- 1State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Institute of Agro-product Safety and Nutrition, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
- 2State Key Laboratory of Grassland Agro-ecosystems, Key Laboratory of Grassland Livestock Industry Innovation, Ministry of Agriculture and Rural Affairs, College of Pastoral Agriculture Science and Technology, Lanzhou University, Lanzhou, China
- 3Department of Plant Pathology, College of Agriculture, University of Sargodha, Sargodha, Pakistan
- 4College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, China
- 5Faculty of Natural Sciences, Plant Cytogenetics and Molecular Biology Group, Institute of Biology, Biotechnology and Environmental Protection, University of Silesia in Katowice, Katowice, Poland
- 6Department of Plant Sciences, Karakoram International University, Gilgit, Gilgit Baltistan, Pakistan
- 7Colin Ratledge Center for Microbial Lipids, School of Agriculture Engineering and Food Science, Shandong University of Technology, Zibo, China
- 8Department of Biotechnology Engineering, Institute of Engineering and Technology, Bundelkhand University, Jhansi, India
Alfalfa plays a significant role in the pasture ecosystems of China’s north, northeast, and northwest regions. It is an excellent forage for livestock, improves soil structure, prevents soil erosion, and has ecological benefits. Presently root rot is a significant threat to the alfalfa productivity because of the survival of the pathogens as soil-borne and because of lack of microbial competition in the impoverished nutrient-deficient soils and resistant cultivars. Furthermore, these regions’ extreme ecological and environmental conditions predispose alfalfa to root rot. Moisture and temperature, in particular, have a considerable impact on the severity of root rot. Pathogens such as Fusarium spp. and Rhizoctonia solani are predominant, frequently isolated, and of major concern. These pathogens work together as disease complexes, so finding a host genotype resistant to disease complexes is challenging. Approaches to root rot control in these regions include mostly fungicides treatments and cultural practices and very few reports on the usage of biological control agents. As seed treatment, fungicides such as carbendazim are frequently used to combat root rot; however, resistance to fungicides has arisen. However, breeding and transgenic approaches could be more efficient and sustainable long-term control strategies, especially if resistance to disease complexes may be identified. Yet, research in China is mainly limited to field investigation of root rot and disease resistance evaluation. In this review, we describe climatic conditions of pastoral regions and the role of alfalfa therein and challenges of root rot, the distribution of root rot in the world and China, and the impact of root rot pathogens on alfalfa in particular R. solani and Fusarium spp., effects of environmental factors on root rot and summarize to date disease management approach.
Introduction
Alfalfa (Medicago sativa L.,) is a perennial legume and has long been utilized as a forage crop due to its high yield, nutritional value and adaptability to wide range of soils and under various climatic conditions (Khoury et al., 2014; Luo et al., 2014; Jiang et al., 2021). In China, alfalfa is mainly grown for feeding grazing livestock (Jiang et al., 2021). Root rot is the major limiting factor to alfalfa production worldwide. It is estimated that the annual yield loss worldwide caused by root rot is 20 to 40% (Wang et al., 2020a). Root rot is a destructive disease complex caused by several plant pathogenic fungi and oomycetes. Among these pathogens, Fusarium spp., and Rhizoctonia solani are the most damaging and frequently occurring root rot pathogens (Anjanappa et al., 2016; Cao et al., 2020; Cai et al., 2021; Jiang et al., 2021; Xu et al., 2021; Zhao et al., 2021). In China, root rot is endemic in major alfalfa producing regions such as northwest China (Gansu, Xinjiang, Qinghai, Shaanxi, and Ningxia Huizu), north China (Hebei, Shanxi, and Inner Mongolia), and northeast China (Heilongjiang, Liaoning, and Jilin), with death rates of more than 60% on severe plots (Fang et al., 2019, 2021). The root-specific symptoms go unreported or are not visible, and plants that show symptoms aboveground do not recover. Root lesions of various sizes and colors (reddish, brownish, and blackish) and browning and weakening of root tips, yellowing and wilting of leaves, slowed plant growth, lower yield, and crop loss are some of the symptoms associated with root rot (Fang et al., 2019). Environmental factors in particular soil temperature and soil moisture have profound effects on the expression and severity of root rot and the consequent productivity of alfalfa (Yinghua et al., 2019; Sharath et al., 2021). In response to root rot, researchers in China have carried out a series of research, focusing mainly on the investigation of the occurrence and severity of the disease, the identification and isolation of the pathogen, the determination of pathogenicity and biological characteristics, evaluation of alfalfa species resistance and cultural, chemical, and biological control (Fang et al., 2021; Zhang et al., 2021). These attempts, however, have only been somewhat successful (Pan et al., 2015; Zhang et al., 2021). Furthermore, it is hard to develop a resistant cultivar resistant to diverse pathogens. Subsequently, farmers rely only on chemical control; nonetheless, fungicide resistance has been recorded. Due to a lack of understanding about the disease and the dire threat root rot poses to the alfalfa in China, there is a pressing need to research the biology, ecology, epidemiology, and management of root rot. This review: (i) describes the climatic conditions of pastoral regions of China and role alfalfa therein and challenges from root rot; (ii) discusses the distribution of root rot in the world and China; (iii) addresses the status of root rot in the world and China mainly focusing Fusarium spp. and R. solani, describes the disease cycle and biological characteristics including the symptoms they caused; and the challenges posed to alfalfa from these pathogens; (iv) addresses the environmental factors affecting the severity of the root rot; (v) addresses the approaches on the disease management made to date using cultural, breeding and transgenic, biological, chemical, gene silencing and editing.
Climatic condition of pastoral regions of China, role of alfalfa there in and the challenges from root rot
The climate of the northwest, north, and northeast region of China, where alfalfa is grown can best be described as “an arid, semi-arid and subhumid climate characterized by plenty of water but low solar radiation in the northeast, with a temperate climate, low precipitation and scarce water in north China and vast areas of low-quality land, abundant solar radiation and thermal resources, scarce water, desertification and salinization in the northwest (Morrison, 2004; Liang et al., 2006). In some locations of the northwest, water is plentiful but unevenly distributed (Su et al., 2004). The soil of these regions has originated from nutrient-poor, ancient parent materials that have been intensively weathered and leached (Delang, 2018). Besides, the soil is not very fertile and generally lacking in phosphorus, resulting in significantly reduced nutrient levels in particular northwest and north region (Wang et al., 2021). Alfalfa is the most famous perennial leguminous forage in the world. The United States is the world’s largest alfalfa producer with a planting area of 9 million hectares, followed by Argentina with a planting area of 6.9 million hectares. China ranks fifth globally with a planting area of 4.7 million hectares (Bao et al., 2016; Gao et al., 2018a; Guo et al., 2019). In China, alfalfa is the most crucial pasture legume in the northwest, northeast, and north regions. In the northwest, alfalfa is sown over an estimated area of 3151.9 thousand hm2, accounting for 66.43% of the country’s total area. North China is the second-largest alfalfa producing area in China, and alfalfa is sown over an estimated area of 1063.4 thousand hm2, accounting for 22.41% of China’s total area. Northeast alfalfa acreage accounted for a small proportion in China, with a planting area of only 234.5 thousand hm2, accounting for 4.94% of China (Feng and Kai, 2014 Shi et al., 2017). In recent years, the Chinese government has gradually allowed more land for forage crops such as alfalfa. Hence, alfalfa has become the most widely used forage legume in China’s integrated farming systems, grazing, and ecological conservation. Besides, the concept of “pasture-based livestock industries” alfalfa has gained attention in recent years, its planting area ranks first among all other forage crops reaching over almost 11 million acres by the end of 2017 (Shi et al., 2017). Compared to other forages and grain legumes like soybeans, alfalfa is the “queen of forage,” providing dairy with digestible protein, fiber, minerals, and vitamins at meager costs. While dairy is a relatively young agricultural business in China, with much lower average yields than in many other countries, boosting alfalfa consumption has been identified as a critical approach for improving milk supply and quality, particularly for big dairy farms. Large dairy farms in China have increased significantly (Qingbin and Yang, 2020). The advantages of alfalfa as a perennial pasture include its outstanding ability to prevent soil and wind erosion. In addition, extensive and deep root system of alfalfa is highly effective for building up organic matter, improving soil structure and soil fertility (Liu et al., 2013; Jun et al., 2014). Additionally, alfalfa is relatively tolerant to water deficit and water loggings. Besides, alfalfa is widely used for livestock and poultry, water and soil conservation, green manure, and disease breaks. However, continuous cropping of alfalfa over time in these regions has caused severe soil water deficit, soil desiccation, and depletion of shallow groundwater (Wang et al., 2021). Besides frequent droughts, soil moisture deficit, erratic precipitation, severe wind, and water erosions, and intensive grazing have made alfalfa vulnerable to root rot. Furthermore, the poor and nutrient-deficient soils, in particular phosphorus, these regions predispose alfalfa plants to root rot because microbial competition with the root rot pathogen in such conditions is generally lacking and, consequently, losses from root rot are exacerbated (Wang et al., 2016; Feng et al., 2020). Besides, cool soil temperature and soil compaction in these regions slows down alfalfa growth and predispose roots to root rot. Alfalfa can survive for ten years or more years; however, once infected by root rot, yield reduction starts even in the third or second year (Fang et al., 2019). Besides, toxins such as mycotoxins and phytotoxins produced by root rot pathogens may also pose a substantial danger to feed quality due to their effects on animal productivity and potentially on human food quality. In addition, increased costs and harmful effects of fungicides are the indirect losses due to root rot (Barbetti et al., 2007). Furthermore, root rot pathogens form pathogen complexes, thereby posing a synergistic influence on the severity of root rot (Gossen et al., 2016; Fang et al., 2019). These pathogen complexes respond differently to fungicides and other management practices, and the root rot they cause requires special treatment measures.
Distribution of root rot in the world and China
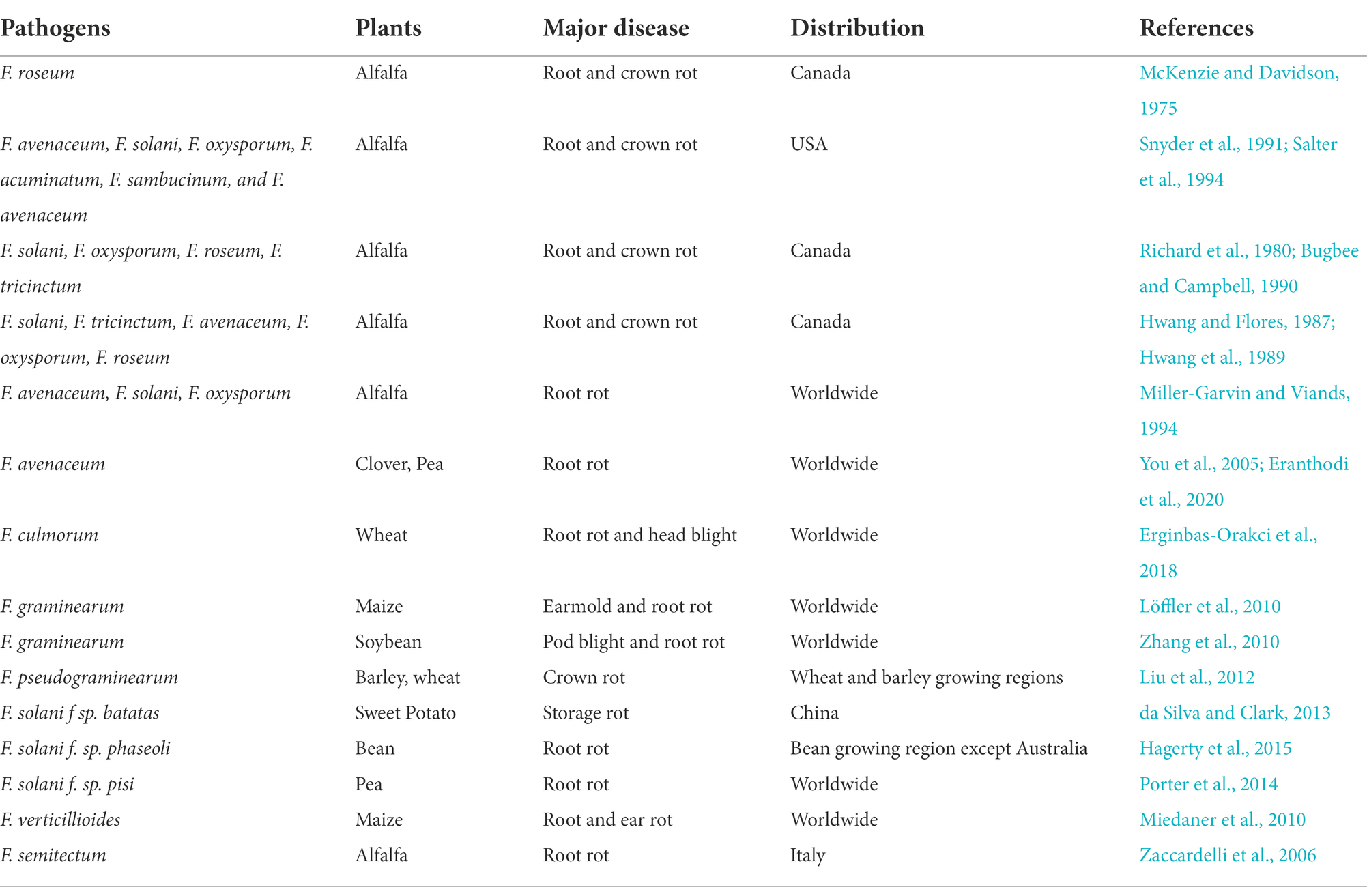
Root rot has seriously affected alfalfa production in the United States, Italy, China, Canada, Australia, Russia, Japan, and Argentina (Akamatsu et al., 2008; Yinghua et al., 2019). In Alberta and British Columbia, root rot of alfalfa was first recognized by McKenzie and Davidson (1975). About 60% production area was affected by root rot (McKenzie and Davidson, 1975). The primary pathogens responsible were R. solani, Phoma sclerotioides, F. roseum, and Phytophthora megasperma. In 1983, the incidence of crown and root rot of alfalfa was recorded in 24 alfalfa-growing areas in southern Alberta, Canada, and the average incidence was 61%, and the highest was 80%. Most of the alfalfa plants were either dead or withered, resulting in reduction of the yield and quality. The main pathogens were F. solani, F. tricinctum, F. avenaceum, F. oxysporum and Pythium irregular. In 1984–1987 a survey of alfalfa fields in the northeast and northwest Alberta, Canada, revealed that F. roseum and F. avenaceum were common pathogens associated with root rot (Hwang and Flores, 1987; Hwang et al., 1989). In contrast, research in Quebec, Canada, in the late 1980s revealed alfalfa fields were severely affected by Phytophthora spp., particularly in low-lying places, since the humid climatic condition was more conducive to proliferation and multiplication of Phytophthora spp. (Richard and Martin, 1991). In 1991, root rot affected alfalfa production in Nevada, United States, and the primary pathogens were Fusarium spp. (Snyder et al., 1991). Later in Wyoming and then in Idaho, United States, Phoma sclerotioides were first identified as the cause of widespread winterkill of alfalfa (Gray et al., 1997; Al-Sadi and Deadman, 2010; Al-Sadi, 2021). Subsequently, the root rot was successively found in all alfalfa-producing areas of the United States (Hollingsworth et al., 2003; Larsen et al., 2007). In 2006, Fusarium semitectum was identified as a major pathogen responsible for causing root rot in Italy (Garrett, 1970; Zaccardelli et al., 2006). Besides many reports on root rot from the United States, Canada, and Italy, root rot has also been reported in New Zealand, Japan, Russia, Australia, India, Brazil, Egypt, Nigeria, Finland, and many other countries (Snyder et al., 1991; Sharath et al., 2021). Hence, numerous root rot pathogens have been shown to have varying degrees of direct involvement in causing root rot disease of alfalfa in the world. Likewise, the distribution of root rot pathogens associated with root rots is influenced by environmental factors such as moisture and temperature. For example, F. pseudograminearum was more widespread during the low-rainfall years in the Pennsylvania of the USA and the low rainfall regions of Australia. Whereas F. culmorum was predominant in the high rainfall areas of eastern Australia and in the cooler and higher altitude areas of Idaho, USA (Smiley and Yan, 2009).
