by cleaving polyproteins (62). RNA dependent RNA polymerase (RdRp) is another nonstructural protein that is vital for viral life cycle (63). These viral protein molecules serve as a novel target to inhibit the viral lifecycle in human host cells.
Basu et al. (64) mentioned that the phytochemical hesperidin (flavonoid) can bind to ACE2 protein. It can also interact with the bound structure of ACE2 protein and spike protein of SARS-CoV2 (64). The binding sites of ACE2 protein for spike protein and hesperidin, are located in different parts of ACE2 protein. Molecular dynamics and docking studies confirmed the conformational change in three-dimensional structure of protein ACE2 due to ligand spike protein. This compound modulates the binding energy of bound structure of ACE2 and spike protein. After all, in the presence of hesperidin, the bound structure of ACE2 and spike protein fragment becomes unstable and lost its viral activity in SARS-CoV-2 infection.
Moreover, natural flavonoids, quercetin, epigallocatechin gallate and gallocatechin gallate (GCG) showed good inhibition properties by binding to the 3CLpro active site and the 3-OH galooyl group, which was required for SARS-CoV inhibitory activity (65). Green tea polyphenols also have SARS-CoV-2 interaction with catalytic residues of major protease (Mpro). Green tea EGCG binds to spike protein by having the highest affinity to SARS-CoV-2 (66). Catechin is another possible therapeutic phenolic compound able to bind to the viral spike protein and ACE2 of the host (67).
In another study, binding of flavonoids, herbacetin, isobavachalcone, quercetin 3-β-d-glucoside and helichristetine was studied by the fluorescence spectroscopy. As a result, flavonols were found to bind to the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) 3CLpro catalytic site and inhibit the virus (68). Chen et al. (69) studied the binding of quercetin-3-β-galactoside to SARS-CoV 3CLpro and found its protease inhibition role (69). Di Petrillo et al. (70) have schematized the interaction between quercetin-3-β-galactoside and the SARS-CoV-2 protease in Figure 3 (70).
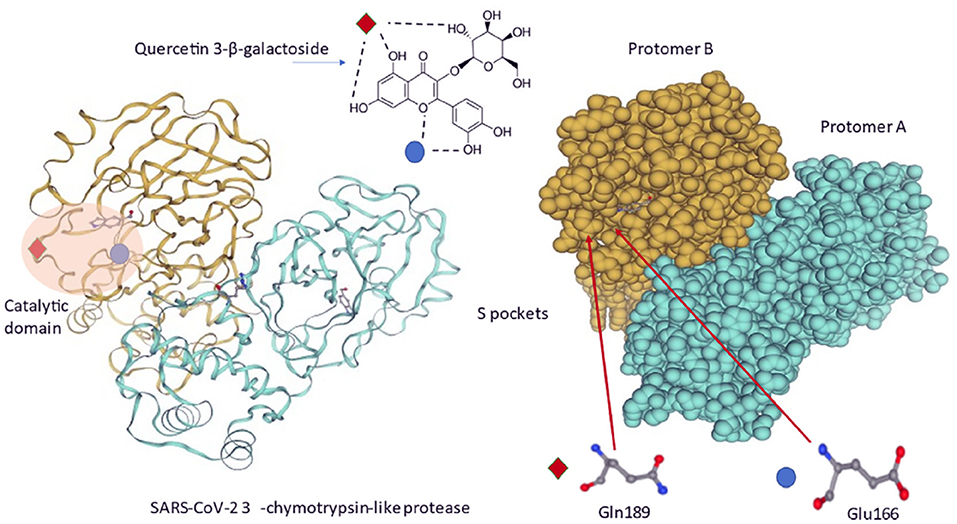
Figure 3. Molecular interactions between quercetin-3-β-galactoside and SARS-CoV-2 3CLpro. Quercetin-3-β-galactoside forms hydrogen bonds specifically with Gln189 and Glu166 amino acids located inside a specific pocket hollowed in 3CLpro surface (70).
The interaction of flavonoids presents in Galla chinensis extract to the structural protein of SARS-CoV was studied by Wang and Liu (71). They observed that the flavonoids could bind to the surface of the spike protein of the virus and prevent their penetration to the cell (71). A recent study with phytochemicals used for the inhibition of SARS-CoV-2 Mpro enzyme indicated that the flavonoid, 5,7-dimethoxyflavanone-4-O-β-d-glucopyranoside, interacts with the target enzyme and this interaction is strengthened by the establishment of five hydrogen bonds with a contribution of hydrophobic interaction, too (72). A flavonolignan, silybin (73) and a phenol phytocompound, 6-gingerol (74) have exhibited higher binding affinity and hydrogen bond interaction with non-structural protein targets in SARS-CoV-2 in comparison to currently used repurposed drugs against SARS-CoV-2. Gingerol also exhibited good binding affinity with SARS-CoV-2 spike glycoprotein. Gingerol formed hydrogen-bonded interaction with those viral proteins but with different amino acid residues. There was also a hydrophobic interaction between them (74). The electronic distribution (DFT study) also provided a clear picture of SARS CoV-2 protein-gingerol interactions and supported the molecular docking results.
Glycyrrhiza (liquorice) active compounds against spike (glycoprotein) and non-structural protein of SARS-CoV-2 were investigated (17, 75). Glyasperin A (flavonoid) had a high interaction with endoribonuclease (NS protein) and a high binding ability with the protein receptor cavity. A covalent binding of luteolin-7-O-glucuronide and chlorogenic acid (phenol) present in O. sanctum to Cys145 of Mpro of SARS-CoV-2 have been mentioned in silico analysis, so this may hinder the virus enzymes (76). Kumar et al. (77) recently reported novel natural metabolites namely, ursolic acid, carvacrol and oleanolic acid as the potential inhibitors against main protease (Mpro) of SARS-CoV-2 by using integrated molecular modeling approaches. They found that three ligands were bound to protease and these chemical molecules had stable and favorable energies causing strong binding with binding site of Mpro protein (77).
Lastly, Singh et al. (78) analyzed the binding affinity of 586 phytochemicals to the viral proteins (glycoprotein spike, Papain-protease, and protease main) and host proteins (ACE2, Importin-subunit a-5, and b-1). Their hydrogen bonds and hydrophobic interaction, as well as the binding energies and interactive amino acid residues, were categorized. Figure 4 shows the binding energy of the most potent phytochemicals. Hetisinone (alkaloid) showed the highest binding energy among all selected phytochemicals. Hetisinone formed 6 H-bonds whereas, control drug, hydroxychloroquine (HCQ), formed 4 H-bonds. This means that the interaction by hydrogen bonds between hetisinone and ACE2 represented a strong interaction than the control drug molecule. The viral-host interaction was disturbed by HCQ binding to the ACE2 receptor (allosteric region), which can inhibit the viral penetration to the human cell (78).
