Irc20 Regulates the Yeast Endogenous 2-μm Plasmid Levels by Controlling Flp1
- Department of Biochemistry, College of Medicine and Health Sciences, United Arab Emirates University, Al-Ain, United Arab Emirates
The endogenous yeast 2-μm plasmid while innocuous to the host, needs to be properly regulated to avoid a toxic increase in copy number. The plasmid copy number control system is under the control of the plasmid encoded recombinase, Flp1. In case of a drop in 2-μm plasmid levels due to rare plasmid mis-segregation events, the Flp1 recombinase together with the cell’s homologous recombination machinery, produce multiple copies of the 2-μm plasmid that are spooled during DNA replication. The 2-μm plasmid copy number is tightly regulated by controlled expression of Flp1 as well as its ubiquitin and SUMO modification. Here, we identify a novel regulator of the 2-μm plasmid, the ATPase, ubiquitin ligase, Irc20. Irc20 was initially identified as a homologous recombination regulator, and here we uncover a new role for Irc20 in maintaining the 2-μm plasmid copy number and segregation through regulating Flp1 protein levels in the cell.
Introduction
The Saccharomyces cerevisiae 2-μm plasmid is an endogenous selfish circular plasmid that utilizes the cell’s own machinery to maintain copy number at 40–60 copies per haploid cell and segregation efficiency close to that of chromosomes (reviewed in Chan et al., 2013; Liu et al., 2014). The faithful segregation of the 2-μm plasmid to the daughter cells is coordinated by the plasmid partitioning system which comprises two plasmid encoded proteins, Rep1 and Rep2 and a cis-acting DNA element STB (Jayaram et al., 2004). Rep1 and Rep2 proteins form a complex at the STB locus, which acts in a manner similar to yeast centromeres where it recruits other host-encoded factors, such as Rsc2 and the cohesin complex (Wong et al., 2002; Yang et al., 2004), and helps in segregating the replicated plasmid into the mother and daughter cells. The copy number control system, on the other hand, ensures the copy number is maintained at 40–60 copies per cell. In rare mis-segregation events of plasmid molecules, the plasmid encoded recombinase Flp1 corrects the copy number by inducing a cut at the Flp recognition target (FRTs) initiating a double rolling circle replication mode which amplifies copy number with the help of the cellular homologous recombination (HR) machinery (Futcher and Cox, 1983; Futcher, 1986).
While innocuous at controlled levels, if the 2-μm plasmid reaches uncontrollable levels, it causes cell toxicities characterized by cold sensitivities and nibbled colony appearance (Chen et al., 2005; Burgess et al., 2007). The activity of Flp1 is tightly regulated at the level of gene expression through Rep1-Rep2 mediated inhibition (Murray et al., 1987), and by SUMO modification to regulate its function and mediate its ubiquitin dependent degradation, all preventing runaway increase in copy number (Xiong et al., 2009). Perturbations in several SUMO and ubiquitin pathway regulators cause increased copy numbers of the 2-μm plasmid, such as mutations in the deSUMOylating enzyme Ulp1 (Dobson et al., 2005), the SUMO conjugating enzyme Ubc9 (Burgess et al., 2007), the SUMO E3 ligases Siz1 and Siz2 (Chen et al., 2005), the SUMO targeted ubiquitin ligase (STUBL) complex Slx5-Slx8 (Burgess et al., 2007), as well as the ubiquitin conjugating enzyme Ubc4 (Sleep et al., 2001).
Irc20 is a recently identified HR regulator. It controls the spontaneous formation of recombination foci (Increased recombination centers) (Alvaro et al., 2007). It was further demonstrated that Irc20 promotes HR, and helps in directing it to the more error-free mode, synthesis dependent strand annealing (SDSA), as well as reducing cross-overs involving long-tracts of DNA (Miura et al., 2012). Irc20 harbors two conserved domains, a SNF2 ATP hydrolysis domain (Flaus et al., 2006), and a RING domain (Freemont, 2000; Richardson et al., 2013), which is characteristic for E3 ubiquitin ligases. Irc20 also contains at least two SIMs, allowing it to target SUMOylated proteins, including itself, hence considered a STUbL (Richardson et al., 2013).
In this study, we uncover a novel function for Irc20 in controlling the HR-dependent increase in 2-μm plasmid levels, and show that mutations in Irc20 affect the proper maintenance of the plasmid. Additionally, we show that Irc20 partly regulates the 2-μm plasmid stability and copy number by controlling the levels of the Flp1 recombinase.
Materials and Methods
Yeast Strains
The strains used in this study are listed in Supplementary Table 1. Gene tagging and deletions were done using single-step PCR mediated gene modifications (Longtine et al., 1998). Point mutations were introduced using in vivo site specific mutagenesis, delitto perfetto (Stuckey et al., 2011). Primers for gene modifications are listed in Supplementary Table 2. Strains lacking the 2-μm plasmid, designated [cir0], were derived from strains containing the 2-μm plasmid, [cir+], by expression of a defective Flp1 recombinase from the plasmid pBIS-GALkFLP-(URA3) (Addgene) (Tsalik and Gartenberg, 1998).
2-μm Plasmid Loss Assay
To monitor plasmid loss rates, [cir0] cells were transformed with pKan4 plasmid lacking flp1 gene (a kind gift from Melanie Dobson), and selected on media containing the antibiotic Geneticin. Cells were then grown in liquid culture without antibiotic for 15 generations, followed by plating of equal amounts on YPD and YPD-Geneticin. Plasmid segregation efficiency was calculated as the ratio of number of colonies grown on geneticin plate (colonies harboring plasmid) to the number of colonies grown on YPD plate (total number of colonies) for irc20Δ and compared to WT strain.
2-μm harboring His3MX6 inserted downstream of the STB locus were used to analyze the plasmid segregation efficiency for plasmids harboring the FLP1 gene. Similar to pKan4, the plasmid segregation efficiency was calculated for Irc20 mutants and compared to WT strain.