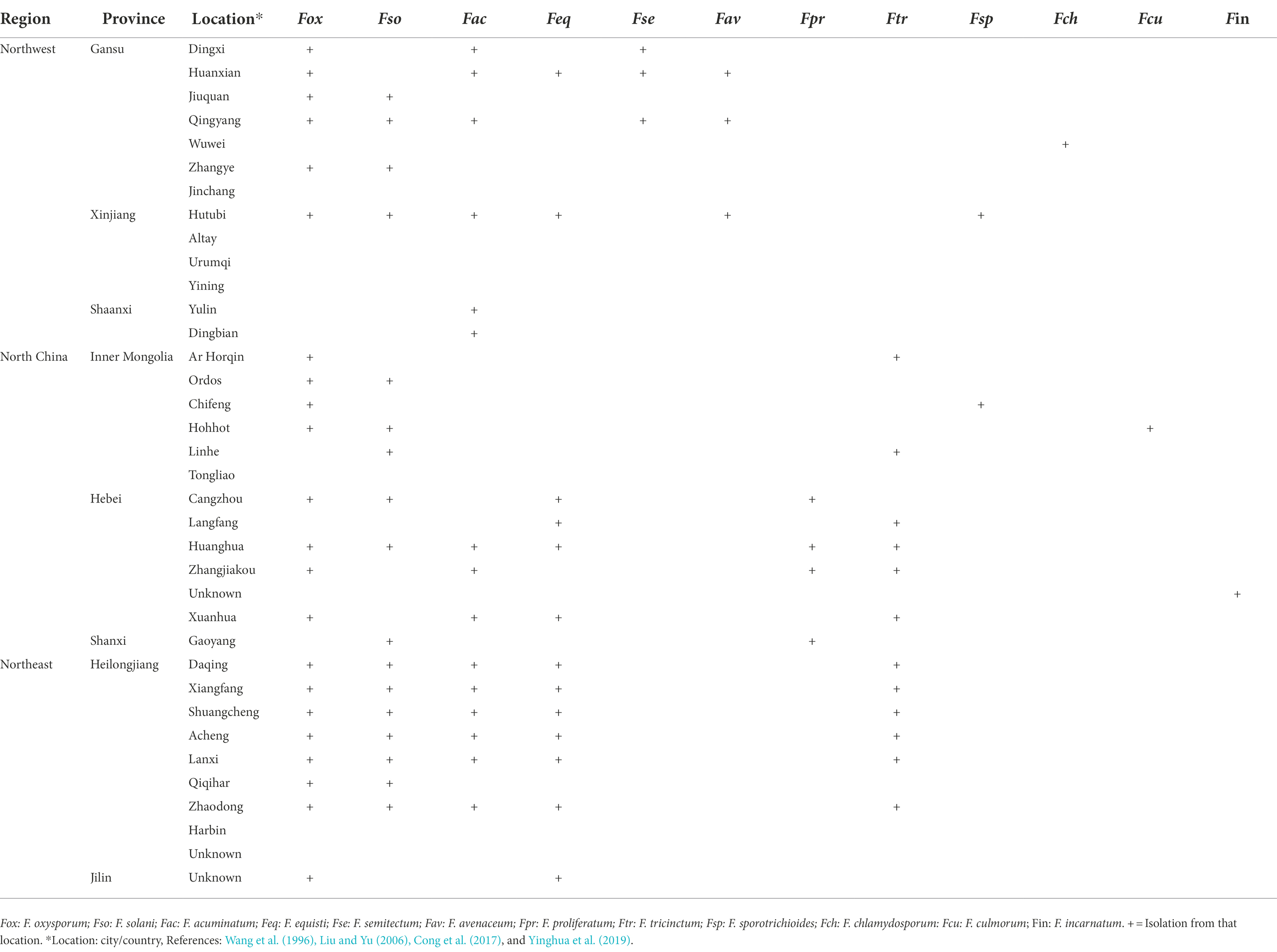
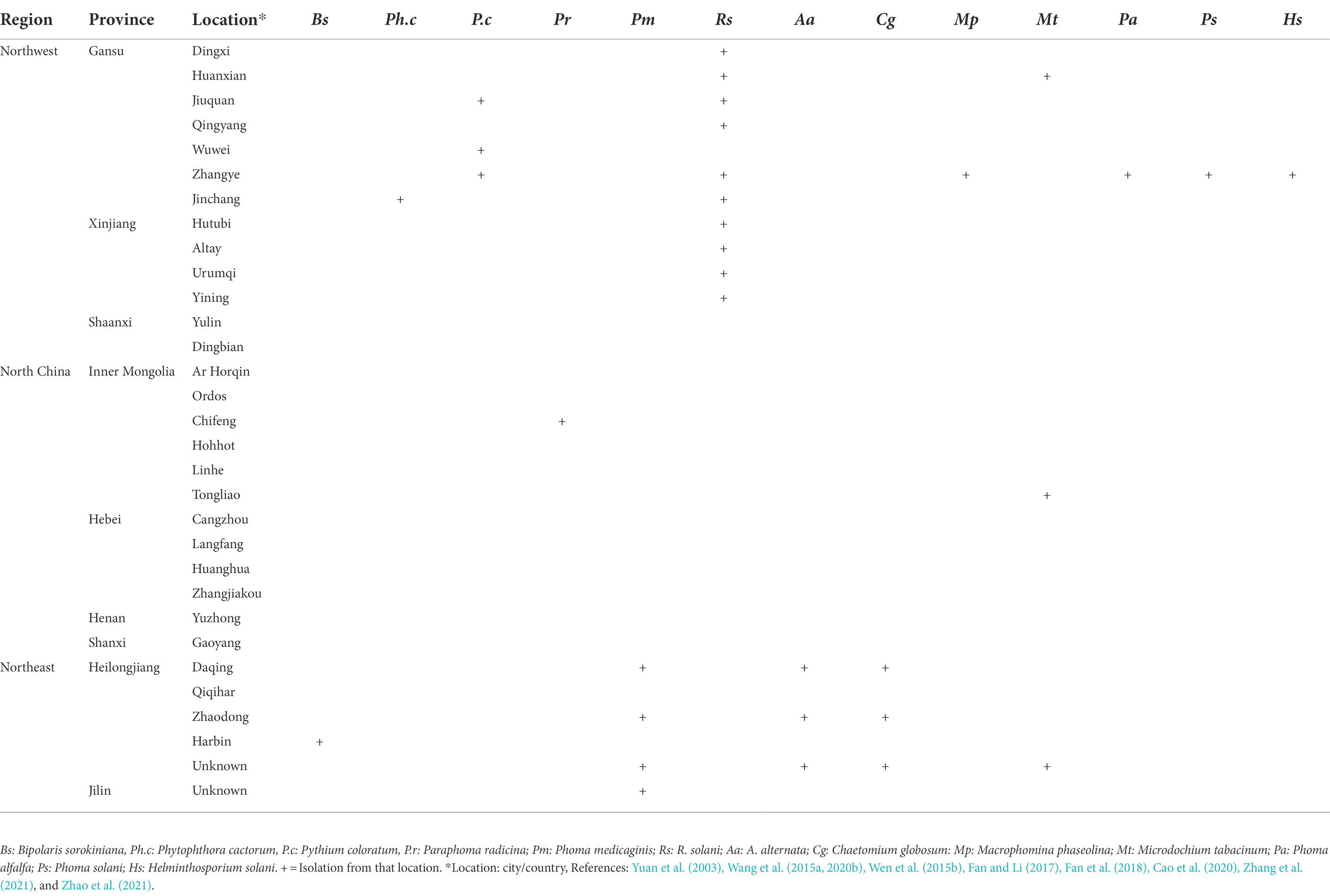
In China, root rot was first recognized by Yao (1989) in Xingjiang and Gansu provinces. Large areas of alfalfa were found to be affected by root rot. Presently, alfalfa root rot has been reported in 11 provinces, with more in the northwest (Xinjiang, Gansu), northeast (Heilongjiang, Jilin), and north region (Inner Mongolia; Figure 1). China’s pastoral areas are concentrated in these regions, particularly the northwest and north regions, where extensive livestock-raising is the leading agricultural enterprise. A range of forages, including alfalfa, is grown in these regions. Root rot is likely to occur in alfalfa in Qinghai, Liaoning, and Tibet, as reported on other legumes (Zhimin, 1996). Still, we could not find any published report on alfalfa root rot in these provinces (Figure 1). Furthermore, there are no published reports of root rot in China’s southern and south regions. These regions are agricultural areas where limited forages are sown. Many pathogens have been shown to have varying degrees of direct involvement in causing root disease. Fusarium spp. and R. solani are predominant and widely distributed root rot pathogens and have been most frequently isolated and described (Yao, 1989; Gang et al., 1996; Li, 2003; Li et al., 2009; Liu et al., 2016; Wang et al., 2016; Cong et al., 2016a; Jiang et al., 2021). Also, these pathogens are serious root rot pathogens in various economically important crops, including grain and forage legumes worldwide (Barbetti and MacNish, 1984; Barbetti et al., 2007; Ajayi-Oyetunde and Bradley, 2016, 2018). The distribution of Fusarium spp. associated with alfalfa root rot in the northeast, north, and northwest region of China is relatively well understood compared to other pathogens, as shown in Table 1. Studies showed that environmental factors, mainly moisture, and temperature, affect root rot (Barbetti et al., 2007). Rainfall in these regions is erratic, resulting in the production of chlamydospores and sclerotia that enable Fusarium spp. and R. solani to survive in prolonged dry periods in these regions. The same distribution trend of these fungi was also noted in the semi-arid, arid, and subhumid regions of the world (Barbetti, 1983; Smiley and Yan, 2009). For example, in Australia, Fusarium spp. is highly distributed in the dry regions where rainfall is erratic. Accordingly, the fungus produces chlamydospores, enabling them to survive more in the dry period than other pathogens (Summerell et al., 2011). In recent years, oomycetes such as Pythium spp. and Phytophthora spp. are causing severe root rot in the irrigated areas of Gansu and Sichuan provinces (Yinghua et al., 2019; Zhang et al., 2020). In addition, Macrophomina phaseolina, Phoma spp., Paraphoma spp. and Microdochium tabacinum have also been recovered from root rot in Gansu province and Inner Mongolia (Kaur et al., 2012; Wang et al., 2015a; Lakhran et al., 2018; Cao et al., 2020; Marquez et al., 2021; Zhao et al., 2021). These fungi in combination with Fusarium spp., R. solani and oomycetes resulting in severe root rot (Min-Quan et al., 2003; Huang et al., 2013; Wen et al., 2015b; Cong et al., 2016b; Zhang et al., 2020; Wang et al., 2020b; Fang et al., 2021). In contrast to northwest and north, the northeast region (Heilongjiang, Jilin) is relatively fertile well-developed agriculture and has plenty of water. From the northeast region, Bipolaris sorokiniana and Alternaria alternata in addition to above mentioned pathogens have also been reported (Li et al., 2019; Jiang et al., 2021).
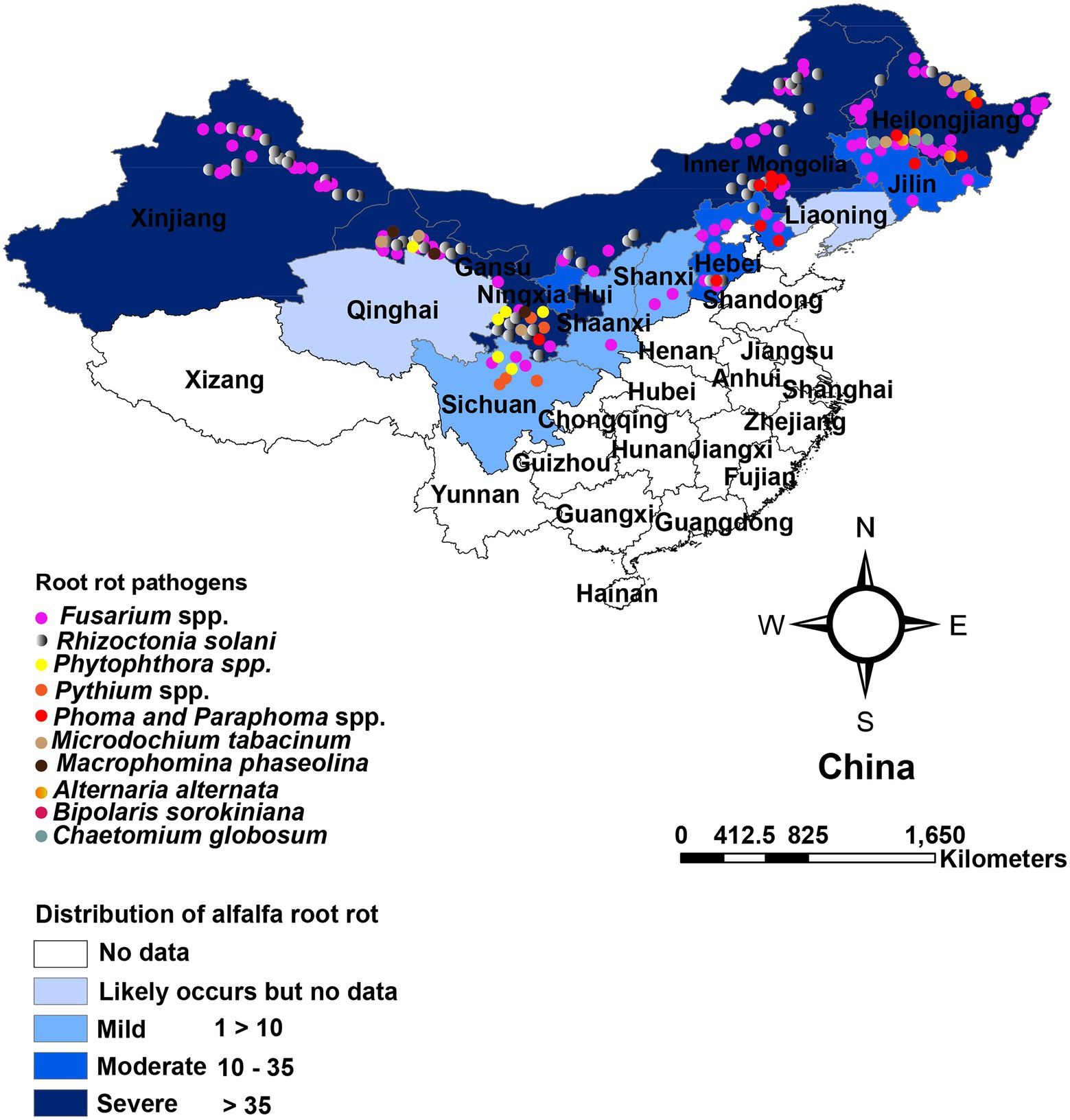
Figure 1. Geographical distribution and affected region in China. Color depth indicates affected provinces, and dots indicate root rot pathogens based on China Academic Journals full-text database (CNKI), Web of sciences (WoS), and other websites.