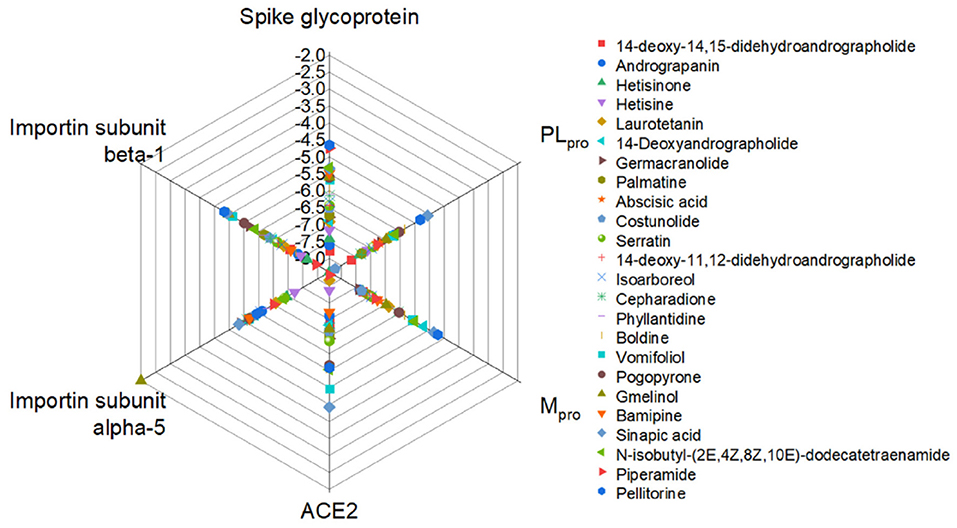
Figure 4. Relative binding energies of phytochemicals with the viral and host proteins (78).
Hepatitis Virus
Manvar et al. (79) identified the anti-Hepatitis C virus (HCV) activity of four different compounds obtained from crude extract of Eclipe alba. These are 3,4-dihydroxybenzoic acid (1), 4-hydroxybenzoic acid (2), luteolin (3), wedelolactone (4) and apigenin (5). They used fluorescence spectroscopy technique and quenching the fluorescence emission of the non-structural HCV protein NS5B (an RNA-dependent RNA polymerase) indicating their ability to bind with the enzyme. In addition to the fluorescence quenching technique, they used cell culture system and found that compounds 3, 4 and 5 bound to the enzyme HCV replicase which resulted with the inhibition of HCV replication (79). In fact, the results showed that these compounds have a dose-dependent synergistic inhibitory effect on the enzyme activity, which confirms that HCV replicase have five different binding sites for small molecules that might bind and block enzyme activity (80). The observed synergistic or additive effect of these compounds against HCV replication suggesting that these compounds are more beneficial when used together in the crude extract (79).
Recently, the possible interaction of quercetin with HCV's non-structural protein was identified in vitro (70). Molecular docking study of Zhong et al. (81) provided that quercetin derivates can interact with NS5B (non-structural protein 5B in HCV) by two magnesium ions as well as establish coordination with residues at the active site (81). Another molecular docking study showed that quercetin can bind and inhibit the NS2 (non-structural protein 2) protease of HCV (82). Its interacting ability with different proteases causes virus replication blocking and reduces HCV-induced reactive oxygen and nitrogen species (ROS/RNS) generation in replicating cells.
Furthermore, quercetin extracted from Guiera senegalensis demonstrated a high anti-Hepatitis B virus (HBV) potential. Data showed that quercetin binds the active site of HBV Polymerase by forming nine hydrogen bonds that stabilizes the quercetin-polymerase complex with an estimated free energy of 7.4 kcal/mol (83). These interactions have important contributions for stabilization of the complexes. Another research suggested that quercetin and its derivatives exert their action by interacting with the M2-1 protein, involved in genome replication and transcription by forming the complex RNA-dependent RNA polymerase (84). Caffeic acid was also found to inhibit HBV by binding to non-structural proteins of the virus (85).
Dengue Virus
Dengue, a mosquito-borne disease, has appeared as a major infectious disease globally. In the study of ul Qamar et al. (86) different phytochemicals were used against the non-structural protein of Dengue virus (DENV), which is the NS2B/NS3 protease having the ability to cleave viral proteins (86). As a phytochemical source, garcinia is a genus of flowering plants in the family Clustaceae native to Asia, America, Australia, tropical and southern Africa, and Polynesia. With the help of molecular docking, (-)-gossypol (polyphenol), mangostenone C (prenylated xanthones), garcidepsidone A (aromatic), 4-hydroxyacetophenone4-O-(6'-O-beta-D-apiofuranosyl)-beta-D-glucopyranoside, demethylcalabaxanthone (flavone glycosides), and mangostanin (natural xanthonoid) were found to be bound deeply inside the active site of DENV NS2B/NS3 protease. They had hydrophobic interactions with catalytic triad of the protease enzyme. Thus, it can be concluded from the study that these gracinia phytochemicals could serve as important inhibitors to inhibit the viral replication inside the host cell.
In addition, cyanidin 3-glucoside, dithymoquinone, and glabridin were predicted to be potent inhibitors against the NS3 protease according to their binding affinity. These ligands showed several non-covalent interactions, including hydrogen bond, hydrophobic interaction, electrostatic interaction, pi-sulfur interactions (Figure 5). After interaction, the C terminal region of NS3 tends to form a helical structure. This deviation from the initial structure resulted in enzyme inhibition (87).
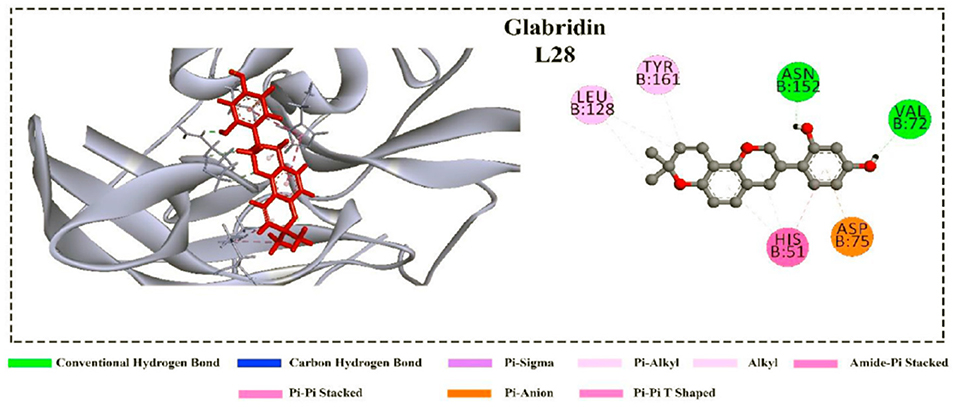
Figure 5. Non-covalent interactions of the glabridin with the NS3 protease (87).
In the other study, kushenol W and kushenol K flavonoids showed not only strong hydrogen binding but also hydrophobic and non-covalent interactions with structurally and functionally active site of dengue proteases (88). At last, kushenol W and kushenol K disrupted non-structural DV protein functions crucial for viral replication, thereby inhibiting DENV infectivity. Moreover, baicalin has a good binding ability with the NS3/NS2B protein of DENV. Recently, Loaizo-cano et al. (6) reviewed 20 different phenolic compounds against DENV and their good binding affinity to viral particles and proteins but didn't mention the interaction mechanism and types of bindings (6).
Other Viruses
Currently novaccines to prevent Zika virus (ZIKV) infection is available. In the work of Byler et al. (89), potential anti-Zika viral agents from 2263 plant-derived secondary metabolites have been investigated in in-silico molecular docking studies. Most of them were polyphenolic compounds (total 1043 of aurones, chalcones, chromones, coumarins, flavonoids, isoflavonoids, lignans, stilbenoids, xanthones, and miscellaneous phenolics). It was shown that the main antiviral molecule targets are several ZIKV non-structural proteins, typically the various enzymes to replicate the virus itself, such as NS2B-NS3 protease, helicase, NS5 methyltransferase. The best binding phytochemical ligands were the polyphenolics with generally two phenolic groups with flexible links to have strong connection to several of the protein targets. For example, rosemarinic acid, cimiphenol, and cimiracemate B showed relatively strong binding energies, thus they inhibit the replication of the virus. They also mentioned that the poorest enzyme binding ligands were found as terpenoids due to their small size, and their paucity of functional groups (89).