Analyzing 2-μm Plasmid Levels
Overnight cell cultures were pelleted and genomic DNA isolated using Wizard Genomic DNA purification kit (Promega). DNA was diluted to a concentration of 5 ng/μl and analyzed by quantitative real time PCR. Levels of 2-μm plasmid were measured using a sequence from the Y subtelomeric element as the reference. Primers for measuring 2-μm DNA and Y subtelomeric element are listed in the primer list. qPCR was carried out with QuantStudio 7 Flex (Applied Biosystems) using SYBRTM green PCR master mix (Applied Biosystems). Each reaction contained 10 μl of SYBR Green Master Mix, 0.1 mM forward and reverse primers, 10 ng genomic DNA, and distilled H2O to a 20 μl final volume. PCR conditions were as follows: 1 cycle at 50°C for 2 min followed by 95°C for 10 min; and 40 cycles, each consisting of 95°C for 15 s, 60°C for 1 min. The fold change of the 2-μm number compared to that of wild-type yeast DNA was calculated by 2–ΔΔCT methods. Five to nine independent colonies were analyzed for each strain, and qPCR done in duplicates or triplicates. Values were compared by the Student t-test.
Extraction of Total Cellular Protein by TCA
Pellets from the yeast strains equivalent to an OD600 of 2.5–5 was collected by centrifugation, washed with 1 ml sterile water. Cells were then resuspended in cold (0.25 M NaOH/1% 2-mercaptoethanol) and incubated on ice for 10 min. 160 μl of 50% TCA was then added and incubated for another 10 min on ice. Precipitated proteins were centrifuged at 14,000 RPM for 10 min, washed with cold acetone and air-dried for 10 min. Proteins were resuspended in 100 μls 2X SDS-PAGE loading buffer, boiled for 5 min at 95°C, and centrifuged at 13,000 RPM for 5 min. 10–15 μls of supernatant were loaded on SDS-PAGE followed by western blotting.
Monitoring Flp1 Levels
The FLP1 gene was cloned into pRS416-Gal-RNQ1-YFP (Addgene; Douglas et al., 2008) using XhoI and BamH1 sites, to replace RNQ1 gene. The resulting vector expresses C-terminal YFP-tagged Flp1 under GAL1-promoter, in which expression is induced when cells are grown in galactose, and is repressed when grown in glucose. This plasmid was transformed into [cir0] strains, and SC-Ura media was used for selection. Cells were grown overnight in SC-Ura broth containing raffinose, subcultured in the same media to get an exponentially growing culture. Cells were then switched to SC-Ura broth containing galactose and allowed to grow for 6 h to induce the expression of Flp1. Cells were then pelleted, resuspended in SC-Ura broth containing glucose, and allowed to grow to monitor the degradation of Flp1. Cells were collected at 45, 90, and 180 min, of total OD600 of 2.5, and whole cell extracts were prepared by TCA method. 10–15 μls of the whole cell extracts were loaded on 8% SDS-PAGE, and western blotting was done using anti-GFP antibody (Abcam). The blots were deprobed and then probed with Tubulin antibody (Abcam). The blots were scanned by Typhoon FLA 9500 and the bands were quantitated using ImageQuant TL software (GE Healthcare). The levels of Flp1 were calculated by comparing the band intensity at the time points indicated with the band intensity at 0 mins, normalized to the loading control.
Southern Blotting for Analyzing 2-μm Species
15 μg of isolated DNA were ran on 1.2% agarose gel in 1X TBE containing 0.75 μg/ml chloroquine at 3 V/cm for 40 h at room temperature. The agarose gel was depurinated by incubating twice for 15 min in 0.25 M HCl, then denatured by incubating twice for 15 min in denaturation buffer (0.5 N NaOH, 1 M NaCl) and finally neutralized by incubating twice for 15 min in neutralization buffer (1 M Tris pH 7.5, 600 mM NaCl). DNA was then transferred to a positively charged nylon membrane overnight in 20X SSC (3 M NaCl, 0.3 Na citrate pH 7.0) by capillary transfer using a wick of Biorad filter paper (Brown, 1999). Membrane was crosslinked using UV, then hybridized with biotinylated probes designed to specifically detect the 2-μm plasmid in hybridization buffer (0.5 M Na2PO4 pH 7.2, 7% SDS, 1 mM EDTA) at 62°C overnight in hybridization oven. Membranes were washed twice for 15 min with 2X SSC, 0.1% SDS at 55°C followed by one wash with 1X SSC, 0.1% SDS at 55°C for 15 min. Detection of biotin labeled probe was done using Chemiluminescent Nucleic Acid Detection Module (Thermo ScientificTM) as per the instructions provided with the kit.
Results
Mutations in Irc20 Promote the Loss of the Highly Stable Endogenous 2-μm Plasmid
We have frequently observed the generation of strains lacking the 2-μm plasmid when irc20 was deleted (Supplementary Figure 1). In order to quantify the 2-μm plasmid segregation efficiency, we used pKan4, a modified form of the endogenous 2-μm plasmid, that is used for analyzing segregation efficiency, where it harbors a KanMX4 selection marker and the partitioning system of the endogenous 2-μm plasmid, Rep1, Rep2, and the STB, but lacks the flp1 gene (Pinder et al., 2013). This plasmid is designed this way because having a functional FLP1 would obscure plasmid mis-segregation defects by correcting decreases in copy number (Sengupta et al., 2001; Pinder et al., 2013; McQuaid et al., 2019). To determine the rate of pKan4 loss, WT and irc20Δ [cir0] strains were transformed with pKan4, and transformants were initially grown in YPD medium containing the antibiotic Geneticin to select for plasmid retention. The proportion of plasmid-containing cells was then determined after 15 generations of growth in a medium that did not select for the retention of the plasmid (YPD), by comparing plating efficiency on solid YPD medium containing or lacking Geneticin. We used rsc2Δ mutant as a positive control for this experiment as it is known to have lower plasmid retention rates because of the role of Rsc2 in the 2-μm plasmid partitioning (Wong et al., 2002; Pinder et al., 2013). To our surprise, the segregation efficiency of the pKan4 plasmid was not reduced in the irc20Δ strain (Figure 1A). The rsc2Δ mutant, however, showed less pKan4 plasmid segregation efficiency, at a similar level to a previous report (Pinder et al., 2013).