Fungal and oomycetes pathogens
Most root rot pathogens are soil-borne and can survive for many years by producing resilient structures such as sclerotia and chlamydospores. Figure 2 depicts the inoculum sources, including the resilient structures and symptoms in addition to root rot caused by these pathogens on alfalfa. Recent reports suggest that these pathogens can also be seed-borne (Kong et al., 2018b). However, there is no report on the relationship between the disease incidence of root rot and pathogen-associated with alfalfa seeds. Hence seed transmission seems unimportant in the etiology of root rot of alfalfa in China. Barbetti (1983) removed fungi from the clover seeds and found no influence on the severity of root rot diseases after sowing (Barbetti, 1983). Among these pathogens, Fusarium spp. and R. solani are accountable for incalculable losses to alfalfa compared to other fungi. The status of these pathogens involved in root rot in China and abroad, biological characteristics and disease cycles are addressed further in the below sections. Other fungi such as Bipolaris sorokiniana, Paraphoma spp. and Phoma spp. that contribute to root rot in alfalfa but are not causing significant losses are shown in Table 2 (de Gruyter et al., 2009, 2010; Moslemi et al., 2018; Jiang et al., 2021).
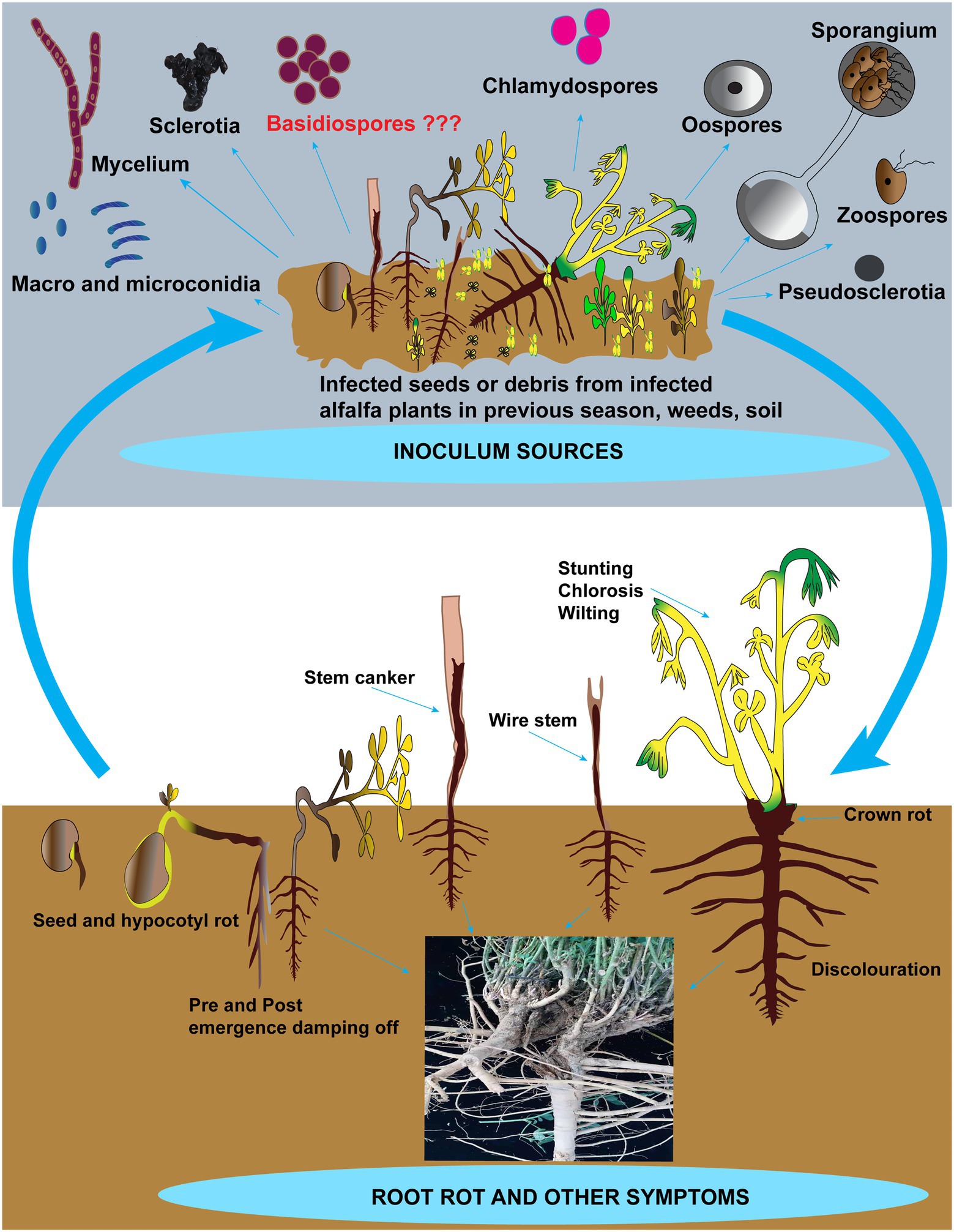
Figure 2. Schematic representation of alfalfa root rot and other symptoms and inoculum sources. Inoculum sources include zoospores, oospores, sporangia, conidia, chlamydospores, sclerotia, mycelia, and basidiospores (not reported). Besides root rot, other symptoms include damping-off, seed and hypocotyl rots, discoloration on roots, crown rot, stunting, chlorosis, wilting, and wire stem.
Fusarium spp.
Fusarium is a cosmopolitan genus that includes filamentous ascomycetes fungi (Sordariomycetes: Hypocreales: Nectriaceae). Link (1809) was the first to describe and identify the genus (Link, 1809). The genus is exceptionally complicated, and its taxonomy has always been contentious due to polyphyletic grouping. Currently, between 100 and 500 species are reported globally (Summerell et al., 2010). The genus includes pathogens that cause severe disease of plants, endophytes, saprophytes and produce several mycotoxins and/or phytotoxins. These mycotoxins and/or phytotoxins can render alfalfa unfit for animal feeding, and some of them may act as virulence factors in enhancing root rot disease (Nedelnik and Repkova, 1997). According to a recent investigation, toxins, including trichothecenes, zearalenone, and fumonisins are not only involved in disease pathogenesis but can also cause human and animal poisoning in China. Besides, these toxins severely affect the germination of alfalfa seeds (Kong et al., 2018a). Fusarium diseases collectively include wilts, rots, blights, and cankers of many horticultural, field, ornamental, forage, and forest crops in natural and agricultural ecosystems (Coleman, 2016). Fusarium root rot caused by Fusarium spp. is harmful in all growing stages of alfalfa, consequently reducing nitrogen fixation ability and longevity, and productivity (Kong et al., 2018a).
Status of Fusarium root rot in China and abroad
Fusarium spp. root rot is the most common root disease of many crops, including alfalfa globally (Table 3). This disease has considerably affected alfalfa in the United States, Canada, New Zealand, Australia, Italy, and India. Countries like Egypt and Japan also reported more than 50% prevalence rates in some areas (Samac et al., 2013). In China, Fusarium root rot is endemic in the north (Inner Mongolia), northeast (Heilongjiang, Jilin), and northwest (Xinjiang, Gansu, Qinghai) regions. The most affected provinces in these regions are Xinjiang, Gansu, Inner Mongolia and Heilongjiang (Fang et al., 2019, 2021). The disease hinders the establishment of the alfalfa stand, reduces yield and forage quality, and shortens the plant’s lifespan (Gang et al., 1996; Wang et al., 2005). In the northwest provinces, i.e., Xingjiang and Gansu, a 60% death rate of alfalfa was recorded. In the northeast region of China, i.e., in Jilin and Heilongjiang provinces, the disease becomes severe in August, with an incidence rate of 20 to 40% and a peak rate of around 92%. Similarly, in north China, i.e., in Inner Mongolia and Hebei, the disease becomes severe from August to October, with an incidence rate of 15 to 30% (Shouyan et al., 2011; Chen et al., 2015b; Cong et al., 2018; Wang et al., 2020a; Jiang et al., 2021). Besides, Fusarium spp. and R. solani co-infection or mixed infections increased root rot disease severity and decreased alfalfa growth and biomass allocation (Li et al., 2009; Wei et al., 2016; Fang et al., 2021). Among the Fusarium spp. F. oxysporum is the most damaging to alfalfa production in China (Fang et al., 2021; Table 1). F. oxysporum is also one of the top ten most economically important fungal pathogens, with over 100 formae speciales (f. spp.) based on host specificity (Edel-Hermann and Lecomte, 2019). Furthermore, F. oxysporum is the host-specific pathogen of alfalfa and other Medicago spp. (Batnini et al., 2020).
Biological characteristics, disease cycle, and damages to alfalfa
Fusarium spp. overwinter on plant debris/residues, seeds, and soils in the form of spores (microconidia and macroconidia), mycelial fragments, and chlamydospores (Cong et al., 2016a,b). The chlamydospores are more durable dormant structures and are regarded as the principal form in which Fusarium spp. survives in the soil for decades. These are enlarged thick-walled vegetative cells and are considered adaptations for survival during unfavorable environmental conditions such as prolonged dry periods (Fang et al., 2019). Chlamydospores usually formed on and in roots showing root symptoms and sometimes present when rot symptoms were not evident (Leath and Kendall, 1978). Once the conditions become favorable, the chlamydospores germinate in response to alfalfa’s root exudates, which contain a wide array of organic compounds, including sugars and amino acids (Fang et al., 2019). A schematic representation of the presumed disease cycle of Fusarium root rot is shown in Figure 3. Infection hyphae from chlamydospores and/or mycelium penetrate the epidermis of rootlets, taproots, and stem base directly or through wounds or injuries and reach to root cortex. Consequently, brown to black necrotic patches are formed around the roots. Then the hypha reaches vascular vessels (xylem) and blocks the water. Finally, the stele of the roots decay, and the collar and the center of the root become hollow and lateral roots rot in large quantities. On the surface of roots, necrotic spots are formed, the tangential face of roots becomes brown or black, and reddish-brown or dark brown stripes/lesions are formed. The roots’ internal and external portions show red-brown discolorations (Berg et al., 2017; Wang et al., 2020a; Li et al., 2021a). In the later stage of the disease, the plants become weak and can be easily removed from the earth. Severely infected plants showed stress-type foliar symptoms such as chlorosis, brown and reddish-brown foliage color, withering of individual twigs, or the whole plant wilting. These symptoms are frequently exacerbated by infrequent rainfall and cool soil temperatures. Besides, Fusarium spp. break cold resistance of alfalfa (Richard et al., 1982) predispose alfalfa to other pathogens. In addition, the taproot is the first root that penetrates the upper soil layer; if it is rotted, then the greatest reduction in plant size can be expected. However, the rotting of lateral roots had little effect on plant size because most plants regenerate new lateral roots to compensate for the loss of the lateral root (Pegg and Parry, 1983; Barbetti and MacNish, 1984). Furthermore, the infected plants dispersed among apparently healthy plants or the affected areas may occur in distinct patches. As a result, alfalfa plantations are sparse in a few years. Once alfalfa root rot occurs in a large area, it will seriously reduce alfalfa production and even need to be replanted (Kong et al., 2018a). The foliar symptoms of root rot in alfalfa vary in the northwest, northeast and north of China, most probably because of particular environmental conditions. Micro and macroconidia are secondary sources of infection to alfalfa and can survive on the surface of contaminated plants and spread to adjacent plants. Hence, chlamydospores play a vital role in the occurrence and circulation of root rot disease. The number and survival of the chlamydospores directly affect the occurrence of the disease and its degree of harm. Furthermore, Fusarium spp. cause severe damage to those alfalfa plants which are already weakened or injured by other abiotic factors, i.e., stress factors. For example low soil temperature and soil compaction predispose alfalfa to Fusarium root rot (Li et al., 2021a). Further, Fusarium spp. can infect any developmental stage of alfalfa; therefore, alfalfa cannot find a chance to escape from the Fusarium spp. Consequently, the alfalfa’s life, the nitrogen-retaining capacity, and the crop’s quality reduce, resulting in the loss of alfalfa’s processing value. Besides, the soluble sugar content of the infected alfalfa plants reduces, resulting in reduced regenerative capacity, slow growth, and reduction of yield. The gradual appearance of sparse plots and plants seriously affects alfalfa production. Further, they can survive in the soil for a long time and accumulate year after year, resulting in a decrease in alfalfa’s resistance to disease, resulting in a longer planting age (Fang et al., 2019). A previous report showed that chlamydospores could survive in the soil for up to 30 years, meaning that infected land cannot be used to replant alfalfa (Fones et al., 2017). Recent reports suggest that Fusarium spp. also infect the alfalfa seeds. For example, 150 Fusarium strains from rotting alfalfa and induced pathogenicity on the alfalfa seeds (Kong et al., 2018a). All the isolated Fusarium strains induced pathogenicity on the germinated alfalfa seeds with varying pathogenic intensities. However, there is still a lack of any relationship between disease incidence of root rot and pathogen-associated with seed. A study showed that removing pathogens from the seeds had no influence on root rot severity after sowing (Barbetti, 1983).