Many phenolic compounds have also been related to antiviral activity against many viruses, such as Herpes simplex virus (HSV), influenza virus (IV), RSV, measles, and rotavirus. These studies demonstrates that flavonoids could be one of the most active compounds against different types of viruses with multiple inhibition mechanisms, such as the inhibition of virus adsorption, entry, binding, and replication by binding both viral and host cell proteins (6). For example, quercetin showed high binding activity on cap-binding site of the PB2 (polymerase basic 2) of influenza viral RNA polymerase. Moreover, it could interact with influenza NA protein (virus surface glycoprotein neuraminidase) and block virus entry at the initial step (90). Flavonoids have non-covalent binding to reverse transcriptase and block RNA synthesis of HSV (7). EGCG also inhibits HSV by binding to their gB, gD, or other envelope proteins (91).
Implications of Dietary Phenolic on Microbiota and Microbial Proteins
Human gastrointestinal tract (GIT) is the home for hundreds of different microorganism species and this microbiota plays crucial roles for healthy human homeostasis, including immune and digestive systems (92). The GIT microbiota is not homogenous but very variable through different part of the system; however, it is one of the most populated microbial habitats on the earth (93, 94). All the commensal microorganisms reside within the mucosal layer of the GIT, except pathogenic ones, which go into epithelial layer by digesting the mucosa (95). The GIT microbiota is shaped by the host factors, one of the most important is the diet (96). The members of GIT microbiota are able to metabolize sugars, especially the indigestible ones (97). Not only the sugars, but also they can metabolize other ingredients of the diet. For instance, dietary phenolic compounds can be transformed by GIT microbiota before absorption and this transformation can modulate their biological activities (98). Very small portion (5–10%) of these phenolic compounds taken with the foods are directly absorbed through the small intestine, while most are transported to the large intestine, especially colon, and there they are metabolized by the microbiota (99). However, the transformation of the phenolics is dependent on the nature of the phenolic compound, as well as the microbial species (100). For example, one of the most-studied and very-well known phenolic compounds, resveratrol, is metabolized by human GIT microbiota, shown both in vitro and in vivo (101). In the study of Bode et al., fecal samples of different volunteers were used to investigate in vitro metabolism of trans-resveratrol. They showed fecal samples completely degraded trans-resveratrol and then metabolites were detected after 2–24 h of fermentation. Furthermore, in the same study, a separate human in vivo study with 12 volunteers was conducted and trans-resveratrol was given orally to the volunteers. After 24 h, a subsequent proportion of trans-resveratrol was found to be metabolized by GIT microbiota and same metabolites found in in vitro study were also detected in the urine samples, which confirms that trans-resveratrol was metabolized into three metabolites, main ones being dihydroresveratrol and lunularin. This study also confirmed the interindividual differences of resveratrol metabolism by GIT microbiota as microbiota can differ between the individuals (101). A pure-culture study investigated the metabolism and transformation of different but related glycosylated phenolic compounds using Lactobacillus acidophilus NCFM (102). They showed that Lactobacillus acidophilus NCFM can use such phenolic compounds and other phytochemicals as carbon source. In this mentioned study, eleven plant glycosides, including salicin, rutin, vanillin, and arbutin, were used and esculin, fraxin, and salicin supplemented to the growth were completely utilized by the bacterium. Vanillin was almost completely utilized and the growth of the bacterium in these phytochemicals reached 0.3 to 1.3 (OD600) in 200-μl cultures in 96-well plates. Furthermore, it is very-well known that dietary phenolic compounds affect both physiology and gene expressions of gut microorganisms. Thus, these effects can result from interactions of phenolic compounds with microbial macromolecules, including proteins. As the phenolic compounds usually interact with food proteins or host proteins, these conjugated complexes should further affect the gut microbiota. Theilmann et al. used transcriptomics to examine which genes are affected by these phytochemicals. Genes related with carbohydrate metabolism were differentially regulated in the phytochemical-supplemented group, with respect to control in which glucose was used as carbon source. In addition, the genes encoding the proteins related with mucus, fibrinogen, and epithelial cell adhesion were also upregulated (102). These proteins may have roles in adhesion to the host components and this upregulation may lead to higher adhesion, which means better colonization in the GIT. Previously, it has been also showed that polyphenols affect the proteome profile of this bacterium, as well as its adhesion to mucus and host cells (103). These studies suggest that plant phenolic compounds can interact either directly to the probiotic proteins or indirectly by altering the gene expressions.
Phenolic compounds can interact not only commensal microorganisms residing in the GIT or probiotics but also pathogen microorganisms so that they exert antimicrobial activities (Figure 6). These compounds especially affect proteins of such organisms, thus inhibiting crucial proteins and microbial growth, leading to antimicrobial activity. The microbial proteins affected by phenolic compounds belong to membrane (efflux system, cell envelope metabolism, and ATP synthase system), anabolic and catabolic reactions, pathogenic and antibiotic-resistance mechanisms, such as toxins and virulence factors transporters and antibiotic-inactivating enzymes, virulence factors, biofilm, and DNA metabolisms. Bacterial cell membrane is a complex system that plays roles as barrier between outside and inside of the cell, regulator of the osmotic, energy, and lipid systems, as well as cell wall maintenance (104). The major antibiotics, including penicillins, cephalosporins, and polymixins, target the bacterial membrane and cell wall, thus disrupting their functions and leading to bacterial death. The studies showed that some phenolic compounds, especially flavonoids can inhibit the efflux pumps present in the bacterial membranes and having roles in drug modulation (105, 106). A study done by Sinsinwar et al. showed that catechin treatment of Staphylococcus aureus led to decreases in the activities of superoxide dismutase and catalase enzymes, which are responsible for anti-oxidant system of the bacteria (107).
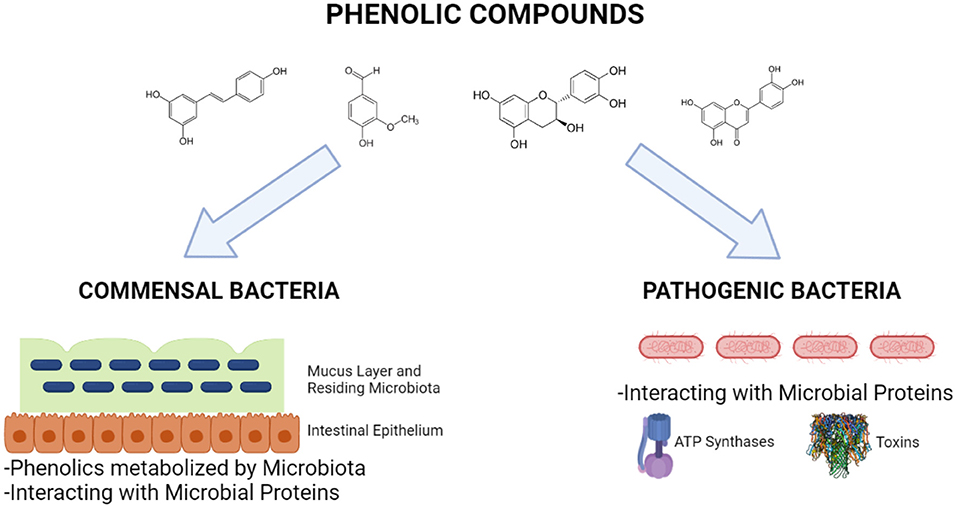
Figure 6. Interactions of phenolic compounds with commensal and pathogenic bacteria (Created with BioRender.com).