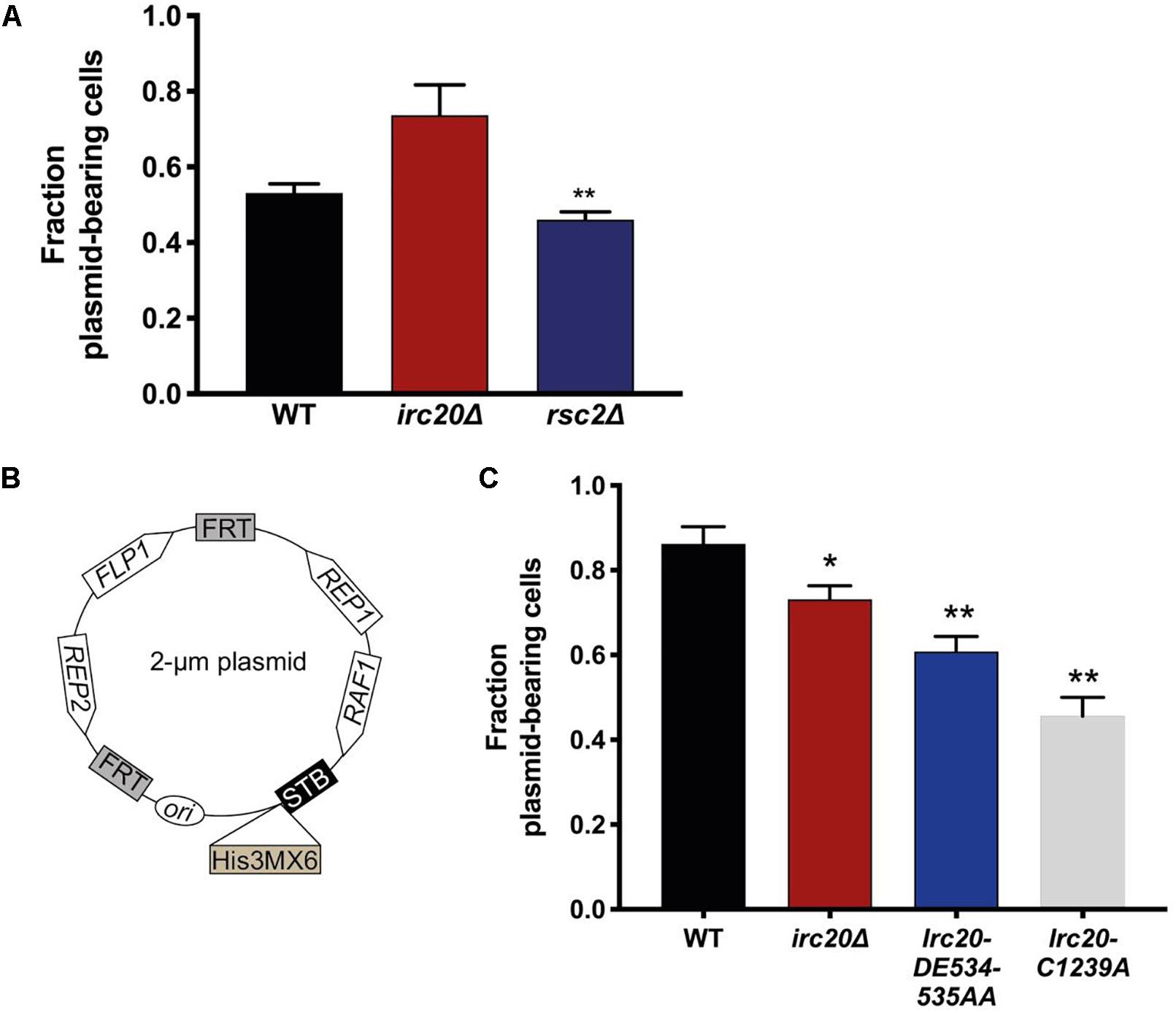
Figure 1. Mutations in Irc20 affect the segregation efficiency of the 2-μm plasmid. (A) The fraction of cells harboring pKan4 plasmid were calculated by counting the number of colonies on Geneticin plates compared to the number of colonies on non-selective plate (YPD) in three different independent cultures. (B) Schematic diagram of p2μm-His3MX6-STB, a modified form of the 2-μm plasmid where a His3MX6 cassette is inserted downstream of the STB locus to allow for selection. (C) The fraction of cells harboring p2μm-His3MX6-STB plasmid were calculated by counting the number of colonies on SD-His plates compared to the number of colonies on non-selective plates (YPD) in three different independent cultures. Values are presented as mean ± SEM. One asterisk shows p-value < 0.05, two asterisks shows p-value < 0.01, as compared to WT.