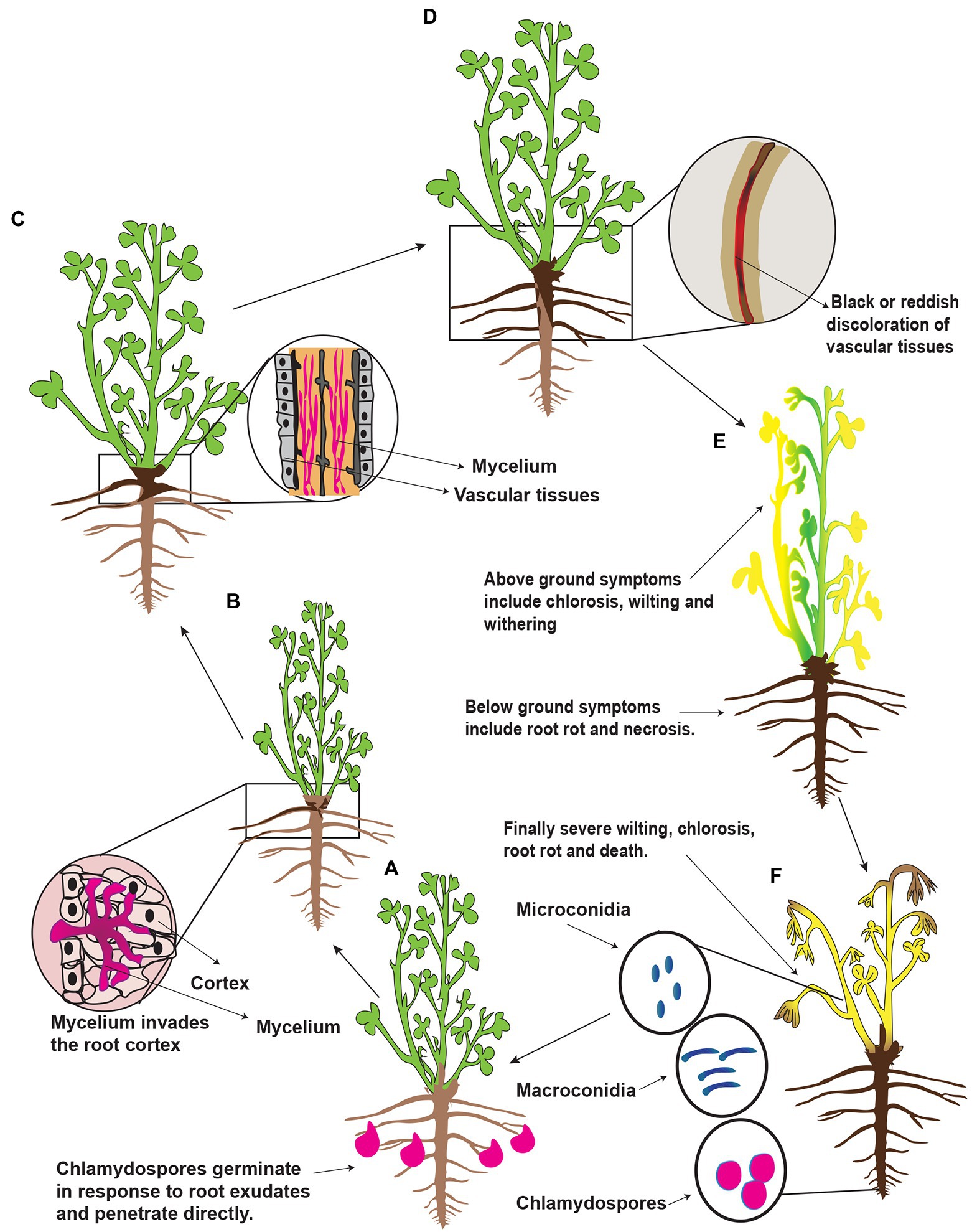
Figure 3. Schematic representation of the presumed disease cycle of Fusarium root rot in China. (A) Alfalfa plant secrete root exudates and in response to exudates Fusarium spp. spores (chlamydospores) germinate and produce infection hypha to penetrate the root epidermis at the root tip. (B) The hypha proliferates in the root cortex and enters into the vascular vessels, i.e., xylem vessels. (C) In the vessels, it grows excessively and causes a blockage; as a result, brown discoloration occurs. (D) First symptoms appear at the base of the stem, and then the symptoms progress upward; as a result, the young leaves withered. (E) Partial chlorosis or complete chlorosis is observed mainly on the mature leaves. (F) Finally, the whole alfalfa plants wilt because of severe root rot followed by death. Fungal spores such as microconidia, macroconidia, and chlamydospores form dead alfalfa plant tissues and remain dispersed in the soil.
Rhizoctonia solani
Rhizoctonia genus was first introduced in 1815 by de Candolle for an unknown fungus severely infecting alfalfa and saffron (De Candole, 1815). According to de Candolle, the basic characteristics of the genus are the presence of sclerotia, mycelia emanating from sclerotia and the association of mycelia with the roots of plants. Moore et al. divided Rhizoctonia like fungi into four genera based on teleomorph, as shown in Figure 4 (Moore, 1987). Later classification of Rhizoctonia has been revised into three main teleomorphic genera (Stalpers and Andersen, 1996; Sneh et al., 2013). One is multinucleated Thanatephorus cucumeris (Frank) Donk (anamorph: R. solani). The other one is a bi-nucleate fungal genus known as Ceratobasidium (anamorph: Ceratorhiza) and the third one is also a multinucleated genus known as Waitea (anamorph: Rhizoctonia zeae; Vilgalys and Cubeta, 1994; Andersen, 1996). All these genera lack clamp connections; however, differences have been found in the moniloid hyphae, sclerotia and dolipore septa (Stalpers and Andersen, 1996). Rhizoctonia solani (synonym: Thanatephorus cucumeris), the most studied and important necrotrophic fungus causing roots rots and other plant diseases, is of most significant interest to plant pathologists (Ajayi-Oyetunde and Bradley, 2016). It was first observed on diseased potato tubers by Kuhn in 1858 (Kühn, 1858). It damages 200 hosts, including cereals, vegetables, agricultural trees, horticultural trees, forest trees, weeds, ornamentals, and forage crops. The important features of R. solani comprise septate hyphae, multinucleate cells in young hyphae, the brown coloration of mature hyphae, right-angled hyphal branching, constriction at the point of branching, dolipore septa that allows unrestricted cell-to-cell movement of cytoplasm, mitochondria and nuclei, production of monilioid cells, and sclerotia of uniform texture. Clamp connections, rhizomorphs, conidia and sexual states other than T. cucumeris and hyphal pigmentations other than brownish mature hyphae have never been observed. This fungus cause root rots, hypocotyl rot, crown rot, stem rot, limb rot, pod rot, stem canker, black scurf, seedling blight, and pre-and post-emergence damping-off in different plants (Stalpers and Andersen, 1996).
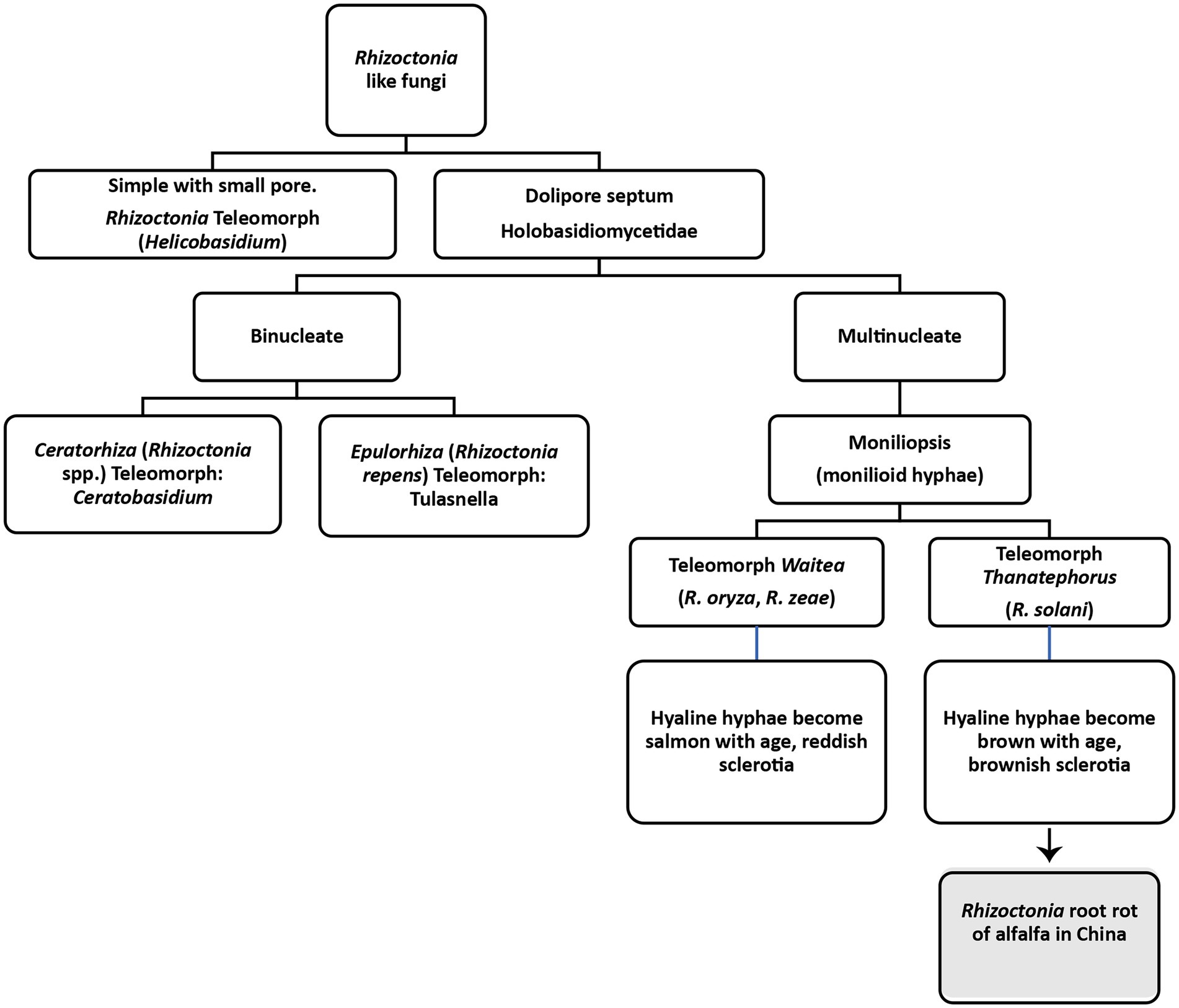
Figure 4. Classification of Rhizoctonia like fungi based on Moore (1987).
Status of Rhizoctonia solani root rot in China and abroad
Root rot caused by R. solani was first reported by Fatemi (1972), who isolated fungus from the rotting roots of alfalfa in Fars province, Iran (Fatemi, 1972). The disease can now be found in many countries of the world including China. The amount of damage it produces in alfalfa varies, but the losses can be significant, even dramatic. Besides, there is considerable diversity in the cultural and colony morphology, host range, molecular and biochemical markers, pathogenicity, virulence, nutritional requirements among isolates of R. solani. Therefore R. solani is considered a species complex consisting of reproductively isolated and non-interbreeding populations. The isolates readily undergo hyphal and cytoplasmic fusion, and exchanging nuclei are grouped in the same anastomosis groups (AGs). In contrast, isolates that fail to achieve hyphal and cytoplasmic fusion and nuclear exchange is considered members of different AGs. R. solani isolates have been divided into 14 AGs (AG-1 to AG-13) and an AG-bridging isolate AG-B1 based on hyphal anastomosis reactions, cultural morphology, pathogenicity/virulence, and DNA homology (Ogoshi, 1987). These AGs are further divided into subgroups based on anastomosis frequency, physiological and morphological characteristics, biomolecular, biochemical, genetic and DNA homology features (Blanco et al., 2018). AGs such as AG-1, AG-2, and AG-4 are mainly associated with stem and root rot diseases in dicots, while AG-8 is associated with root rots in monocots. In general, AG-1, AG-2, AG-3, and AG-4 cause severe diseases in plants globally, whereas the remaining AGs are less harmful and have limited geographic distribution (Eken and Demirci, 2003; Li et al., 2009; Zhang et al., 2021). AGs responsible for severe root rot of alfalfa across the world are; AG-1 and AG-4 in the United States (Vincelli and Herr, 1992), AG-1, -2, -3, -4, -5, and 10 in Turkey (Eken and Demirci, 2003), AG-4 in Iran (Balali and Kowsari, 2004), AG-1 to -10 in Saudi Arabia (Alkherb et al., 1997) and AG-11, -8, and -6 in Australia (Anderson et al., 2013; Oladzad et al., 2019). The anastomosis group of R. solani causing rot diseases of economically important crops, including alfalfa, are highlighted in Table 4. In China, root rot of alfalfa is caused by AG-1, AG-2, AG-4, and AG-5; however, so far, no binucleate Rhizoctonia spp. have been reported to cause root rot of alfalfa in China (Tables 2, 4). In addition, the occurrence frequency of AG-2 and AG-5 was higher than AG-1 and AG-4. Additionally, the pathogenicity of each anastomosis group was also significantly different and AG-2 had the highest pathogenicity on alfalfa (Li et al., 2009). In 2015, the pathogenicity of six isolates of R. solani was checked on 14 alfalfa varieties. All the isolates resulted in lower germination rates. Besides, many seedlings died before emergence and a large percentage of seedlings died after emergence due to root rot (Guo et al., 2019). Recently, a study was conducted to determine whether any host resistance to R. solani exists among the alfalfa varieties. A considerable variation in disease responses among the alfalfa varieties was observed, with the range of disease indices of shoots from 23 to 94%, roots from 31 to 98%, and reductions in dry weight of shoots from 35 to 96% and roots from 2 to 99% (Zhang et al., 2021).