Bacterial energetics is mainly driven by ATPase activity, which is a complex protein structures responsible for ATP production. Other than some known antibiotics or antibacterial compounds, phenolics can also be inhibiting agents of ATPase system. For example, resveratrol, piceatannol, quercitrin, and quercetin inhibit ATPase belonging to Escherichia coli but in different degrees (108). Among these, the ATPase activity was completely inhibited by piceatannol, thus can lead to bacterial death; however, this study was performed in vitro using purified ATPase or membrane. Furthermore, Chinnam et al. also investigated the inhibitory effect of flavonoids on E. coli ATPase (109). In their study, seventeen polyphenols were used and E. coli growth, as well as inhibitory effect on purified ATPase activity were investigated. They found polyphenols used in the study caused complete, partial, slight, or no inhibition on ATPase activity. For instance, morin, silymarin, baicalein, silibinin, epicatechin, rimantadine HCl, and amantidin completely inhibited the ATPase activity, while hesperidin, chrysin, kaempferol, diosmin, apigenin, genistein, and rutin led to partial inhibition. On the other hand, luteolin, daidzein, and galangin showed insignificant inhibition. The authors stated that presence of number of hydroxyl groups can be important for the inhibition (109).
Pathogenic bacteria produce virulence factors, which are various molecules promoting disease state by invading and attacking host cells. They include endotoxins (found within the cell) or exotoxin (released to the outside of the cells). Some phenolic compounds target and inhibit such virulence factors. For example, flavone and luteolin, which are flavonoids, were found to reduce production of staphyloxantin, a carotenoid pigment produced by some strains of Staphylococcus aureus, and acting as a virulence factor (110). Furthermore, flavone also inhibited gene expression of α-hemolysin, causing red blood cell lysis by disrupting the cell membrane, investigated using qRT-PCR technique (110). Another study done by Stockovic et al. showed that methanolic extract of Phlomis fruticosa L. (Jerusalem sage) inhibited the expression of staphyloxantin (111). Another common pathogen is Listeria monocytogenes, which produces a virulence factor called listeriolysin O. This bacterium causes gastroenteritis, meningitis, and abortions and listeriolysin allows this bacterium to escape phagocytosis by the host immune system and also disrupt the vacuole of the host cells. A study done by Wang et al. indicated that fisetin, a plant flavonol present in many plants including strawberries, apples, persimmons, onions and cucumbers, inhibits listeriolysin O–induced hemolysis, suggesting that fisetin antagonizes the hemolytic activity and protects the red blood cells by directly or indirectly interacting with the toxin (112). Furthermore, fisetin also facilitates the elimination of L. monocytogenes by macrophages by blocking bacterial escape into the cytosol. These studies implicated that those phenolic compounds inhibit the pathogenic bacteria by interacting with their proteins that are crucial for bacterial growth or pathogenesis. However, more molecular investigations are required to enlighten how exactly phenolic compounds interact and inhibit microbial proteins.
Concluding Remarks
Phenolic compounds are abundant bioactive compounds in human diet and used as natural colorants and preservatives in foods. Despite the valuable biological activities of phenolics, such as antioxidant, antimicrobial, anticancer, antiallergenic, and anti-inflammatory effects, little is known about the health-related activity of phenolic-protein conjugates. Thus, in the last decade there is a growing interest on the functionalization of proteins with phenolic compounds. The interaction of food polyphenolics and proteins, as well as its effect on the functional properties including antioxidant and antimicrobial properties related to the chemistry of their binding, are reviewed in this paper.
Protein-phenolic interactions take place as covalent and non-covalent interactions. Protein interaction with polyphenolics-flavonoids and phenolic acids is influenced by their hydrophobicity, molecular weight, conformational configurations, amino acid composition and sequence. Type of phenolics has also significant effect on the protein-phenolic interactions and biological properties. However, very few studies in the literature provided an insight about the type of protein-polyphenolic interaction mechanism in the matrix and most of the studies focused only on the reciprocal consequences of the interactions, instead of the mechanisms. These reciprocal interactions between proteins and phenolics result in a change in the nutritional, functional, biological characteristics of both proteins and polyphenolics, and thus needs to be deeply investigated.
The main interaction mechanism between the phenolic acids and proteins was the electrostatic attraction. Depending on the temperature, both non-covalent complexations and covalent conjugates can lead to increased antioxidant capacity. Moreover, covalent conjugated forms significantly reduce the digestibility of some food proteins, therefore have an indirect effect on allergen activity. On the other hand, protein-phenolic interactions have direct effect on viral inhibition. These interactions are generally of non-covalent origin and occur between phenolic and non-structural proteins of the viruses. Many phenolics form hydrogen-bonded interactions with those viral proteins and increased number hydrogen bonds result in more stable proteins, which leads to more effective antiviral capacity. In addition to the antioxidant and antiviral activity, dietary phenolic compounds affect both physiology and gene expressions of gut microorganisms as a result of the interaction with microbial proteins. This interaction is not only with commensal microorganism in the gastrointestinal track but also with pathogen microorganisms; so that, they exert antimicrobial activities by inhibiting crucial proteins and microbial growth of such organisms.
Overall, studies on the antioxidant and antimicrobial activities of protein-phenolic conjugates are very limited. Due to the altered structures of both proteins and phenolics, systematic structure-functionality relationships of various conjugates should be investigated together with in vitro and in vivo studies to fully evaluate the biological activities of protein- phenolic conjugates.
Author Contributions
HY and BS: conceptualization and methodology. HY, BS, HC, and TO: writing—original draft preparation. HY, BS, HC, and EC: writing—review and editing. EC: supervision. All authors have read and agreed to the published version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Laura A, Moreno-Escamilla JO, Rodrigo-García J, Alvarez-Parrilla E. Phenolic compounds. In: Postharvest physiology and biochemistry of fruits and vegetables. Sawston: Woodhead Publishing. (2019). p. 253–271.
2. Ozdal T, Yalcinkaya IE, Toydemir G, Capanoglu E. Polyphenol-Protein Interactions and Changes in Functional Properties and Digestibility In: Melton L, Shahidi F, Varelis PBT-E of FC, editors. Oxford: Academic Press (2019). p. 566–77.