To reconcile the discrepancies in the 2-μm plasmid segregation efficiency, we constructed a new 2-μm plasmid harboring all the plasmid encoded genes, including FLP1, and a His3MX6 cassette (which allows the cells to grow on media lacking Histidine) inserted downstream of the STB locus (Figure 1B). To determine the rate of p2μm-His3MX6-STB loss, WT and irc20Δ [cir0] strains were transformed with the plasmid and were initially grown in SD-His medium to select for the plasmid retention. The proportion of plasmid-containing cells was determined as discussed earlier. Using this construct, we were able to detect lower plasmid segregation efficiency in the irc20Δ mutant strain as compared to WT (Figure 1C). To identify the functional domain in Irc20 responsible for plasmid loss, we introduced point mutations in the ATPase domain and in the RING finger domain, Irc20-DE534-535AA and Irc20-C1239A, respectively, that abolishes either activity (Richardson et al., 2013). These point mutants were transformed with the p2μm-His3MX6-STB and tested for plasmid retention, and similar to irc20Δ showed lower plasmid segregation efficiency than WT. The 2-μm plasmid segregation efficiency was even lower when cells harbor point mutations in either the ATPase or the ubiquitin ligase domain of Irc20, than in the irc20Δ mutant. The maximum reduction in plasmid segregation efficiency was seen in the ubiquitin ligase mutant (Irc20-C1239A), where the fraction of cells bearing the plasmid reached 0.5 as compared to WT where it was 0.85. It is worth-noting that the pKan4 and the p2μm-His3MX6-STB differ in the plasmid background, where pKan4 contains bacterial maintenance sequences and p2μm-His3MX6-STB does not. This accounts for the difference in plasmid segregation efficiencies between both in WT strain, as introduction of DNA sequences in the endogenous 2-μm plasmid is known to affect its segregation efficiency (Wong et al., 2002; Pinder et al., 2013).
Irc20 Controls Copy Number Amplification of the 2-μm Plasmid by Regulating the Flp1 Protein Levels in the Cell
The elevation of the 2-μm plasmid in several SUMO pathway mutants, such as siz1Δsiz2Δ double mutant, and STUbLs, such as slx5Δ and slx8Δ, have already been reported and is considered to be responsible for the nibbled colony appearance and cold sensitivities observed in these mutants (Chen et al., 2005). Even though irc20Δ does not show any growth sensitivities, there seems to be a link between Irc20 and the plasmid amplification system, specifically Flp1, underlying the defective segregation efficiency of the 2-μm plasmid observed in irc20 mutants. To understand this further, we measured the levels of the endogenous 2-μm plasmid by isolating DNA from WT and irc20Δ strains, and quantified by real time PCR using 2-μm specific primers normalized to a genomic control region, Y-subtelomeric regions. Our results show 3 to 4-fold higher 2-μm plasmid levels in irc20Δ mutant (Figure 2A). We next measured whether the elevation of 2-μm plasmid levels in irc20Δ was dependent on the HR pathway, since the Flp1 mediated copy number amplification of the 2-μm plasmid depends on the cellular HR machinery (Futcher, 1986; Xiong et al., 2009). This was done by analyzing the 2-μm plasmid levels in irc20Δrad52Δ and in the irc20Δmre11Δ double mutants. We observed a significant reduction in the endogenous 2-μm levels when loss of the irc20 gene was coupled with deletions in mre11 and rad52 HR factors. The reduction was more pronounced in double mutants with rad52, possibly reflecting some residual HR events occurring in the absence of Mre11. These results indicate that the amplification of the 2-μm copy number in irc20Δ mutant is due to improperly regulated HR events.
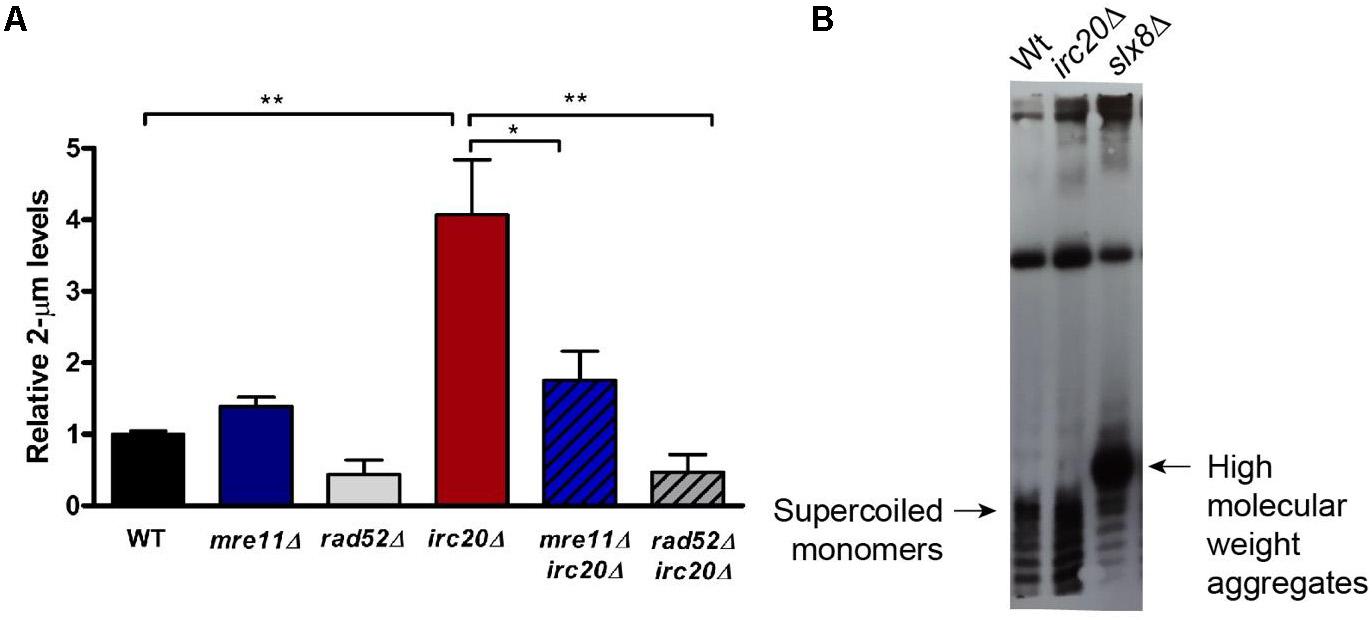
Figure 2. The absence of Irc20 increases the endogenous 2-μm plasmid levels without forming HMW aggregates. (A) The levels of endogenous 2-μm plasmids in WT, irc20Δ, mre11Δ, rad52Δ, mre11Δirc20Δ, and rad52Δirc20Δ mutants were measured by real-time PCR using primers specific to the 2-μm plasmid DNA sequence and quantified relative to Y-subtelomeric regions. irc20Δ mutant showed 4-fold higher levels of the 2-μm plasmid compared to WT. Double mutants of irc20 with HR repair factors, mre11 and rad52, showed significantly reduced 2-μm plasmid levels compared to single irc20Δ mutant. Five to nine independent cultures were analyzed and values are presented as mean ± SEM. Statistical significance was calculated by two-tailed Student t-test for unpaired samples. One asterisk shows p-value < 0.05 and two asterisks show p-value < 0.01, as compared to WT or irc20Δ as indicated. (B) Southern blot analysis of DNA isolated from WT, irc20Δ and slx8Δ using biotinylated probes specific to 2-μm sequence. The hybridized probes were then visualized using Chemiluminescent Nucleic Acid Detection Module (Thermo ScientificTM).