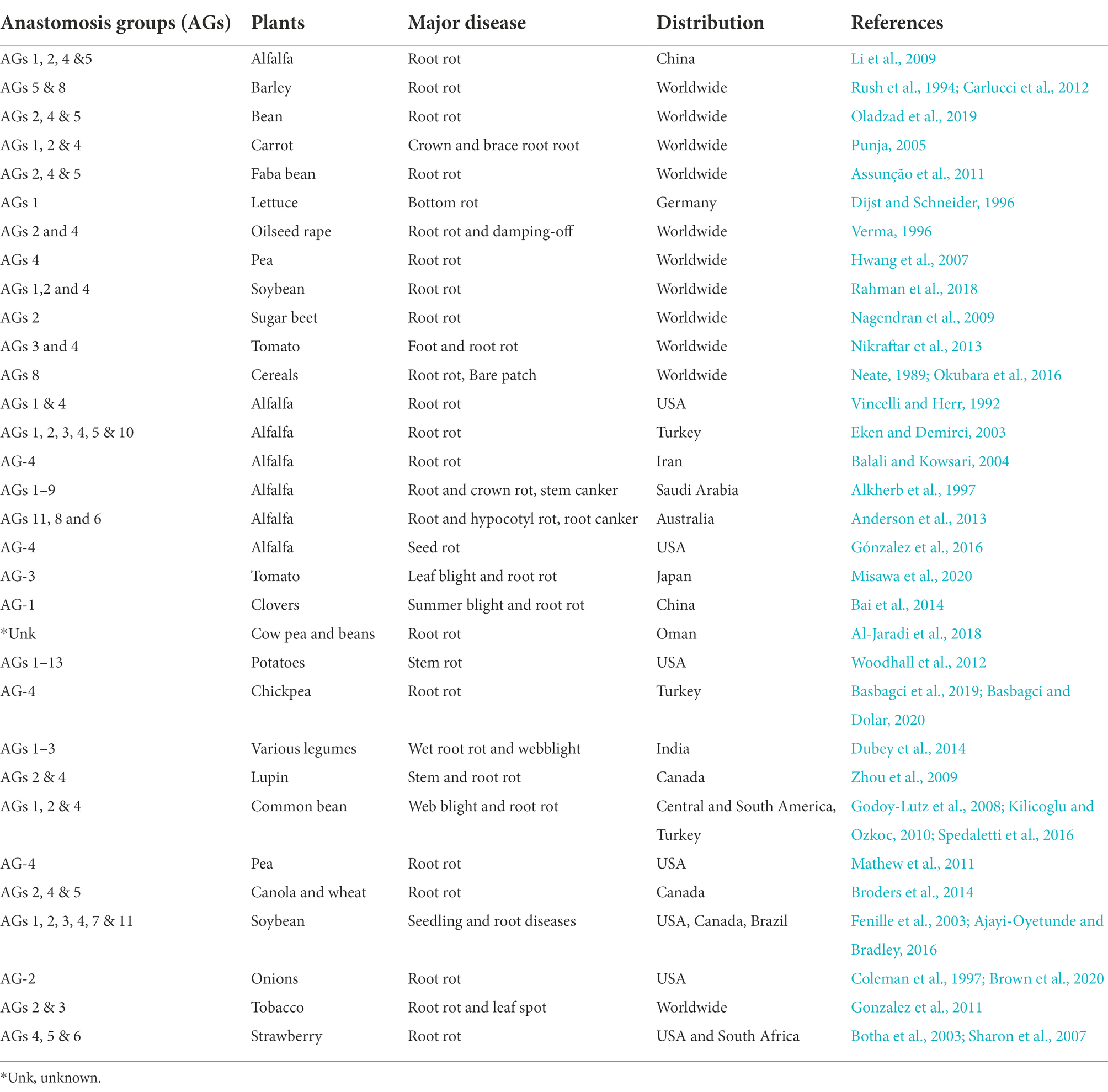
Table 4. Economically important plants including alfalfa affected by anastomosis groups (AGs) of Rhizoctonia solani root rots.
Biological characteristics, disease cycle, and damage to alfalfa
Rhizoctonia solani doesn’t produce vegetative or asexual spores, e.g., conidia. The role of sexual spores, e.g., basidiospores, as an inoculum source for the alfalfa root rot disease is unknown. In addition, the R. solani is a facultative parasite and can easily compete with other soil born saprophytes. In order to survive in the soil, it develops sclerotia, a long-lasting structures or propagules (Williamson-Benavides and Dhingra, 2021). These nutrient-independent propagules are formed from the undifferentiated hyphae or monilioid cells. When the conditions become favorable, sclerotia germinate and mycelia are formed. These mycelia are attracted to alfalfa roots in response to the root exudates. A schematic representation of the presumed disease cycle of Rhizoctonia root rot is shown in Figure 5. Upon reaching to roots, hypha grows along with the epidermal cells and forms appressoria to penetrate the alfalfa tissues by infection pegs. Hence, sclerotia are considered as the primary inoculum of root rot. The pathogen is also considered necrotrophic and produces extracellular hydrolytic enzymes to kill its host in advance of colonization (Ajayi-Oyetunde and Bradley, 2018). Unfortunately, there is little information regarding the pathogenicity of R. solani on alfalfa, whether the discoloration and necrotic of the alfalfa roots are the results of certain toxic or enzymes secreted by R. solani. If the environmental conditions are favorable, the sclerotia germinate and form hyphae, enter the root cortex, and continuously grow inside and on the surface of alfalfa roots. Thereby, longitudinal blackish lesions appear on the roots and at advanced stages, the roots become decay and rot. Also, the crown area becomes dark brown or black (Zhang et al., 2021). Foliar symptoms include yellow or reddish color or wilt leaves. R. solani also infect seeds and usually, infected seeds don’t germinate and if they germinate, the seedlings are killed before or after emergence. Sclerotia forms, again, thereby completing the disease cycle and can remain viable for several years under harsh environmental conditions such as temperature, starvation, desiccation, chemicals, and severe radiation. According to previous research, the sclerotia of R. solani can remain viable in soil without a host for 8 to 10 years and as the primary inoculum. In addition, the pathogen also can survive in the form of mycelium in the plant debris. The mycelia and sclerotia are spread by irrigation water, rain and floods to other alfalfa fields (Garibaldi et al., 2021).
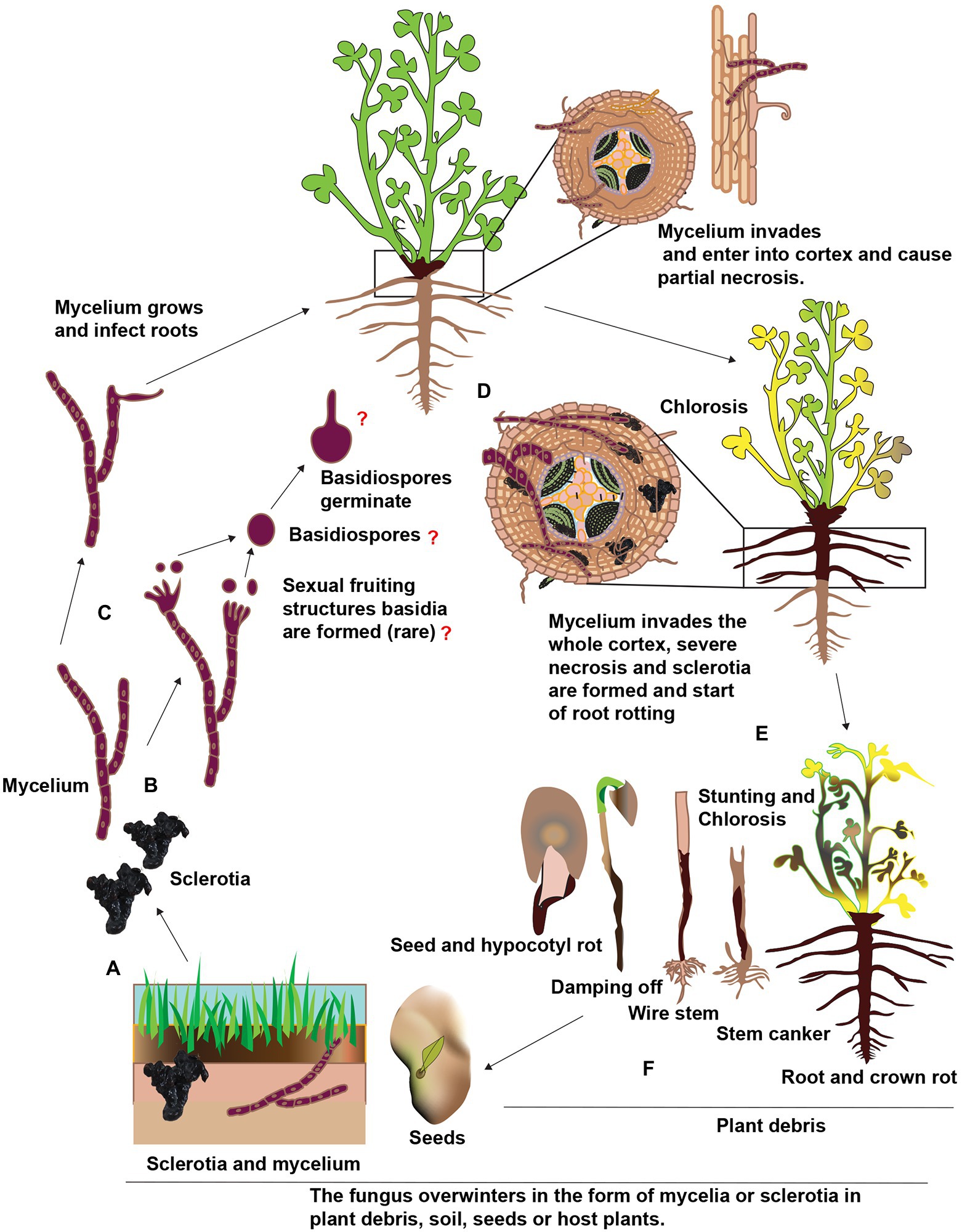
Figure 5. Schematic representation of the presumed disease cycle of Rhophitulus solani root rot in China. (A), The fungus overwinters in the plant debris and seeds in the form of mycelium and in the soil as sclerotia and mycelium. (B,C) The young hyphae germinate and develop under favorable conditions, sexual fruiting structures basidia and basidiospores are rare. (D) The mycelium penetrates roots near the soil line and colonized in inter and intracellular spaces. (E) The mycelium proliferates further in the cortex ultimately results in necrosis and sclerotia are formed in and on infected tissues and disintegration and acute rotting of roots. (F) Above ground symptoms, include chlorosis, blights, stunting and finally death, the fungus also infects seeds and seedlings and also causing damping-off.
Oomycetes
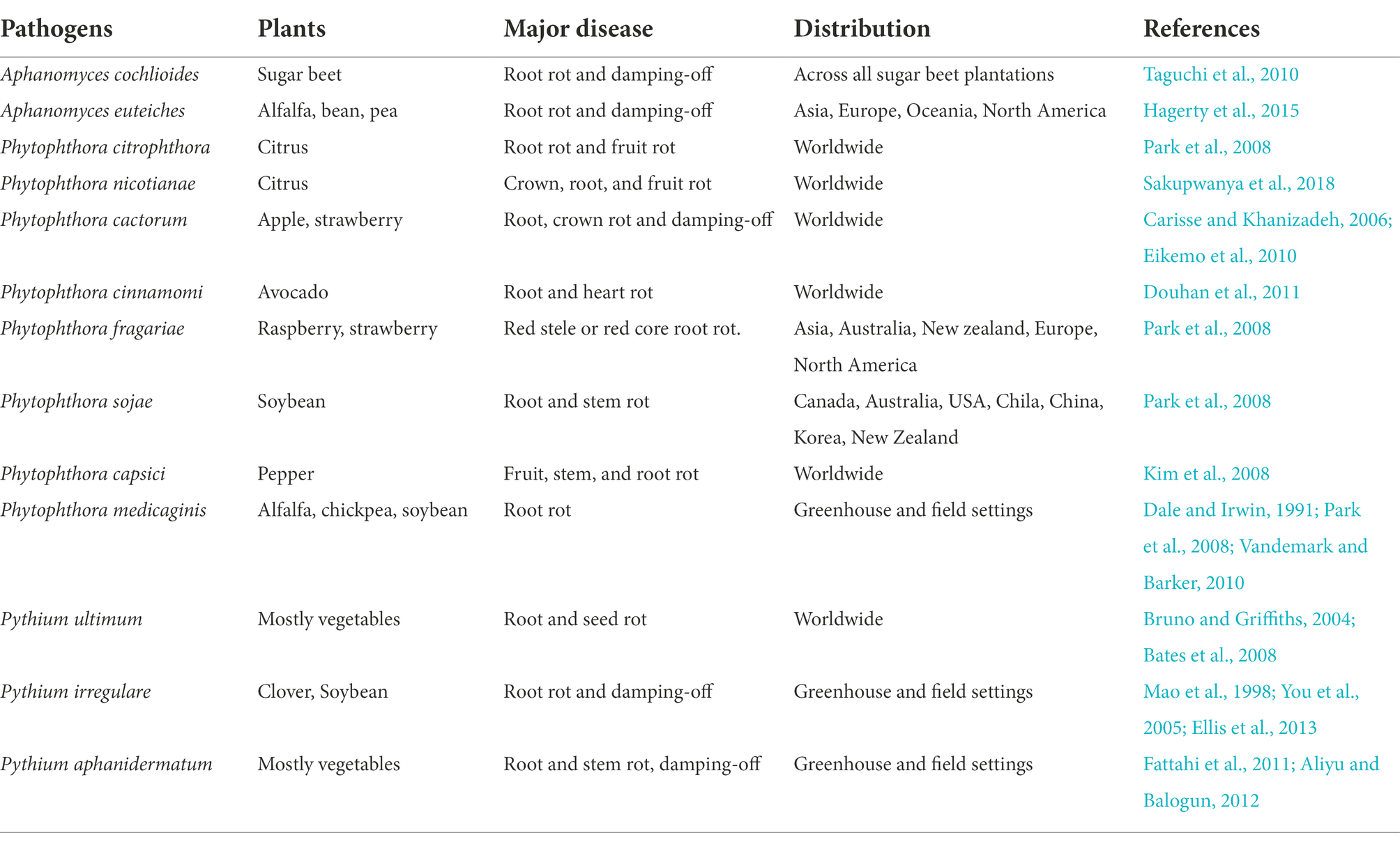
Oomycetes (syn. Peronosporomycete), often known as “water molds,” are a group of hundreds of organisms (between 600 and 1,500 species; Dick et al., 1999). They were assumed to be closely related to the kingdom Fungi for a long time because of their similar ecological and morphological traits (Harper et al., 2005). However, they are now thought to be phylogenetically distinct from fungi, with diatoms, chromophyte algae, and other heterokont protists being their closest relatives. Therefore, they have been placed in a separate kingdom, Stramenopiles, consisting of the most devastating plant pathogen. They cause seedling blights, damping-off, foliar blights, downy mildew, and root rot. In contrast to fungi, their cells walls contain cellulose and have tubular mitochondrial cristae and are vegetative diploid (Van West et al., 2003). They have the potential to survive both in aquatic and terrestrial environments. Oomycetes such as Aphanomyces spp., Pythium spp., and Phytophthora spp. are causing severe root rot of alfalfa growing regions in the world (Table 5). They survive in the soil in the form of rigid, resistant structures called oospores. Oospores germinate directly by producing germ tubes in response to chemical signals from the alfalfa host or proliferate as sporangia (Harper et al., 2005). Zoospores with heterokont flagella (one tinsel and one whiplash) inside the sporangia are formed. Zoospores are released from sporangia and swim through water-filled soil pores with the help of flagella. Previous reports suggest that soil moisture significantly affect the incidence and severity of root rot caused by oomycetes (Erwin, 1954; Faris and Sabo, 1981; Gray et al., 1983). Also, the disease becomes more severe when the soil remains wet for ten days or longer (Kuan and Erwin, 1980). Water saturation predisposes alfalfa to oomycetes by increasing root damage and exudation of nutrients like amino acids and sugars that boost the chemotactic attraction of zoospores to roots (Gray et al., 1983). Once a zoospore reaches the root surface of alfalfa, it loses both flagella, encysts and germinate by forming a germ tube. Hyphae derived from the germ tube directly penetrate the root epidermis and colonize roots. The hyphae then differentiate into antheridia and oogonia within the roots, forming oospores. Oospores survive in the soil for many years in the absence of alfalfa. Both Pythium spp. and Phytophthora spp. are a severe problem only in the irrigated and/or flood irrigated alfalfa-growing areas. Aphanomyces spp. have been recognized as a severe root rot pathogen of legumes in several American states and other legumes-growing regions of the world, notably in Europe (Gaulin et al., 2007). We could not find a published report of root rot of alfalfa caused by Aphanomyces spp. in China. However, there are reports on root rot of other legumes caused by Aphanomyces spp. (Zhimin, 1996). In many studies, oomycetes were found to form pathogens complexes with R. solani and Fusarium spp., and inflicting severe damage to alfalfa roots causing root rot and damping-off, resulting in reduced yields, decreased winter survival, and shortened stand life (Hancock, 1983; Berg et al., 2017).