3. Carocho M, Morales P, Ferreira ICFR. Antioxidants: reviewing the chemistry, food applications, legislation and role as preservatives. Trends Food Sci Technol. (2018) 71:107–20. doi: 10.1016/j.tifs.2017.11.008
4. Grosso G. Dietary antioxidants and prevention of non-communicable diseases. Antioxidants. (2018) 7:94. doi: 10.3390/antiox7070094
5. Cianciosi D, Forbes-Hernández TY, Regolo L, Alvarez-Suarez JM, Navarro-Hortal MD, Xiao J, et al. The reciprocal interaction between polyphenols and other dietary compounds: impact on bioavailability, antioxidant capacity and other physico-chemical and nutritional parameters. Food Chem. (2022) 375:131904. doi: 10.1016/j.foodchem.2021.131904
6. Loaiza-Cano V, Monsalve-Escudero LM, Filho C da SMB, Martinez-Gutierrez M, Sousa DP de. Antiviral role of phenolic compounds against dengue virus: a review. Biomolecules. (2020) 11:11. doi: 10.3390/biom11010011
7. Kapoor R, Sharma B, Kanwar SS. Antiviral phytochemicals: an overview. Biochem Physiol. (2017) 6:7. doi: 10.4172/2168-9652.1000220
8. Jakobek L. Interactions of polyphenols with carbohydrates, lipids and proteins. Food Chem. (2015) 175:556–67. doi: 10.1016/j.foodchem.2014.12.013
9. Zhang Q, Cheng Z, Wang Y, Fu L. Dietary protein-phenolic interactions: characterization, biochemical-physiological consequences, and potential food applications. Crit Rev Food Sci Nutr. (2021) 61:3589–615. doi: 10.1080/10408398.2020.1803199
10. Quan TH, Benjakul S, Sae-leaw T, Balange AK, Maqsood S. Protein–polyphenol conjugates: Antioxidant property, functionalities and their applications. Trends Food Sci Technol. (2019) 91:507–17. doi: 10.1016/j.tifs.2019.07.049
11. Chu Q, Bao B, Wu W. Mechanism of interaction between phenolic compounds and proteins based on non-covalent and covalent interactions. Med Res. (2018) 2:180014. doi: 10.21127/yaoyimr20180014
12. Liu F, Ma C, Gao Y, McClements DJ. Food-grade covalent complexes and their application as nutraceutical delivery systems: a review. Compr Rev Food Sci Food Saf. (2017) 16:76–95. doi: 10.1111/1541-4337.12229
13. Sui X, Sun H, Qi B, Zhang M, Li Y, Jiang L. Functional and conformational changes to soy proteins accompanying anthocyanins: focus on covalent and non-covalent interactions. Food Chem. (2018) 245:871–8. doi: 10.1016/j.foodchem.2017.11.090
14. He W, Xu H, Lu Y, Zhang T, Li S, Lin X, et al. Function, digestibility and allergenicity assessment of ovalbumin–EGCG conjugates. J Funct Foods. (2019) 61:103490. doi: 10.1016/j.jff.2019.103490
15. Wu S, Zhang Y, Ren F, Qin Y, Liu J, Liu J, et al. Structure–affinity relationship of the interaction between phenolic acids and their derivatives and β-lactoglobulin and effect on antioxidant activity. Food Chem. (2018) 245:613–9. doi: 10.1016/j.foodchem.2017.10.122
16. Zhang Q, Cheng Z, Chen R, Wang Y, Miao S, Li Z, et al. Covalent and non-covalent interactions of cyanidin-3-: O -glucoside with milk proteins revealed modifications in protein conformational structures, digestibility, and allergenic characteristics. Food Funct. (2021) 12:10107–20. doi: 10.1039/D1FO01946E
17. Sinha SK, Prasad SK, Islam MA, Gurav SS, Patil RB, AlFaris NA, et al. Identification of bioactive compounds from Glycyrrhiza glabra as possible inhibitor of SARS-CoV-2 spike glycoprotein and non-structural protein-15: a pharmacoinformatics study. J Biomol Struct Dyn. (2021) 39:4686–700. doi: 10.1080/07391102.2020.1779132
18. Ren C, Xiong W, Li J, Li B. Comparison of binding interactions of cyanidin-3-O-glucoside to β-conglycinin and glycinin using multi-spectroscopic and thermodynamic methods. Food Hydrocoll. (2019) 92:155–62. doi: 10.1016/j.foodhyd.2019.01.053
19. Xiao J, Kai G A review of dietary polyphenol-plasma protein interactions: characterization, influence on the bioactivity, and structure-affinity relationship. Crit Rev Food Sci Nutr. (2012) 52:85–101. doi: 10.1080/10408398.2010.499017
20. Dubeau S, Samson G, Tajmir-Riahi HA. Dual effect of milk on the antioxidant capacity of green, Darjeeling, and English breakfast teas. Food Chem. (2010) 122:539–45. doi: 10.1016/j.foodchem.2010.03.005
21. Kanakis CD, Hasni I, Bourassa P, Tarantilis PA, Polissiou MG, Tajmir-Riahi HA. Milk β-lactoglobulin complexes with tea polyphenols. Food Chem. (2011) 127:1046–55. doi: 10.1016/j.foodchem.2011.01.079
22. Chanphai P, Bourassa P, Kanakis CD, Tarantilis PA, Polissiou MG, Tajmir-Riahi HA. Review on the loading efficacy of dietary tea polyphenols with milk proteins. Food Hydrocoll. (2018) 77:322–8. doi: 10.1016/j.foodhyd.2017.10.008
23. Dobreva MA, Green RJ, Mueller-Harvey I, Salminen JP, Howlin BJ, Frazier RA. Size and molecular flexibility affect the binding of ellagitannins to bovine serum albumin. J Agric Food Chem. (2014) 62:9186–94. doi: 10.1021/jf502174r
24. Xiao JB, Huo JL, Yang F, Chen XQ. Noncovalent interaction of dietary polyphenols with bovine hemoglobin in vitro: Molecular structure/property–affinity relationship aspects. J Agric Food Chem. (2011) 59:8484–90. doi: 10.1021/jf201536v
25. Wu X, Lu Y, Xu H, Lin D, He Z, Wu H, et al. Reducing the allergenic capacity of β-lactoglobulin by covalent conjugation with dietary polyphenols. Food Chem. (2018) 256:427–34. doi: 10.1016/j.foodchem.2018.02.158
26. Seczyk Ł, Swieca M, Kapusta I, Gawlik-Dziki U. Protein–phenolic interactions as a factor affecting the physicochemical properties of white bean proteins. Molecules. (2019) 24:408. doi: 10.3390/molecules24030408