We next tested whether the copy number amplification in irc20Δ is associated with the formation of high molecular weight aggregates (HMW) as seen in several SUMO pathway mutants (Xiong et al., 2009). The 2-μm plasmid forms isolated from WT and irc20Δ mutant were analyzed by Southern blotting using biotinylated probes specific to the 2-μm sequence. Despite the increased copy number observed in irc20Δ mutant, we did not observe aberrant species of the 2-μm plasmid like those observed in the slx8Δ mutant (Figure 2B). This is, however, consistent with the lack of growth defects and cold sensitivities in the irc20Δ mutant.
Since HR only participates in copy number amplification initiated by Flp1 incision at the FRTs, we tested whether there could be an increased Flp1 activity in the absence of Irc20. To test this, the FLP1 gene was cloned into a centromeric plasmid (pRS416), under the GAL1-promoter with a YFP-tag at the C-terminus (Figure 3A). The repressible GAL1-promoter allows for shutting off expression, to monitor the existing levels of a protein without interference from newly expressed ones. This plasmid was transformed into WT, irc20Δ, Irc20-DE534-535AA and Irc20-C1239A [cir0] strains and were grown overnight in raffinose containing media. To induce Flp1 expression, the cells were grown for 6 h in galactose, then transferred to glucose-containing medium, to switch off expression and monitor the degradation of Flp1 in the presence or absence of Irc20. We observed less degradation of Flp1 in cells lacking Irc20 (Figures 3B,C). The levels of Flp1 reduced to 31–34% by 45 min in wild-type and Irc20-DE534-535AA mutant following switching to glucose media, whereas it remained at 57–58% levels in the irc20Δ and Irc20-C1239A mutants. This suggests that the increased copy number of the 2-μm plasmid in irc20Δ mutant could be, at least partly, due to increased Flp1 levels. We next measured the steady state Flp1 levels in WT and irc20Δ using a Myc tagged Flp1 to understand how the degradation rate reflects on the steady state level of Flp1 when expressed from its native promoter. Similar to a previous report (Chen et al., 2005), we found that even though C-terminal tagging of Flp1 does not affect its activity in vitro (Chen and Rice, 2003), the Flp1 tagging reduces the accumulation of 2-μm plasmid in irc20Δ (1.5-fold) compared to unmodified 2-μm plasmid (4-fold) (Supplementary Figure 2A). However, even though 2-μm plasmid levels were elevated in irc20Δ, we found similar Flp1 levels in both WT and irc20Δ when whole cell extracts were analyzed (Supplementary Figure 2B). This is likely because the expression of Flp1 from its native promoter is rapidly repressed when the 2-μm plasmid levels rise. Altogether, Irc20 seems to play a role in facilitating the degradation of Flp1 in the cell, and acts as an additional control over Flp1 to prevent the toxic increase in 2-μm plasmid copy number.
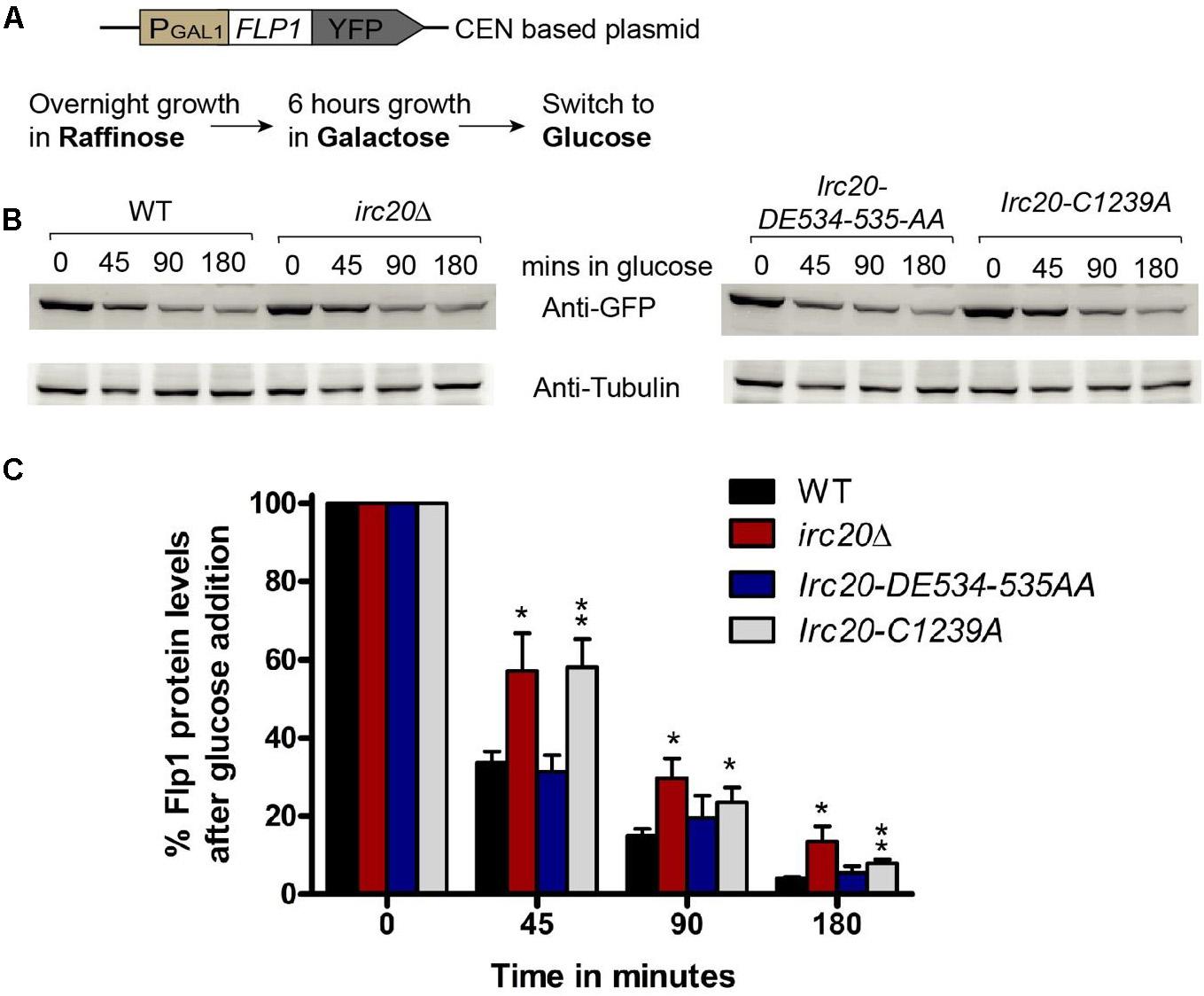