Phytophthora spp.
The genus Phytophthora consists of more than 100 species, and the majority of them are aggressive plant pathogens that cause extensive losses in agricultural, horticultural and forage crops (Cai et al., 2021). Phytophthora means “plant destroyer,” a term coined in the 19th century when they destroyed potato fields of Ireland, causing the Great Irish Famine (Yang et al., 2017). Erwin first described Phytophthora root rot of alfalfa in California, United States. He observed the disease as causing severe root rot in less than three years old alfalfa plants in high rainfall, heavily irrigated, poorly drained soils (Erwin, 1954). Initially, he named the pathogen P. cryptogea; however, later, the pathogen was classified as P. megasperma. Phytophthora root rot is prevalent in almost all alfalfa-growing regions of the United States, Canada, and Australia (Kuan and Erwin, 1980; Faris and Sabo, 1981; Rogers, 1981; Gray et al., 1983, 2004). Among the Phytophthora spp. i.e., P. infestans, P. megasperma, P. citrophthora, P. cactorum, P. cinnamomic, P. fragariae, P. sojae, P. capsici, P. nicotianae, and P. medicaginis are causing severe root rot including forages worldwide (Steinmetz et al., 2020). In China, P. cactorum has been reported to cause severe root rot. In 2018, 2 years old alfalfa plants in Jinchang, Gansu province, were severely infected by P. cactorum. There were red to dark-brown discolorations in the taproots. Affected plants have wilting shoots with decaying and rotting taproots and lateral roots. Besides sporangia, chlamydospores, and oospores were also recovered (Cai et al., 2021). The symptoms of alfalfa root rot were similar to symptoms on alfalfa crops caused by Phytophthora in other countries (Erwin, 1954; Marks and Mitchell, 1970; Rogers, 1981). The disease cycle begins with zoospores which move freely in water and contact with the tips of rootlets. The lesions on the roots become yellowish and then brownish and later turn to dark brown or black, often with halo margins. The size and type of root lesions depend on the duration of wet soil conditions, alfalfa genotypes, or both. In severe conditions, lateral and tap roots are rotted, and foliar symptoms such as yellowing, stunting and wilting appear (Marks and Mitchell, 1970).
Pythium spp.
The genus Pythium contains more than 200 described species, and at least 10–15 species are causing damping-off and root rots in various agricultural, horticultural and forage crops (Beckerman, 2010; Berg et al., 2017; Wang et al., 2020a). Symptoms on alfalfa caused by Pythium are like Phytophthora that cause root rots; however, root tips become necrotic in the early infection. Furthermore, the entire primary roots become black and rotting moves upward to the stem (Zhang et al., 2020). Four important Pythium spp. such as P. ultimum, P. irregulare, P. aphanidermatum, and P. myriotylum have been reported from alfalfa fields and are the most common root rot causing pathogens in the world (Williamson-Benavides and Dhingra, 2021). During 2017 and 2019, 30 to 80% of alfalfa plants in Gansu province stunted, wilted, and dried. Likewise, irregular brown necrotic lesions were observed on the taproots. In addition, the lateral roots showed brown discoloration and were poorly developed, necrotic, and rotted. Morphological characters of sporangia, oogonia and antheridia, were identified. Morphological characters and molecular identification suggested that the pathogen was P. coloratum (Zhang et al., 2020).
Effects of environmental factors on root rot
The occurrence, severity, and prevalence of root rot are affected by primary infection sources, management practices, environmental factors, and alfalfa cultivars’ ability to resist disease (Williamson-Benavides and Dhingra, 2021; Fahad et al., 2021a,b). Among environmental factors, such as soil moisture and soil temperature, play a significant role in the occurrence, severity and prevalence of root rot (Jat and Ahir, 2013). Root rot of alfalfa caused by oomycetes is not generally severe in alfalfa growing regions of China. Oomycetes root rot is higher in locations with excessive rainfall, heavily irrigated or poorly drained soil or flood irrigated (Liang et al., 2006; Cai et al., 2021). For example, in the southwest region (Sichuan province), which has comparative more rainfall than northwest (Xingjiang and Gansu) and north (Inner Mongolia) of China, oomycetes are causing severe rot of legume crops. Besides, the soil of this region is wetter and less dry than the northwest, northeast and north region of China. Oomycetes produce oospores which germinate into hyphae under high soil moisture conditions (generally in compacted soil). Furthermore, oomycetes such as Phytophthora spp. and Pythium spp. are causing the most severe root rotting over the range of temperatures 17–23°C. Moreover, oomycetes root rot develops in alfalfa when the soil remains excessively moist for about ten days or longer (Karppinen et al., 2020). Many researchers postulated that excessive soil moisture cause lack of oxygen that predispose alfalfa to oomycetes root rot (Fahad et al., 2021c,d). Hence the incidence of root rot caused by oomycetes increased with the increase in soil water content, and the lower soil moisture could reduce the development of root rot disease (Fahad et al., 2020; Cai et al., 2021). However, oomycetes become most aggressive against alfalfa when a cool, wet spring is followed by an early, warm, dry summer (Yinghua et al., 2019). Wong et al. (1984) checked how soil moisture interacted with the pathogenicity of Pythium spp., Phytophthora spp. Fusarium spp. and Rhizoctonia spp. both alone or in combination. These fungi and their combination caused root disease over the range of soil moisture conditions. The most severe root rotting occurred at 65% water holding capacity and less at 45% (Wong et al., 1984). It indicates that high soil moisture favor the growth of oomycete pathogens. Soil moisture and soil temperature also influences the Phoma and Paraphomra root rot of alfalfa. For example, Paraphoma and Phoma root rots of alfalfa are favored by moderate soil temperatures (15–21°C) and soil moistures (60–70%; Cao et al., 2020; Deb et al., 2020). In 2014, seven cultivars of alfalfa were grown in seven different fields in the Chifeng county in Inner Mongolia. The variability of disease incidence of Paraphoma root rot was extremely high and the potential reason was that soil moisture content at these fields varied due to the uneven terrains (Cao et al., 2020). Furthermore, the peak period of root rot also varies according to changes in humidity and temperature in the alfalfa-growing regions of China. For example, in the north region of China, the peak period for root rot disease is the first week of August, whereas the peak period for the disease in the southwest region is in mid-September (Cao et al., 2008). Fusarium spp. and R. solani are causing more severe root rot in the drier soils and lower rainfall regions such as northeast, northwest and north of China. However, both fungi can cause severe root rot at high soil temperature (24–32°C) and soil moisture (70–80%; Ajayi-Oyetunde and Bradley, 2018). In addition, the number of infections’ propagules of these fungi varies seasonally. After January, it began to rise, peaking in May and June, and then began to drop from July through December (Wong et al., 1984). Similarly, each anastomosis groups (AGs) of R. solani require certain temperature to cause root rot (Kousik et al., 1995). Furthermore, Fusarium spp. and R. solani also infect alfalfa at cold temperatures (15–18°C), which slows down alfalfa seedlings’ growth (Coleman, 2016; Wang et al., 2020a). Winter survival of alfalfa depends on the accumulations of food reserves in the roots and crown. However, both Fusarium spp. and R. solani reduce the cold resistance of alfalfa and predispose alfalfa to winterkill by affecting the accumulation of food reserves (Hwang et al., 1989). Research reports on other legumes showed that even changes in the soil pH affected the alfalfa survival and root rot disease severity caused by Fusarium spp. Phythium spp. and R. solani. These pathogens responded to pH differently, and alfalfa resistance to individual pathogens also varied depending on the amount of lime added (Barbetti, 1990). Furthermore, these fungi infect alfalfa mostly in spring and temperature ranging from 15 to 25°C was found to be optimum for infection of alfalfa (Yuan et al., 2003). Alfalfa root rot caused by Bipolaris spp. is favored in dry and warm soil and a temperature range from 15 to 25°C. Also, when alfalfa plants are grown under stress conditions such as warm and less moist soil, the root rot caused by Bipolaris spp. become severe (Acharya et al., 2011). Moreover, the erratic weather conditions herald a rise in mean temperatures as well as other natural disasters like droughts, floods, and storms. These circumstances are anticipated to put alfalfa under constant stress, which is anticipated to encourage the pathogens that cause root rot to become more active (Fahad et al., 2021e,f). Furthermore, soil of alfalfa growing regions of China is nutrient-poor, compact, and not very fertile and generally lacks phosphorus, resulting in significantly reduced nutrient levels (Wang et al., 2021). Previous studies showed that nutrient-poor and leached soils impose nutrition stress on crops; as a result, natural resistance to root rot disease reduced (Graham, 1983). By adding appropriate nutrients to alfalfa-growing regions, the root rot disease severity can be reduced because plant nutrients promote growth and enhance disease tolerance to root rot. Especially phosphate should be added to alfalfa fields, which can enhance host resistance by stimulating the production of phytoalexins against the virulence factors of root rot pathogen (Barbetti et al., 2007). Besides the activities of antagonists that restrict root rot, pathogens and other soil microflora can also be affected by soil nutrients. Hence, soil nutrients influence disease severity by changing root physiology and host resistance and by influencing the interaction between the host and the pathogen and/or antagonist, each of which can be influenced independently by the availability of nutrients. In China, the interaction of soil nutrients with root rot pathogens of alfalfa is currently unknown, however, with greater knowledge, root rot can be managed by altering soil nutrients. In addition, temperature and moisture also affect mycelia growth, sporulation, germ tube growth, and spore germinations. For example, the appropriate temperature range for Fusarium spp. to produce spores is from 25 to 32°C and the appropriate temperature range for R. solani to produce vegetative structures such as mycelia is 20–30°C and to produce overwintering structure such as sclerotia is 25°C. Similarly, sporangia of P. megasperma germinated indirectly by releasing zoospores in the flooded soil at 8–24°C (Pfender et al., 1976). Furthermore, each pathogen in the root rot complex has its specific optimal developmental conditions. For example, the optimum temperatures for the mycelia growth of F. equiseti and F. proliferatum were 20 and 28°C, respectively, (Kong et al., 2018b). Even the developmental conditions vary for each strain of same species. For example, the optimum temperatures for the mycelial growth of different F. oxysporum strains, D19-2 and B1-63 were 26 and 28°C, respectively (Kong et al., 2018b) Besides, soil pH also influences the developmental phases of root rot pathogens. For example, the optimum pH for the mycelial growth of F. equiseti and F. proliferatum was 6 and 10, while the pH for sporulation and spore germination was 8.0 and 7.0, respectively (Kong et al., 2018b). Similarly, alternating light and dark are important for mycelia growth, and light is important for sporulation. Spores do not germinate when the relative humidity is lower than 75% (Pan et al., 2015). As previously stated, among the environmental factors, soil moisture, and soil temperature significantly impact the root rot disease of alfalfa. More research is required to understand how soil moisture, soil temperature, and other environmental factors such as pH affect root rot of alfalfa in natural pastures.
Management strategies
Many strategies have been evaluated to manage the root rot in recent years. They are explained below.