27. Haslam E. Plant Polyphenols: Vegetable Tannins Revisited. Cambridge: CUP Archive, pp. 154–195. (1989).
28. García-Estévez I, Ramos-Pineda AM, Escribano-Bailón MT. Interactions between wine phenolic compounds and human saliva in astringency perception. Food Funct. (2018) 9:1294–309. doi: 10.1039/C7FO02030A
29. Yildirim-Elikoglu S, Erdem YK. Interactions between milk proteins and polyphenols: Binding mechanisms, related changes, and the future trends in the dairy industry. Food Rev Int. (2018) 34:665–97. doi: 10.1080/87559129.2017.1377225
30. Zhan F, Yang J, Li J, Wang Y, Li B. Characteristics of the interaction mechanism between tannic acid and sodium caseinate using multispectroscopic and thermodynamics methods. Food Hydrocoll. (2018) 75:81–7. doi: 10.1016/j.foodhyd.2017.09.010
31. Buchner N, Krumbein A, Rohn S, Kroh LW. Effect of thermal processing on the flavonols rutin and quercetin. Rapid Commun Mass Spectrom. (2006) 20:3229–35. doi: 10.1002/rcm.2720
32. Xu H, Zhang T, Lu Y, Lin X, Hu X, Liu L, et al. Effect of chlorogenic acid covalent conjugation on the allergenicity, digestibility and functional properties of whey protein. Food Chem. (2019) 298:125024. doi: 10.1016/j.foodchem.2019.125024
33. Rawel HM, Meidtner K, Kroll J. Binding of selected phenolic compounds to proteins. J Agric Food Chem. (2005) 53:4228–35. doi: 10.1021/jf0480290
34. Abdollahi K, Ince C, Condict L, Hung A, Kasapis S. Combined spectroscopic and molecular docking study on the pH dependence of molecular interactions between β-lactoglobulin and ferulic acid. Food Hydrocoll. (2020) 101:105461. doi: 10.1016/j.foodhyd.2019.105461
35. Stojadinovic M, Radosavljevic J, Ognjenovic J, Vesic J, Prodic I, Stanic-Vucinic D, et al. Binding affinity between dietary polyphenols and β-lactoglobulin negatively correlates with the protein susceptibility to digestion and total antioxidant activity of complexes formed. Food Chem. (2013) 136:1263–71. doi: 10.1016/j.foodchem.2012.09.040
36. Pham LB, Wang B, Zisu B, Adhikari B. Covalent modification of flaxseed protein isolate by phenolic compounds and the structure and functional properties of the adducts. Food Chem. (2019) 293:463–71. doi: 10.1016/j.foodchem.2019.04.123
37. Prigent SVE, Gruppen H, Visser AJWG, Van Koningsveld GA, De Jong GAH, Voragen AGJ. Effects of non-covalent interactions with 5-O-caffeoylquinic acid (chlorogenic acid) on the heat denaturation and solubility of globular proteins. J Agric Food Chem. (2003) 51:5088–95. doi: 10.1021/jf021229w
38. Carnovale V, Britten M, Couillard C, Bazinet L. Impact of calcium on the interactions between epigallocatechin-3-gallate and β-lactoglobulin. Food Res Int. (2015) 77:565–71. doi: 10.1016/j.foodres.2015.08.010
39. Le Bourvellec C, Renard C. Interactions between polyphenols and macromolecules: Quantification methods and mechanisms. Crit Rev Food Sci Nutr. (2012) 52:213–48. doi: 10.1080/10408398.2010.499808
40. Kamiloglu S, Tomas M, Ozdal T, Capanoglu E. Effect of food matrix on the content and bioavailability of flavonoids. Trends Food Sci Technol. (2021) 117:15–33. doi: 10.1016/j.tifs.2020.10.030
41. Baba WN, McClements DJ, Maqsood S. Whey protein–polyphenol conjugates and complexes: Production, characterization, and applications. Food Chem. (2021) 365:130455. doi: 10.1016/j.foodchem.2021.130455
42. Guan H, Zhang W, Sun-Waterhouse D, Jiang Y, Li F, Waterhouse GIN, Li D. Phenolic-protein interactions in foods and post ingestion: Switches empowering health outcomes. Trends Food Sci Technol. (2021) 118:71–86. doi: 10.1016/j.tifs.2021.08.033
43. Li Y, He D, Li B, Lund MN, Xing Y, Wang Y, et al. Engineering polyphenols with biological functions via polyphenol-protein interactions as additives for functional foods. Trends Food Sci Technol. (2021) 110:470–82. doi: 10.1016/j.tifs.2021.02.009
44. Shahidi F, Senadheera R. Encyclopedia of food chemistry: protein–phenol interactions. Encycl Food Chem. (2018) 2:532–8. doi: 10.1016/B978-0-08-100596-5.21485-6
45. Jiang L, Liu Y, Li L, Qi B, Ju M, Xu Y, et al. Covalent conjugates of anthocyanins to soy protein: Unravelling their structure features and in vitro gastrointestinal digestion fate. Food Res Int. (2019) 120:603–9. doi: 10.1016/j.foodres.2018.11.011
46. Zhou SD, Lin YF, Xu X, Meng L, Dong MS. Effect of non-covalent and covalent complexation of (–)-epigallocatechin gallate with soybean protein isolate on protein structure and in vitro digestion characteristics. Food Chem. (2020) 309:125718. doi: 10.1016/j.foodchem.2019.125718
47. Hu M, Du X, Liu G, Huang Y, Qi B, Li Y. Sodium alginate/soybean protein-epigallocatechin-3-gallate conjugate hydrogel beads: evaluation of structural, physical, and functional properties. Food Funct. (2021) 12:12347–61. doi: 10.1039/D1FO03099J
48. Parolia S, Maley J, Sammynaiken R, Green R, Nickerson M, Ghosh S. Structure – Functionality of lentil protein-polyphenol conjugates. Food Chem. (2022) 367:130603. doi: 10.1016/j.foodchem.2021.130603
49. Hanuka Katz I, Okun Z, Parvari G, Shpigelman A. Structure dependent stability and antioxidant capacity of strawberry polyphenols in the presence of canola protein. Food Chem. (2022) 385:132630. doi: 10.1016/j.foodchem.2022.132630
50. Qie X, Cheng Y, Chen Y, Zeng M, Wang Z, Qin F, et al. In vitro phenolic bioaccessibility of coffee beverages with milk and soy subjected to thermal treatment and protein–phenolic interactions. Food Chem. (2022) 375:131644. doi: 10.1016/j.foodchem.2021.131644
51. Pu P, Zheng X, Jiao L, Chen L, Yang H, Zhang Y, et al. Six flavonoids inhibit the antigenicity of β-lactoglobulin by noncovalent interactions: a spectroscopic and molecular docking study. Food Chem. (2021) 339:128106. doi: 10.1016/j.foodchem.2020.128106
52. Pan T, Wu Y, He S, Wu Z, Jin R. Food allergenic protein conjugation with plant polyphenols for allergenicity reduction. Curr Opin Food Sci. (2022) 43:36–42. doi: 10.1016/j.cofs.2021.10.002
53. Jiang J, Zhang Z, Zhao J, Liu Y. The effect of non-covalent interaction of chlorogenic acid with whey protein and casein on physicochemical and radical-scavenging activity of in vitro protein digests. Food Chem. (2018) 268:334–41. doi: 10.1016/j.foodchem.2018.06.015
54. Qie X, Chen W, Zeng M, Wang Z, Chen J, Goff HD, et al. Interaction between β-lactoglobulin and chlorogenic acid and its effect on antioxidant activity and thermal stability. Food Hydrocoll. (2021) 121:107059. doi: 10.1016/j.foodhyd.2021.107059
55. Guo Y, Bao YH, Sun KF, Chang C, Liu WF. Effects of covalent interactions and gel characteristics on soy protein-tannic acid conjugates prepared under alkaline conditions. Food Hydrocoll. (2021) 112:106293. doi: 10.1016/j.foodhyd.2020.106293