Figure 3. Loss of Irc20 ubiquitin ligase activity increases the levels of Flp1. (A) WT, irc20Δ, Irc20-DE534-535AA and Irc20-C1239A [cir0] mutants transformed with a centromeric plasmid (pRS416) expressing YFP-tagged Flp1 under GAL1-promoter were grown for 6 h in media containing 2% galactose to induce expression of Flp1 and then transferred to media containing 2% glucose to monitor the degradation of Flp1. (B) Whole cell extracts of cells grown for 45, 90, and 180 min in media containing glucose were analyzed for Flp1 levels by western blotting using anti-GFP antibody. (C) Band intensities were normalized to Tubulin band signals, and quantified as a percentage of signals before glucose addition. Values are the average of at least four replicate experiments, and are presented as mean ± SEM. Statistical significance was calculated by two-tailed Student t-test for unpaired samples. One asterisk shows p-value < 0.05 and two asterisks show p-value < 0.01, as compared to WT.
Discussion
Irc20 was identified in a screen for genes causing increased recombination centers when deleted (Alvaro et al., 2007). The absence of Irc20 was shown to cause a general reduction in SDSA and crossover recombination, suggesting a regulatory role for Irc20 in recombination (Miura et al., 2012). Irc20 has two conserved domains, a SNF2 ATP hydrolyzing domain and a RING domain characteristic for E3 ubiquitin ligases, in addition to at least two SIMs, suggesting a role as a STUbL. E3 ubiquitin ligases including STUbLs play a regulatory role on various proteins in diverse cellular pathways.
Here we expand the cellular role of Irc20 to include the proper maintenance of the endogenous 2-μm plasmid. First, we show that mutations in irc20 frequently caused the loss of the otherwise highly stable 2-μm plasmid, in a manner dependent on Flp1 expression, and not through the control of the Rep1-Rep2-STB pathway. In addition to a role in plasmid segregation, we show that despite the lack of cell sensitivities associated with increased 2-μm plasmid levels, irc20Δ mutant increased 2-μm plasmid levels 4-folds compared to the WT. Since Flp1 overexpression causes an amplification of 2-μm plasmid copy number (Reynolds et al., 1987; Xiong et al., 2009), we checked the role of Irc20 in regulating Flp1 protein levels. We monitored the YFP-tagged Flp1 levels in WT and irc20Δ after expressing from a repressible promoter and our results showed higher levels of Flp1 in irc20Δ, even though the steady state levels of the protein is not affected when expressed from its native promoter. This could be because of the autoregulatory nature of 2-μm plasmid genes, where the expression of Flp1 is rapidly repressed when the 2-μm plasmid levels rise, due to an increase in Rep1-Rep2 complex that act as repressor at the Flp1 promoter (Murray et al., 1987; Reynolds et al., 1987; Som et al., 1988). Altogether, our data suggests that Irc20 fine-tunes Flp1 levels, thereby controlling the amplification of the 2-μm plasmid copy number.