Cultural approach
The cultural approach makes conditions unfavorable to the root rot pathogens to reduce root rot disease severity (Sumner, 1994). The conditions for the root rot pathogens can be made unfavorable for root rot pathogen in many ways, such as soil pH and soil nutrients adjustment, fallowing, composting, timely grazing, biofumigation, sanitation, mixed cropping, intercropping and crop rotation. Fallowing is to keep the area fallow for several years before cultivation of alfalfa, however, most of the root rot pathogens are soil borne, the inoculum remains even after fallowing (Barbetti and MacNish, 1984). Bio-fumigation and sanitation in combination can also effectively reduce the survival structure of root rot pathogen such as microsclerotia and sclerotia in the soil (Wang et al., 2014). However, production losses from fallowing and bio-fumigation cannot be bearable for farmers. Inoculating seeds with rhizobia or BCAs can also reduce the root rot severity of other legume crops. For example, Rhizobium trifolii significantly reduced root rot of clover caused by F. avenaceum in glasshouse studies (Wong et al., 1984). However, if the soil is acidic or contains a high concentration of available aluminum, in that case it negatively affects the rhizobia growth and survival and interferes with the legume-rhizobia symbiosis by affecting rhizobia attachment to roots (Tabares-da Rosa et al., 2019). As outlined above, the soil of pastoral areas is poor nutrients deficient in phosphates. Therefore, the effect of soil nutrition and soil pH is probably so significant to manage root rot. The productivity of alfalfa can be improved through the application of phosphate. However, this may enhance soil acidity in many areas making conditions less favorable for the growth of alfalfa. Besides, the addition of lime to manipulate soil pH should also be further investigated as lime influences alfalfa and root rot pathogens. In a previous study, pathogen complexes associated with root rot such as Fusarium spp., Pythium spp., and R. solani responded differently to the addition of lime (Barbetti, 1990). It indicates that these pathogen complexes can be broken by altering soil pH resulting reduction of root rot severity. Furthermore, dense planting should be avoided to reduce the disease spread. Also, avoid planting too deep because it takes more time to emerge from the soil, increasing the chances of getting infected by soil pathogens (Hwang et al., 2002). In addition, leftovers, stubbles, infected plants, debris and weeds should be burned to reduce the disease inoculum. In addition, the application of green organic manures, farmyard manure and organic fertilizers, including composts, green manures and animal manures, reduce the root rots and promote the growth of beneficial soil microbes significantly (Wiggins and Kinkel, 2005). Moreover, the amount and time of application of inorganic fertilizers are also crucial for disease development (Summers, 1998). The application of nitrogenous fertilizers should be discouraged because they make the plants succulent, and pathogens get more chances to infect the plants (Stone et al., 2003). Alfalfa seedlings are more susceptible to Pythium spp. and R. solani following seed germination; hence high-quality seeds which can rapidly germinate should be sown. Excessive irrigation, compaction and poor drainage of alfalfa fields should be avoided because these conditions favor oomycetes. Besides, judicious application of fungicides in combination with appropriate cultural practices can significantly reduce the disease severity. Soil compaction in the alfalfa fields is also a major problem. Continuous cropping of alfalfa, application of inorganic fertilizers and reduction in the use of green or animal manures cause soil compaction (Stone et al., 2003). Increased soil compaction affects aeration, porosity, and water retention capacity while increasing bulk density. Plant growth, biomass, and yield are reduced as a result, the incidence and severity of root rot diseases rise (Williamson-Benavides and Dhingra, 2021). Multiyear continuous cropping of alfalfa allows root rot pathogens to continue their uninterrupted disease cycle, resulting in their perpetuation and multiplication. However, rotating with unrelated crops or related crops of differential disease susceptibility such as clovers, cereals and soybeans, many root rot pathogens having a narrow host range can be controlled successfully (Curl, 1963). However, crop rotations are not effective against soil-borne pathogens, especially against Fusarium spp. and R. solani. These fungi produce survival structures such as chlamydospores and sclerotia and survive for several years. Besides, most of the pathogens on alfalfa are common to other rotational legumes and/or crop species. For example, R. solani is a common pathogen of alfalfa and rice. Similarly, Pythium spp. and Fusarium spp. can cross infect between pasture legumes such as alfalfa and clovers and/or crop species such as barley, wheat and oat (Sivasithamparam, 1993). However, host-mediated selection occurs in the pathogen isolates and if that is, then root rot diseases can be limited to a certain extent. Besides, grazing and harvesting could be timely to reduce the impact of root rot, especially at the seedling stage when plants are more vulnerable to root rot (Barbetti, 1990). For example, in Inner Mongolia (North China), alfalfa is harvested in autumn in some areas. Studies showed that autumn harvest reduces organic residues in the roots; as a result, plants become weak to face winter and predispose to Fusarium root rot (Couture et al., 2002). In conclusion, cultural practices are ineffective against root rot in susceptible alfalfa cultivars. Root rot can only be managed effectively when resistant cultivars are grown besides appropriate cultivation and cultural practices and judicious use of fungicides.
Breeding, transgenic, gene silencing, and editing approach
Breeding approach
Currently, 77 alfalfa cultivars have been registered in China. Among these, 36 cultivars were bred through breeding programs, 17 were introduced from other countries, five domesticated from wild ecotypes, and 19 were collected from the regional/breeding programs (Zhang et al., 2021). These commercial alfalfa cultivars acquired by farmers in China are created and sold by seed corporations and marketed as pests resistant. However, so far, the breeders could not develop a resistant cultivar against alfalfa root rot. There are seven main reasons which limit the selection of alfalfa-resistant cultivars: (i) Alfalfa is a perennial plant that is primarily cross-pollinated, and several factors influence its self-fertility; (ii) Alfalfa is autotetraploid; the breeding and selection factors are different from diploid plant species; (iii) Genetic complexity of root rot pathogens. There are varieties with resistance to a single pathogen; however, developing varieties with resistance to multiple pathogens which cause alfalfa root rot is challenging; (iv) Pathogens vary in different environmental conditions in the saturated soil; for example, oomycetes become dominant and in the dried soil Fusarium spp. and R. solani becomes dominant; (v) Alfalfa cultivars cannot adapt to different environmental conditions; (vi) Pathogen evolution is speedy compared to breeding, which takes 20–30 years to discover resistant markers and new cultivars; and (vii) Lack of understanding of resistance mechanisms in alfalfa roots to root rot pathogens. Therefore, there is a delay in utilizing breeding approaches to develop root rot-resistant cultivars compared to other crops. In China, selective breeding, cross-breeding, male-sterile line breeding, space breeding, biotechnology-assisted breeding, transgenic technology, and molecular marker technology are all employed to improve resistance in cultivars against many diseases, but not against root rot. For example, researchers have tried to develop disease-resistant varieties using molecular marker technology, i.e., Random Amplified Polymorphism (RAPD) technique and Bulked Segregation Analysis (BSA) were used to study molecular markers linked to resistance genes against a brown spot disease in five Medicago species (Gui and Yuan, 2002). Furthermore, R-gene mediated resistance is race-specific, and resistance to root rot is quantitatively inherited. However, alfalfa resistance can be improved by stacking or pyramiding major R genes/QTLs for multiple pathogens associated with root rot (Fuchs, 2017). We could not find a published report on the pyramiding R genes/QTLs (quantitative trait loci) against root rot in China. It indicates that studies regarding QTLs mapping of alfalfa to uncover genetic architectures related to root rot are in preliminary stages. In other countries, pyramiding R genes/QTLs have achieved resistance to root rot. For example, QTLs were mapped in various plants that contributed to high-level resistance against root rot caused by Phytophthora and Pythium spp. Though QTL mapping has been a powerful technique in identifying genomic regions associated with root rot resistance traits in bi-parental populations (Ramalingam et al., 2020). However, QTL mapping has limitations because QTL cannot detect natural variations in diverse genetic backgrounds due to low allelic diversity and recombination rates in bi-parental populations. To overcome these limitations, genome-wide association study (GWAS) is widely used to evaluate broader genetic diversity and inquire greater quantities of recombination due to the evolutionary history of natural populations. For example, a candidate gene encoding F-box protein was identified in alfalfa, using GWAS, as a negative regulator of resistance to root rot caused by Aphanomyces spp. (Bonhomme et al., 2014). There is also a high-resolution NGS SNP data developed in the Medicago truncatula HapMap Project1 which involved sequencing of 288 Medicago accessions by Illumina technology (Stanton-Geddes et al., 2013). Recently, to mitigate GWAS and QTL mapping limitations, these two QTL analysis approaches are being combined, providing complementary, robust, and vigorous assays to uncover the genetic basis underlying complex traits. For example, the genetic architecture of Aphanomyces root rot resistance in lentils has been dissected by linking QTL Mapping and GWAS (Ma et al., 2020). Recently scientists are trying to examine the genetic architecture of root rot disease resistance in other legume crops by QTL Mapping and GWAS. These studies will highlight the accumulation of favorable haplotypes in the most resistant accessions against root rot disease (Ma et al., 2020). Most research in China has concentrated on screening germplasm accessions and commercial cultivars in greenhouses and fields only against strains of Fusarium spp. and R. solani, which could serve as possible sources of resistance. For example, in a field screening assay, 20 alfalfa cultivars were inoculated with several F. oxysporum isolates, four F. acuminatum isolates, and five F. semitectum isolates. Only seven cultivars showed resistance to Fusarium spp., including Verla, Derful, Ameristand 201, Caoyuan 2, Sitel Algongum, Tumu 2, and Gannong 2. The rest of the cultivars were susceptible to Fusarium spp. (Quan, 2003). Thirty alfalfa cultivars were tested in another screening assay against three Fusarium species: F. solani, F. semitectum, and F. camptoceras. Only three varieties, “Delilande,” “Xinjiangdaye” and “Muxuwang” were resistant to F. semitectum with disease index 18.11, 19.93 and 19.96, respectively. With a disease index of 25.30 to 39.95, seven varieties were found to be disease tolerant. The disease index ranged from 40.55 to 59.56 for eighteen varieties. The remaining varieties were susceptible, with disease index ranging from 61.7 to 68.6. These findings indicate that no alfalfa variety is immune or resistant to alfalfa root rot (Ding et al., 2011). In a recent study, about 68 alfalfa varieties were screened for resistance against R. solani (Zhang et al., 2021). Among these, three varieties (Gannong 9, Trifecta and Common), originating from three different countries, exhibited a high level of resistance, with disease indices of shoots and roots and reductions in dry weight of shoots and roots being all ≤ 40%. In addition, five varieties (7%) showed resistance, 15 (22%) were moderately resistant, and the remaining ones exhibited susceptibility. In addition to screening alfalfa varieties against root rot pathogens, intensive efforts have also been dedicated to elucidating the defensive response to the pathogen invasion. For example, in a study, alfalfa plants were inoculated with Fusarium spp. In the resistant varieties of alfalfa, enzyme phenylalanine aminase (PAL) was more active than in the sensitive varieties whereas other defensive enzymes such as superoxide dismutase (SOD), peroxidase (POD), polyphenol oxidase (PPO) were more active in the susceptible varieties than the resistant varieties (Fang et al., 2003; Quan et al., 2003). These studies offer valuable resistance sources for breeding programs to develop alfalfa varieties with improved resistance to root rot pathogens and for facilitating the identification of molecular mechanisms underlying the resistant varieties to this pathogen. Unfortunately, most the studies regarding alfalfa disease resistance use a single strain or multiple strains of a fungal pathogen; it’s impossible to tell whether the cultivars chosen are disease-resistant in general to the diversity of pathogens in the field conditions.
Transgenic approaches
Transgenic approaches are widely employed to manage root rot disease of other crops. In a study, an alfalfa seed antibacterial peptide-encoding gene (alfAFP), was fused to the C-terminal of the rice chitinase-encoding gene and transferred into tobacco. In transgenic tobacco plants, the recombinant protein improved resistance to F. solani. Even 30 days after being inoculated with F. solani, transgenic lines did not show root rot (Azam et al., 2018). Pathogenesis-related proteins (PRs) are widely distributed in plants, including alfalfa, and are essential in defense responses. Their expression is regulated by specific hormone signaling pathways (Li et al., 2021b). These PRs proteins have different functions, i.e., production of hydrolytic enzymes such as glucanases and chitinases, which lysis cell wall components (Sunpapao and Pornsuriya, 2016), thaumatin-like and osmotin-like proteins which weaken cell walls and permeabilized plasma membranes (Rather et al., 2015), antimicrobial peptides (Nawrot et al., 2014) and RNAse activities to degrade pathogens RNA (Tang et al., 2019). Hence, applying reverse genetics technology involving gene overexpression and gene silencing (e.g., RNAi) has enabled the rapid functional characterization of PR genes. For example, when the PR5 gene was overexpressed in the M. truncatula, the resistance responses such as Abscisic acid (ABA) production and signaling and reactive oxygen species (ROS) were high after inoculation with A. euteiches. Besides, disease resistance against A. euteiches was linked to the lignification and production of a small GTPase (20-O-methylisoliquiritigenin) which regulated ROS (Badis et al., 2015). A recent study showed Recombinant PnPR10-3 functions as an RNase in vitro exhibited strong antifungal effects on Fusarium species (F. oxysporum, F. solani, and F. verticillioides; Tang et al., 2019). On the other hand, reverse genetics has been found to overcome the limits of traditional breeding approaches. However, producing transgenic cultivars resistant to numerous pathogens that cause root rot remains difficult. Fortunately, the recent advent of transcriptomics and next-generation sequencing technologies offers the potential to identify genes involved in root rot resistance on a broader scale (Song et al., 2016).
Genes silencing and editing approach
Currently, host-induced gene silencing (HIGS) is being used, whereby the host produces double-stranded RNA molecules (dsRNA) that target pathogen genes and are processed into short interfering RNA molecules (siRNAs; Perez et al., 2021). Pathogens procure these siRNAs upon infection; consequently, their target genes are silenced. HIGS has been successfully applied against viral, pests, parasitic plants, and fungi. The main advantage of this method is to silence the pathogen genes without introducing new proteins into food and food products. To date, HIGS has been successfully used against mildews, rusts, and wilting diseases of agriculturally important crops. Recent discoveries of gene-editing technology have made it possible to target pathogenesis-related genes. CRISPR/Cas9 editing has been recently used to inhibit the infection caused by pathogens (Gupta et al., 2021). For example, the pathogen R. solani activates the OsSWEET11 sugar transporter in the infected rice tissues to acquire sugar molecules for its nutrition. However, when the sugar transporter, OsSWEET11 was knock-out using CRISPR-Cas9, it was found that rice crops became less susceptible to rice sheath blight disease as compared to OsSWEET11 overexpressing and wild-type plants (Gao et al., 2018b). These modern approaches may help to deploy resistance against root rot rapidly. However, no transgenic anti-root rot alfalfa cultivars have been developed in China and even globally. Besides, these approaches face public and political distrust (Fones et al., 2020).