56. Guo K, Zhou G, Lok US, Wang X, Jiang L. Improving interface-related functions and antioxidant activities of soy protein isolate by covalent conjugation with chlorogenic acid. J Food Meas Charact. (2022) 16:202–13. doi: 10.1007/s11694-021-01148-6
57. Hao L, Sun J, Pei M, Zhang G, Li C, Li C, et al. Impact of non-covalent bound polyphenols on conformational, functional properties and in vitro digestibility of pea protein. Food Chem. (2022) 383:132623. doi: 10.1016/j.foodchem.2022.132623
58. Liu J, Yong H, Yao X, Hu H, Yun D, Xiao L. Recent advances in phenolic–protein conjugates: Synthesis, characterization, biological activities and potential applications. RSC Adv. (2019) 9:35825–40. doi: 10.1039/C9RA07808H
59. Fu S, Wu C, Wu T, Yu H, Yang S, Hu Y. Preparation and characterisation of chlorogenic acid-gelatin: a type of biologically active film for coating preservation. Food Chem. (2017) 221:657–63. doi: 10.1016/j.foodchem.2016.11.123
60. Ali M, Keppler JK, Coenye T, Schwarz K. Covalent whey protein–rosmarinic acid interactions: a comparison of alkaline and enzymatic modifications on physicochemical, antioxidative, and antibacterial properties. J Food Sci. (2018) 83:2092–100. doi: 10.1111/1750-3841.14222
61. Nag A, Banerjee R, Chowdhury RR, Krishnapura Venkatesh C. Phytochemicals as potential drug candidates for targeting SARS CoV 2 proteins, an in silico study. VirusDisease. (2021) 32:98–107. doi: 10.1007/s13337-021-00654-x
62. Mengist HM, Fan X, Jin T. Designing of improved drugs for COVID-19: Crystal structure of SARS-CoV-2 main protease Mpro. Signal Transduct Target Ther. (2020) 5:1–2. doi: 10.1038/s41392-020-0178-y
63. Elfiky AA. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: an in silico perspective. J Biomol Struct Dyn. (2021) 39:3204–12. doi: 10.1080/07391102.2020.1761882
64. Basu A, Sarkar A, Maulik U. Molecular docking study of potential phytochemicals and their effects on the complex of SARS-CoV2 spike protein and human ACE2. Sci Rep. (2020) 10:1–15. doi: 10.1038/s41598-020-74715-4
65. Nguyen NK, Nguyen PB, Nguyen HT, Le PH. Screening the optimal ratio of symbiosis between isolated yeast and acetic acid bacteria strain from traditional kombucha for high-level production of glucuronic acid. LWT Food Sci Technol. (2015) 64:1149–55. doi: 10.1016/j.lwt.2015.07.018
66. Ghosh R, Chakraborty A, Biswas A, Chowdhuri S. Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors–an in silico docking and molecular dynamics simulation study. J Biomol Struct Dyn. (2021) 39:4362–74. doi: 10.1080/07391102.2020.1779818
67. Jena AB, Kanungo N, Nayak V, Chainy GBN, Dandapat J. Catechin and curcumin interact with S protein of SARS-CoV2 and ACE2 of human cell membrane: insights from computational studies. Sci Rep. (2021) 11:1–14. doi: 10.1038/s41598-021-81462-7
68. Jo S, Kim H, Kim S, Shin DH, Kim M. Characteristics of flavonoids as potent MERS-CoV 3C-like protease inhibitors. Chem Biol Drug Des. (2019) 94:2023–30. doi: 10.1111/cbdd.13604
69. Chen L, Li J, Luo C, Liu H, Xu W, Chen G, et al. Binding interaction of quercetin-3-β-galactoside and its synthetic derivatives with SARS-CoV 3CLpro: Structure–activity relationship studies reveal salient pharmacophore features. Bioorg Med Chem. (2006) 14:8295–306. doi: 10.1016/j.bmc.2006.09.014
70. Di Petrillo A, Orrù G, Fais A, Fantini MC. Quercetin and its derivates as antiviral potentials: a comprehensive review. Phyther Res. (2022) 36:266–78. doi: 10.1002/ptr.7309
71. Wang X, Liu Z. Prevention and treatment of viral respiratory infections by traditional Chinese herbs. Chin Med J (Engl). (2014) 127:1344–50. doi: 10.3760/cma.j.issn.0366-6999.20132029
72. Gurung AB, Ali MA, Lee J, Farah MA, Al-Anazi KM. Unravelling lead antiviral phytochemicals for the inhibition of SARS-CoV-2 Mpro enzyme through in silico approach. Life Sci. (2020) 255:117831. doi: 10.1016/j.lfs.2020.117831
73. Pandit M, Latha N. In silico studies reveal potential antiviral activity of phytochemicals from medicinal plants for the treatment of COVID-19 infection. Res Sq. (2020) 1:22687. doi: 10.21203/rs.3.rs-22687/v1
74. Rathinavel T, Palanisamy M, Palanisamy S, Subramanian A, Thangaswamy S. Phytochemical 6-Gingerol–A promising Drug of choice for COVID-19. Int J Adv Sci Eng. (2020) 6:1482–9. doi: 10.29294/IJASE.6.4.2020.1482-1489
75. Gupta S, Singh AK, Kushwaha PP, Prajapati KS, Shuaib M, Senapati S, et al. Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies. J Biomol Struct Dyn. (2021) 39:4334–45. doi: 10.1080/07391102.2020.1776157
76. Anand AV, Balamuralikrishnan B, Kaviya M, Bharathi K, Parithathvi A, Arun M, et al. Medicinal plants, phytochemicals, and herbs to combat viral pathogens including SARS-CoV-2. Molecules. (2021) 26:1775. doi: 10.3390/molecules26061775
77. Kumar A, Choudhir G, Shukla SK, Sharma M, Tyagi P, Bhushan A, et al. Identification of phytochemical inhibitors against main protease of COVID-19 using molecular modeling approaches. J Biomol Struct Dyn. (2021) 39:3760–70. doi: 10.1080/07391102.2020.1772112
78. Singh P, Chauhan SS, Pandit S, Sinha M, Gupta S, Gupta A, et al. The dual role of phytochemicals on SARS-CoV-2 inhibition by targeting host and viral proteins. J Tradit Complement Med. (2022) 12:90–9. doi: 10.1016/j.jtcme.2021.09.001
79. Manvar D, Mishra M, Kumar S, Pandey VN. Identification and evaluation of anti hepatitis C virus phytochemicals from Eclipta alba. J Ethnopharmacol. (2012) 144:545–54. doi: 10.1016/j.jep.2012.09.036
80. Hang JQ, Yang Y, Harris SF, Leveque V, Whittington HJ, Rajyaguru S, et al. Slow binding inhibition and mechanism of resistance of non-nucleoside polymerase inhibitors of hepatitis C virus. J Biol Chem. (2009) 284:15517–29. doi: 10.1074/jbc.M808889200
81. Zhong D, Liu M, Cao Y, Zhu Y, Bian S, Zhou J, et al. Discovery of metal ions chelator quercetin derivatives with potent anti-HCV activities. Molecules. (2015) 20:6978–99. doi: 10.3390/molecules20046978
82. Sajitha Lulu S, Thabitha A, Vino S, Mohana Priya A, Rout M. Naringenin and quercetin–potential anti-HCV agents for NS2 protease targets. Nat Prod Res. (2016) 30:464–8. doi: 10.1080/14786419.2015.1020490
83. Parvez MK, Rehman MT, Alam P, Al-Dosari MS, Alqasoumi SI, Alajmi MF. Plant-derived antiviral drugs as novel hepatitis B virus inhibitors: cell culture and molecular docking study. Saudi Pharm J. (2019) 27:389–400. doi: 10.1016/j.jsps.2018.12.008
84. Teixeira TSP, Caruso ÍP, Lopes BRP, Regasini LO, de Toledo KA, Fossey MA, et al. Biophysical characterization of the interaction between M2-1 protein of hRSV and quercetin. Int J Biol Macromol. (2017) 95:63–71. doi: 10.1016/j.ijbiomac.2016.11.033