The reduced turnover of Flp1 when irc20 is mutated is similar to that observed in siz1Δsiz2Δ, slx5Δ and slx8Δ which affect the SUMOylation and/or the subsequent ubiquitin dependent degradation of Flp1 (Ma et al., 2019). The resulting increase in copy number and formation of HMW aggregates is, however, different in case of irc20 mutants. The increase in 2-μm plasmid copy number observed in irc20Δ mutant is not as high as that in other SUMO pathway mutants (10 to 30-folds) (Xiong et al., 2009), and the increased Flp1 induced recombination does not lead to the formation of HMW aggregates. We thus speculate that improper resolution of daughter plasmids during the Flp1-dependant amplification of copy number is not solely due to increased Flp1 levels or its deficient SUMOylation, but due to an additional role for Siz1, Siz2, Slx5, and Slx8 in regulation of HR. Several HR proteins are known to be SUMOylated during HR repair, both facilitating the assembly of repair foci, and inducing a Slx5-Slx8 control on HR protein levels (Psakhye and Jentsch, 2012). The increase in Flp1-induced nicks concomitant with a lack of regulatory control over HR, would lead to the aberrant processing of the replicated plasmids into monomers in daughter cells, thus HMW aggregate formation. This is consistent with that slx5Δ and slx8Δ still produces toxic 2-μm plasmid forms even when Flp-SUMO is expressed, despite being attenuated in strand cleavage (Ma et al., 2019). The reason would be deficient regulation on HR proteins. In the case of irc20 mutants, however, the increase in Flp1 protein levels is associated with a reduction in HR, thus would only cause a small increase in 2-μm plasmid levels. A similar modest increase in copy number without significant growth sensitivities was previously reported for the ubiquitin conjugating enzyme Ubc4 (Sleep et al., 2001).
The Flp1-dependent increase in 2-μm copy number in irc20Δ does not explain, and somewhat contradicts the low 2-μm plasmid segregation efficiency observed in the irc20Δ mutant. We can only speculate that the imprecise repair in the absence of Irc20 (Miura et al., 2012), causes mutations in the regions surrounding the FRTs possibly the STB, thus affecting the integrity of the partitioning system, in a manner dependent on Flp1-induced incision, and HR repair. Altogether, we believe that Irc20 regulates the copy number of the 2-μm plasmid through regulating Flp1 levels, possibly by assisting its degradation. Irc20 also seems to be important in the precise recombination, and its absence could lead to mutations affecting the plasmid’s segregation efficiency.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
AH obtained the funding and conceived and supervised the research. DJ designed and performed the experiments and analyzed the data with help from JC throughout. Experiments in Figures 2B, 3 and Supplementary Figure 2 were performed by JC as well as plasmid cloning and some strain constructions. DJ wrote the manuscript with contribution from JC and AH. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the funding from United Arab Emirates University-UPAR; and Terry Fox Foundation [to AH].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Fransesca Storici (Georgia Institute of Technology) for delitto perfetto plasmids and protocols and Melanie Dobson (Dalhousie University) for pKan4 plasmid. We would like to thank Akela Ghazawi and Suneesh Kaimala for help with the Southern blotting.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2020.00221/full#supplementary-material
References
Alvaro, D., Lisby, M., and Rothstein, R. (2007). Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 3:e228. doi: 10.1371/journal.pgen.0030228
Brown, T. (1999). Analysis of DNA Sequences by Blotting and Hybridization. Hoboken, NJ: John Wiley & Sons.
Burgess, R. C., Rahman, S., Lisby, M., Rothstein, R., and Zhao, X. (2007). The Slx5-Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol. Cell. Biol. 27, 6153–6162. doi: 10.1128/mcb.00787-07
Chan, K. M., Liu, Y.-T., Ma, C.-H., Jayaram, M., and Sau, S. (2013). The 2 micron plasmid of Saccharomyces cerevisiae: a miniaturized selfish genome with optimized functional competence. Plasmid 70, 2–17. doi: 10.1016/j.plasmid.2013.03.001
Chen, X. L., Reindle, A., and Johnson, E. S. (2005). Misregulation of 2 microm circle copy number in a SUMO pathway mutant. Mol. Cell Biol. 25, 4311–4320. doi: 10.1128/mcb.25.10.4311-4320.2005
Chen, Y., and Rice, P. A. (2003). The role of the conserved Trp330 in Flp-mediated recombination. Functional and structural analysis. J. Biol. Chem. 278, 24800–24807. doi: 10.1074/jbc.m300853200
Dobson, M. J., Pickett, A. J., Velmurugan, S., Pinder, J. B., Barrett, L. A., Jayaram, M., et al. (2005). The 2 microm plasmid causes cell death in Saccharomyces cerevisiae with a mutation in Ulp1 protease. Mol. Cell Biol. 25, 4299–4310. doi: 10.1128/mcb.25.10.4299-4310.2005
Douglas, P. M., Treusch, S., Ren, H. Y., Halfmann, R., Duennwald, M. L., Lindquist, S., et al. (2008). Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc. Natl. Acad. Sci. U.S.A. 105, 7206–7211. doi: 10.1073/pnas.0802593105
Flaus, A., Martin, D. M., Barton, G. J., and Owen-Hughes, T. (2006). Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 34, 2887–2905. doi: 10.1093/nar/gkl295
Freemont, P. S. (2000). RING for destruction? Curr. Biol. 10, R84–R87. doi: 10.1016/S0960-9822(00)00287-6
Futcher, A. B. (1986). Copy number amplification of the 2 micron circle plasmid of Saccharomyces cerevisiae. J. Theor. Biol. 119, 197–204. doi: 10.1016/s0022-5193(86)80074-1