85. Wang GF, Shi LP, Ren YD, Liu QF, Liu HF, Zhang RJ, et al. Anti-hepatitis B virus activity of chlorogenic acid, quinic acid and caffeic acid in vivo and in vitro. Antiviral Res. (2009) 83:186–90. doi: 10.1016/j.antiviral.2009.05.002
86. ul Qamar T, Mumtaz A, Ashfaq UA, Azhar S, Fatima T, Hassan M, et al. Computer aided screening of phytochemicals from Garcinia against the dengue NS2B/NS3 protease. Bioinformation. (2014) 10:115. doi: 10.6026/97320630010115
87. Rahman MM, Biswas S, Islam KJ, Paul AS, Mahato SK, Ali MA, et al. Antiviral phytochemicals as potent inhibitors against NS3 protease of dengue virus. Comput Biol Med. (2021) 134:104492. doi: 10.1016/j.compbiomed.2021.104492
88. Tahir ul Qamar M, Maryam A, Muneer I, Xing F, Ashfaq UA, Khan FA, et al. Computational screening of medicinal plant phytochemicals to discover potent pan-serotype inhibitors against dengue virus. Sci Rep. (2019) 9:1–16. doi: 10.1038/s41598-018-38450-1
89. Byler KG, Ogungbe IV, Setzer WN. In-silico. screening for anti-Zika virus phytochemicals. J Mol Graph Model. (2016) 69:78–91. doi: 10.1016/j.jmgm.2016.08.011
90. Gansukh E, Nile A, Kim DH, Oh JW, Nile SH. New insights into antiviral and cytotoxic potential of quercetin and its derivatives–a biochemical perspective. Food Chem. (2021) 334:127508. doi: 10.1016/j.foodchem.2020.127508
91. Isaacs CE, Wen GY, Xu W, Jia JH, Rohan L, Corbo C, et al. Epigallocatechin gallate inactivates clinical isolates of herpes simplex virus. Antimicrob Agents Chemother. (2008) 52:962–70. doi: 10.1128/AAC.00825-07
92. Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut microbiota in health and disease. Physiol Rev. (2010) 90:859–904. doi: 10.1152/physrev.00045.2009
93. Hakansson A, Molin G. Gut microbiota and inflammation. Nutrients. (2011) 3:637–82. doi: 10.3390/nu3060637
94. Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci. (1998) 95:6578–83. doi: 10.1073/pnas.95.12.6578
95. Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J. (2017) 474:1823–36. doi: 10.1042/BCJ20160510
96. Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. (2016) 14:20–32. doi: 10.1038/nrmicro3552
97. Zoetendal EG, Raes J, Van Den Bogert B, Arumugam M, Booijink CCGM, Troost FJ, et al. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. (2012) 6:1415–26. doi: 10.1038/ismej.2011.212
98. Selma M V, Espin JC, Tomas-Barberan FA. Interaction between phenolics and gut microbiota: role in human health. J Agric Food Chem. (2009) 57:6485–501. doi: 10.1021/jf902107d
99. Cardona F, Andrés-Lacueva C, Tulipani S, Tinahones FJ, Queipo-Ortuño MI. Benefits of polyphenols on gut microbiota and implications in human health. J Nutr Biochem. (2013) 24:1415–22. doi: 10.1016/j.jnutbio.2013.05.001
100. D'Archivio M, Filesi C, Varì R, Scazzocchio B, Masella R. Bioavailability of the polyphenols: status and controversies. Int J Mol Sci. (2010) 11:1321–42. doi: 10.3390/ijms11041321
101. Bode LM, Bunzel D, Huch M, Cho GS, Ruhland D, Bunzel M, et al. In vivo and in vitro Metab trans-resveratrol by Hum gut microbiota. Am J Clin Nutr. (2013) 97:295–309. doi: 10.3945/ajcn.112.049379
102. Theilmann MC, Goh YJ, Nielsen KF, Klaenhammer TR, Barrangou R, Abou Hachem M. Lactobacillus acidophilus metabolizes dietary plant glucosides and externalizes their bioactive phytochemicals. MBio. (2017) 8:e01421–17. doi: 10.1128/mBio.01421-17
103. Celebioglu HU, Delsoglio M, Brix S, Pessione E, Svensson B. Plant polyphenols stimulate adhesion to intestinal mucosa and induce proteome changes in the probiotic Lactobacillus acidophilus NCFM. Mol Nutr Food Res. (2018) 62:1700638. doi: 10.1002/mnfr.201700638
104. Donadio G, Mensitieri F, Santoro V, Parisi V, Bellone ML, De Tommasi N, et al. Interactions with microbial proteins driving the antibacterial activity of flavonoids. Pharmaceutics. (2021) 13:660. doi: 10.3390/pharmaceutics13050660
105. Maia GL de A, Falcão-Silva V, dos S, Aquino PGV, Araújo-Júnior JX de, Tavares JF, et al. Flavonoids from Praxelis clematidea RM King and Robinson modulate bacterial drug resistance. Molecules. (2011) 16:4828–35. doi: 10.3390/molecules16064828
106. Farooq S, Wahab A, Fozing CDA, Rahman A, Choudhary MI. Artonin I inhibits multidrug resistance in S taphylococcus aureus and potentiates the action of inactive antibiotics in vitro. J Appl Microbiol. (2014) 117:996–1011. doi: 10.1111/jam.12595
107. Sinsinwar S, Vadivel V. Catechin isolated from cashew nut shell exhibits antibacterial activity against clinical isolates of MRSA through ROS-mediated oxidative stress. Appl Microbiol Biotechnol. (2020) 104:8279–97. doi: 10.1007/s00253-020-10853-z
108. Dadi PK, Ahmad M, Ahmad Z. Inhibition of ATPase activity of Escherichia coli ATP synthase by polyphenols. Int J Biol Macromol. (2009) 45:72–9. doi: 10.1016/j.ijbiomac.2009.04.004
109. Chinnam N, Dadi PK, Sabri SA, Ahmad M, Kabir MA, Ahmad Z. Dietary bioflavonoids inhibit Escherichia coli ATP synthase in a differential manner. Int J Biol Macromol. (2010) 46:478–86. doi: 10.1016/j.ijbiomac.2010.03.009
110. Lee JH, Park JH, Cho MH, Lee J. Flavone reduces the production of virulence factors, staphyloxanthin and α-hemolysin, in Staphylococcus aureus. Curr Microbiol. (2012) 65:726–32. doi: 10.1007/s00284-012-0229-x
111. Stojković D, Gašić U, Drakulić D, Zengin G, Stevanović M, Rajčević N, et al. Chemical profiling, antimicrobial, anti-enzymatic, and cytotoxic properties of Phlomis fruticosa L. J Pharm Biomed Anal. (2021) 195:113884. doi: 10.1016/j.jpba.2020.113884
112. Wang J, Qiu J, Tan W, Zhang Y, Wang H, Zhou X, et al. Fisetin inhibits Listeria monocytogenes virulence by interfering with the oligomerization of listeriolysin O. J Infect Dis. (2015) 211:1376–87. doi: 10.1093/infdis/jiu520
Keywords: food phenolics, protein-phenolic interaction, antiviral, antioxidant activity, phytochemicals, microbial protein (MP)
Citation: Yilmaz H, Gultekin Subasi B, Celebioglu HU, Ozdal T and Capanoglu E (2022) Chemistry of Protein-Phenolic Interactions Toward the Microbiota and Microbial Infections. Front. Nutr. 9:914118. doi: 10.3389/fnut.2022.914118
Received: 06 April 2022; Accepted: 25 May 2022;
Published: 01 July 2022.
Edited by:
A. M. Abd El-Aty, Cairo University, EgyptReviewed by:
Ryszard Amarowicz, Institute of Animal Reproduction and Food Research (PAS), PolandKebede Taye Desta, National Agrobiodiversity Center, South Korea
Copyright © 2022 Yilmaz, Gultekin Subasi, Celebioglu, Ozdal and Capanoglu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Esra Capanoglu, capanogl@itu.edu.tr
 Hilal Yilmaz
Hilal Yilmaz Busra Gultekin Subasi
Busra Gultekin Subasi Hasan Ufuk Celebioglu1
Hasan Ufuk Celebioglu1  Tugba Ozdal
Tugba Ozdal Esra Capanoglu
Esra Capanoglu