Futcher, A. B., and Cox, B. S. (1983). Maintenance of the 2 microns circle plasmid in populations of Saccharomyces cerevisiae. J. Bacteriol. 154, 612–622. doi: 10.1128/jb.154.2.612-622.1983
Jayaram, M., Mehta, S., Uzri, D., Voziyanov, Y., and Velmurugan, S. (2004). Site-specific recombination and partitioning systems in the stable high copy propagation of the 2-micron yeast plasmid. Prog. Nucleic Acid. Res. Mol. Biol. 77, 127–172. doi: 10.1016/s0079-6603(04)77004-x
Liu, Y. T., Sau, S., Ma, C. H., Kachroo, A. H., Rowley, P. A., Chang, K. M., et al. (2014). The partitioning and copy number control systems of the selfish yeast plasmid: an optimized molecular design for stable persistence in host cells. Microbiol. Spectr. 2. doi: 10.1128/microbiolspec.PLAS-0003-2013
Longtine, M. S., McKenzie, A. III, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., et al. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. doi: 10.1002/(sici)1097-0061(199807)14:10<953::aid-yea293>3.0.co;2-u
Ma, C. H., Su, B.-Y., Maciaszek, A., Fan, H.-F., Guga, P., and Jayaram, M. (2019). A Flp-SUMO hybrid recombinase reveals multi-layered copy number control of a selfish DNA element through post-translational modification. PLoS Genet. 15:e1008193. doi: 10.1371/journal.pgen.1008193
McQuaid, M. E., Polvi, E. J., and Dobson, M. J. (2019). DNA sequence elements required for partitioning competence of the Saccharomyces cerevisiae 2-micron plasmid STB locus. Nucleic Acids Res. 47, 716–728. doi: 10.1093/nar/gky1150
Miura, T., Yamana, Y., Usui, T., Ogawa, H. I., Yamamoto, M. T., and Kusano, K. (2012). Homologous recombination via synthesis-dependent strand annealing in yeast requires the Irc20 and Srs2 DNA helicases. Genetics 191, 65–78. doi: 10.1534/genetics.112.139105
Murray, J. A., Scarpa, M., Rossi, N., and Cesareni, G. (1987). Antagonistic controls regulate copy number of the yeast 2 mu plasmid. Embo J. 6, 4205–4212. doi: 10.1002/j.1460-2075.1987.tb02768.x
Pinder, J. B., McQuaid, M. E., and Dobson, M. J. (2013). Deficient sumoylation of yeast 2-micron plasmid proteins Rep1 and Rep2 associated with their loss from the plasmid-partitioning locus and impaired plasmid inheritance. PLoS One 8:e60384. doi: 10.1371/journal.pone.0060384
Psakhye, I., and Jentsch, S. (2012). Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151, 807–820. doi: 10.1016/j.cell.2012.10.021
Reynolds, A. E., Murray, A. W., and Szostak, J. W. (1987). Roles of the 2 microns gene products in stable maintenance of the 2 microns plasmid of Saccharomyces cerevisiae. Mol. Cell Biol. 7, 3566–3573. doi: 10.1128/mcb.7.10.3566
Richardson, A., Gardner, R. G., and Prelich, G. (2013). Physical and genetic associations of the Irc20 ubiquitin ligase with Cdc48 and SUMO. PLoS One 8:e76424. doi: 10.1371/journal.pone.0076424
Sengupta, A., Blomqvist, K., Pickett, A. J., Zhang, Y., Chew, J. S., and Dobson, M. J. (2001). Functional domains of yeast plasmid-encoded Rep proteins. J. Bacteriol. 183, 2306–2315. doi: 10.1128/jb.183.7.2306-2315.2001
Sleep, D., Finnis, C., Turner, A., and Evans, L. (2001). Yeast 2 microm plasmid copy number is elevated by a mutation in the nuclear gene UBC4. Yeast 18, 403–421. doi: 10.1002/yea.679
Som, T., Armstrong, K. A., Volkert, F. C., and Broach, J. R. (1988). Autoregulation of 2 micron circle gene expression provides a model for maintenance of stable plasmid copy levels. Cell 52, 27–37. doi: 10.1016/0092-8674(88)90528-4
Stuckey, S., Mukherjee, K., and Storici, F. (2011). In vivo site-specific mutagenesis and gene collage using the delitto perfetto system in yeast Saccharomyces cerevisiae. Methods Mol. Biol. 745, 173–191. doi: 10.1007/978-1-61779-129-1_11
Tsalik, E. L., and Gartenberg, M. R. (1998). Curing Saccharomyces cerevisiae of the 2 micron plasmid by targeted DNA damage. Yeast 14, 847–852. doi: 10.1002/(sici)1097-0061(19980630)14:9<847::aid-yea285>3.0.co;2-9
Wong, M. C., Scott-Drew, S. R., Hayes, M. J., Howard, P. J., and Murray, J. A. (2002). RSC2, encoding a component of the RSC nucleosome remodeling complex, is essential for 2 microm plasmid maintenance in Saccharomyces cerevisiae. Mol. Cell Biol. 22, 4218–4229. doi: 10.1128/mcb.22.12.4218-4229.2002
Xiong, L., Chen, X. L., Silver, H. R., Ahmed, N. T., and Johnson, E. S. (2009). Deficient SUMO attachment to Flp recombinase leads to homologous recombination-dependent hyperamplification of the yeast 2 microm circle plasmid. Mol. Biol. Cell 20, 1241–1251. doi: 10.1091/mbc.e08-06-0659
Yang, X. M., Mehta, S., Uzri, D., Jayaram, M., and Velmurugan, S. (2004). Mutations in a partitioning protein and altered chromatin structure at the partitioning locus prevent cohesin recruitment by the Saccharomyces cerevisiae plasmid and cause plasmid missegregation. Mol. Cell Biol. 24, 5290–5303. doi: 10.1128/mcb.24.12.5290-5303.2004
Keywords: homologous recombination, Irc20, ubiquitin ligase, SUMO, Flp1, 2-μm plasmid
Citation: Jalal D, Chalissery J and Hassan AH (2020) Irc20 Regulates the Yeast Endogenous 2-μm Plasmid Levels by Controlling Flp1. Front. Mol. Biosci. 7:221. doi: 10.3389/fmolb.2020.00221
Received: 23 February 2020; Accepted: 07 August 2020;
Published: 19 November 2020.
Edited by:
Cesare Indiveri, University of Calabria, ItalyReviewed by:
Román González-Prieto, Leiden University Medical Center, NetherlandsYuqi Wang, Saint Louis University, United States
Copyright © 2020 Jalal, Chalissery and Hassan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ahmed H. Hassan, ahmedh@uaeu.ac.ae
 Deena Jalal
Deena Jalal Jisha Chalissery
Jisha Chalissery Ahmed H. Hassan
Ahmed H. Hassan