Biological approach
Concerns regarding the chemicals have recently grown among the general public. Chemicals are wreaking havoc on the ecosystem. Researchers are increasingly focused on biocontrol agents (BCAs), and there have been several success stories about the use of biological agents thus far. Competition for nutrients or space, antibiosis, induce host resistance and lytic enzyme production are the recognized mechanisms by which BCAs control root rot (Fravel, 2005). BCAs are becoming more popular, and however, in the case of forage crops, their use is still limited in China. The reason might be survival, growth, adaptation, and establishment of biological agents in the fragile pastures ecosystems of China is challenging. To date, only a few fungal and bacterial BCAs have been used to manage root rot. So far, among the fungal BCAs, saprophytic fungi Trichoderma spp. and arbuscular mycorrhizal fungi Glomus spp. have been used against root rot. T. harzianum, T. koningii and Glomus mosseae were tested against Fusarium spp. i.e., F. solani, F. oxysporum, F. semidaricum, and F. solani and Microdochium tabacinum. Besides significantly reducing root rot, they also enhanced alfalfa growth and nutrient uptake (Li et al., 2018; Zeng et al., 2019). In another experiment, Glomus mosseae and Rhizobium (Sinorhizobium medicae) combined effects on the root rot caused by Microdochium tabacinum have significantly reduced root rot besides helping alfalfa to uptake water and nutrients, specifically phosphorus (Gao et al., 2018a). Bacterial BCAs, Bacillus spp., Pseudomonas spp., and Actinomycetes are also widely used to manage alfalfa root rot (Xiao et al., 2002; Knezevic et al., 2021). In 2009, about 91 actinomycetes were isolated from 10 soil samples in Chifeng Inner Mongolia through the gradient dilution separation method. Most of them have significantly reduced root rot caused by Fusarium solani, F. oxysoporum and F. avenaceum (Wang et al., 2010). In another study, Bacillus subtilis subsp. spizizenii (MB29) was evaluated against F. semitectum. The strain effectively reduced the mycelia growth of F. semitectum. Furthermore, in vivo test, MB29 significantly reduced root rot, producing a disease control efficiency of 43.41% (Wen et al., 2015a). In another study, cultural filtrates of Ochrobactrum intermedium strain (I-5) significantly reduced the spore production, germination, and mycelia growth of F. tricinctum. In addition, a 10% filtrate of the strain reduced root rot by >73% in repeated experiments. Besides, the strain enhanced invertase, urease, cellulose, and neutral phosphatase activity in the rhizosphere soil and reduced root rot-related soil quality damage. Also promoted the growth of alfalfa without causing apparent damage to plants (Li et al., 2021a,c). Currently, antagonists which are endophytes have also been used to manage root rot. Endophytes are more protected than free-living (rhizospheric) (Lugtenberg et al., 2016) BCAs because they inhabit the internal tissues of plants without causing disease, forming a close symbiotic relationship. For example, seeds are the reservoir of endophytes that protect plants from root rot. They can infiltrate the host systems without exposing them to pathogens. Plant health and vigor, as well as root persistence through rotation, can be improved with the use of endophytes (White et al., 2019). Chen et al. (2015a) isolated 363 strains of endophytes from alfalfa fields in Hebei, Inner Mongolia, and Ningxia provinces of China. These strains include fungi, bacteria, and actinomycetes. Among these strains, three endophytic bacterial strains, e.g., NA NX51R-5, NA NX90R-8, and NA NM1S-1, showed strong biocontrol capability with > 50% effectiveness against F. oxysporum under in vitro and pot experiments. The strains NA NM1S-1 and NA NX51R-5 were identified as Bacillus spp. while the strain NA NX90R-8 was Pseudomonas spp.. There are also some drawbacks to employing BCAs against root rot pathogens. In most cases, a single BCAs is employed to fight against a single pathogen (Raupach and Kloepper, 1998). This may sometimes account for inconsistent field performances even though their efficacy was quite good under controlled conditions (Nicot, 2011). This variability of efficacy is generally due to environmental variations (changing soil temperatures and moisture) in the field, a lack of ecological competence (such as the ability to survive and colonize), intrinsic characteristics of the antagonistic microbe (such as variability in the production of required metabolites or enzymes), and/or an unstable quality of the formula (Bardin et al., 2015). In addition, efficacy may be reduced due to diversity in sensitivity of pathogens to biocontrol agents with the presence of less sensitive isolates in the natural populations of plant pathogens (Fravel, 2005). Hence single BCA is not active in all environmental conditions especially against the pathogen complexes of root rot. More attention should be paid to the application of a mixture of BCAs that can better cope with environmental changes during the growing season and defend against pathogen complexes associated with root rot. Increased genetic diversity of BCAs may allow them to stay longer in the rhizosphere and utilize a wide array of antagonistic activities against root rot pathogens.
Chemicals, phytochemicals, and elicitor approach
Though many management approaches have been attempted to combat root rot, chemical control still remains the primary method for managing root rot. Fungicides are used as seed treatment and soil application to protect the alfalfa from root rot (Min et al., 2002). For example, Fludioxonil, hexaconazole, tebuconazole, propiconazole, difenoconazole, vitavax, carbendazim, captan, metalaxyl, thiophanate methyl, shenqinmycin mefenoxam and mancozeb alone or in combinations were applied as seed treatment and/or as a soil application to control root rot of various crops including alfalfa in China. Carbendazim was the most widely used fungicide in China to control alfalfa root rot (Liu et al., 2016). A study of 20 years ago showed that carbendazim reduced the Fusarium spp. (Min et al., 2002). However, in a recent study, Fusarium spp. were found to be resistant or intermediately sensitive to carbendazim, suggesting that carbendazim has failed to protect against root rot and resistance against carbendazim has emerged (Jiang et al., 2021). Recently in China mixture of fungicides, each having different modes of action, are being used to control root rot of many crops, including alfalfa. For example, seed treatment with the mixture of fludioxonil (phenylpyroles group) and difenoconazole (trizol group) in the 1:4 ratio demonstrated the best control efficiency at seedling and adult stages against root rot caused by B. sorokiniana (Wei et al., 2020). Fludioxonil inhibits glucose phosphorylation, while difenoconazole inhibits the C14-demethylase enzyme that participates in the ergosterol production of root rot pathogens (Wei et al., 2020). In the United States, metalaxyl and mefenoxam were widely used as a seed treatment or soil application to combat alfalfa root rot. These fungicides specifically target the ribosomal RNA polymerases of pathogens. However, resistance in oomycetes to these fungicides had emerged (Hwang et al., 1993). Quinone outside inhibitor (QoI) fungicides such as azoxystrobin and pyraclostrobin have also been widely used to manage alfalfa disease caused by root rot pathogens worldwide. These fungicides block the electron transport at the mitochondrial cytochrome oxidase bc1 complex, affecting respiration (Venancio et al., 2003). However, resistance to these fungicides has emerged in some pathogens due to a mutation at a target binding site (Bartlett et al., 2002). However, combined with other fungicides, these are still effective against root rot. For example, fungicides azoxystrobin and tebuconazole reduced 50 to 90% root rot caused by R. solani over three years when applied as a soil drench at 76 to 304 g a.i./ha and 250 g a.i./ha, respectively. In other countries, fungicides are combined with BCAs to control root rot. For example, a combination of azoxystrobin applied at 76 g a.i./ha and the Bacillus isolate MSU-127 reduced the crown and root rot disease and increase the yield remarkably (Kiewnick et al., 2001).
Besides, numerous phytochemicals, for example, steroids, tannins, flavonoids and alkaloids, have demonstrated antimicrobial activities against root rot (Lee et al., 2001). A product Biotos from Gaultheria spp. not only controlled root rot disease but also increase the yield. In another study, different concentrations of aqueous Chenopodium album extracts have been used to control root rot disease caused by F. solani. About 6% C. album extract reduced Fusarium root rot incidence from 47.49 to 28.25% (Abu-Tahon et al., 2021). The medicinal plants, i.e., Prosopis africana, Anacardium occidentale and Nigella sativa leaf and/whole plant parts extracts, have been assayed against root rot disease caused by M phaseolina, observing inhibition of its growth. Various alkaloids, saponins, tannins, flavonoids, anthraquinones, octadecadienoic acid, pentadecanoic acid, 1,2,3,4, butaneteterol, octadecanoic acid and linoleic acid were found in these extracts. In addition, Lippia gracilis oil extracts were found to suppress root rot disease.
Elicitors are natural or synthetic compounds, which induce systemic acquired resistance (SAR) and protect alfalfa from bacterial, fungal, and viral pathogens. The elicitors such as benzothiadiazole (BTH), chitosan (CHT), phenylalanine (PHE), and salicylic acid (SA), have been applied to control root rot in other plants (Pawlowski et al., 2016). The use of elicitors to stimulate the defensive system of alfalfa against root rot is currently unknown in China. In conclusion, since carbendazim and many fungicides are not species-specific, therefore it is likely that treated seeds though may protect the seeds from root rot pathogens but also may eliminate the keystone fungal species (Zotti et al., 2020). Fungicides applications as seed treatments can also affect endophytes which promote plant growth and protect plants from root rot pathogens. Besides, seed treatment could lead to loss of seed germination and reduction in early seedling development. If applied in high concentration can affect the plant metabolism. Usually, seeds are coated with fungicides and stored for long periods that results phytotoxicity (Lamichhane et al., 2020). The other main issue is that in many studies showed that fungicides significantly reduced the nodule formation in legume crops and affect their symbiotic association with mycorrhizal fungi (Mårtensson, 1992). In contrast to seed treatment, soil application is not detrimental to endophytes or root pathogens however it can disrupt the carbon and nitrogen cycling in soil, soil respiration and affect not target organisms. Furthermore, soil or seed application with one fungicide may be effective against single pathogen involve in root rot complex but not against diversity of pathogens. Combination of two or more fungicides having different mode of action could reduce the root rot effectively though it would be unbearable to farmers to bear costs of fungicides. The best strategy would be the combination of fungicide with BCAs because not only the amount of fungicide will be lowered, but pollution can also be reduced. Besides, both BCAs and fungicides both reduce the risk of the occurrence of fungicide resistance and improve the reliability of disease control compared with that provided using a biocontrol agent/fungicide alone.
Conclusion, issues, and future perspectives
In conclusion, due to the involvement of diverse pathogens, the management of alfalfa root rot is exceptionally challenging. Fusarium spp. and R. solani are playing a dominant role involving other pathogens in causing root rot in different regions of China. However, large-scale isolation, systematic identification, and pathogenicity evaluation of the pathogens causing alfalfa root rot are still lacking in all major alfalfa-growing regions of China. Several alfalfa breeding lines or cultivars that show partial resistance to root rot have been screened. On the other hand, resistant responses are quantitative and governed by several genes. It is challenging to develop a cultivar that could resist diverse pathogens. Furthermore, alfalfa is a cross-pollinated autotetraploid with different breeding and selection requirements than diploid plants and pathogens with genetic complexities. Also, the disease severity resulting from root rot pathogens varies with location and years due to alterations in climatic (moisture and temperature), soil factors (compaction), host and pathogens factors. Besides, it is tough to assess the yield losses because roots are rarely examined unless foliar symptoms appear. In addition, most of the studies on the root rot pathogens are conducted in controlled environmental conditions. The capacity of a pathogen to cause root rot in alfalfa under such controlled conditions is not directly related to what happens to alfalfa in the pastures. Hence information of the controlled conditions environments experiments cannot be reliably correlated to pastures. Besides fungal and oomycetes pathogens, there is also a need to assess other soil-borne pathogens such as nematodes. All these data obtained will be used to control the root rot rationally. Moreover, investigations on alfalfa resistance QTLs are limited, and the genes that cause alfalfa resistance are unknown. There is a need to focus on research into alfalfa’s disease resistance mechanism against various pathogenic strains of the pathogens and look for the broad-spectrum and specificity of alfalfa against these pathogenic strains. In addition, identification of resistance genes to breed alfalfa resistant varieties to root rot and promote the sustainable production of alfalfa is also required. Furthermore, because most studies regarding alfalfa disease resistance use a single strain, it’s impossible to tell whether the cultivars chosen are disease-resistant in general. To comprehensively investigate disease resistance and screen a broad spectrum of varieties, there is a need to standardize disease resistance evaluation criteria for different pathogenic species and understand alfalfa resistance mechanisms to the strains within and between pathogens species. Furthermore, alfalfa’s pathogenic mechanisms, particularly its molecular basis, is still unknown. Future research should include genome sequencing and comparative analysis of different strains within and within species of the pathogen that causes alfalfa root rot and transcriptomics investigations of the genes expressed by different strains during the infection of alfalfa. Currently, farmers rely solely on fungicides as seed treatments and/or soil sprays, but pathogen populations have developed resistance to fungicides. Biological control agents, such as arbuscular fungi, Bacillus and Trichoderma spp. have recently been utilized to prevent alfalfa root rot. On the other hand, these biological control agents present challenges in terms of field survival, proliferation, growth, and adaptability. Furthermore, each pathogen that causes root rot produces distinct spores, all of which contribute to the disease’s occurrence and spread. The number of spores and their ability to survive directly impact the disease’s prevalence and severity. The ideal climatic conditions for the formation and germination of many types of spores, on the other hand, are unknown. Simultaneously, studying spore production and germination mechanisms and their major regulatory factors is required to offer a theoretical foundation for developing innovative alfalfa root rot prevention and control approaches. In the future, high-throughput phenotyping and genotyping technologies, genomic methods, genome-wide investigations, transcriptomics, and next-generation sequencing techniques will make it possible to find root rot resistant cultivars and better understand root rot pathogen pathogenesis. These techniques will also aid in the discovery of genes linked to alfalfa resistance or susceptibility to root rot pathogens.
Author contributions
AA and MM: writing original draft and software and figure preparations. MAS, MKS, BH, SN, and BK: collecting literatures and tables preparations and editing. XF: validation, formal analysis, and finalized the review. LZ: supervision, project administration, resources, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the High-talent Introduction and Continuous Training Fund to LZ (grant no: 10300000021LL05) and Discipline Construction Funds (grant no: 10407000019CC2213G), supported by Zhejiang Academy of Agricultural Sciences (ZAAS) and State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products (10417000022CE0601G/029).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Abu-Tahon, M. A., Isaac, G. S., and Mogazy, A. M. (2021). Protective role of fat hen (Chenopodium album L.) extract and gamma irradiation treatments against Fusarium root rot disease in sunflower plants. Plant Biol. 23, 497–507. doi: 10.1111/plb.13229
Acharya, K., Dutta, A. K., and Pradhan, P. (2011). Bipolaris sorokiniana’(Sacc.) Shoem.: The most destructive wheat fungal pathogen in the warmer areas. Aust. J. Crop. Sci. 5:1064. doi: 10.3316/informit.044224283000719
Ajayi-Oyetunde, O. O., and Bradley, C. A. (2016). Identification and characterization of Rhizoctonia Species associated with soybean seedling disease. Plant Dis. 101, 520–533. doi: 10.1094/PDIS-06-16-0810-RE
Ajayi-Oyetunde, O., and Bradley, C. (2018). Rhizoctonia solani: taxonomy, population biology and management of Rhizoctonia seedling disease of soybean. Plant Pathol. J. 67, 3–17. doi: 10.1111/ppa.12733
Akamatsu, H. O., Chilvers, M. I., and Peever, T. L. (2008). First report of spring black stem and leaf spot of alfalfa in Washington State caused by Phoma medicaginis. Plant Dis. 92:833. doi: 10.1094/PDIS-92-5-0833A
Aliyu, T. H., and Balogun, O. S. (2012). Effect of Pythium aphanidermatum on two cultivars of pepper Capsicum spp. Nigerian J. Agric. Food Environ. 8, 64–69.
Al-Jaradi, A., Al-Mahmooli, I., Janke, R., Maharachchikumbura, S., Al-Saady, N., and Al-Sadi, A. M. (2018). Isolation and identification of pathogenic fungi and oomycetes associated with beans and cowpea root diseases in Oman. PeerJ 6:e6064. doi: 10.7717/peerj.6064
Alkherb, S. M., Aldoss, A. A., and Elhussieni, S. (1997). Identification of Anastomosis groups of Rhizoctonia solani Kuhn causing Alfalfa Root Canker in Riyadh area of the Kingdom of Saudi Arabia. J. King Saudi Univ. Agric. Sci. 9, 337–340.
Al-Sadi, A. M. (2021). Bipolaris sorokiniana-induced black point, common root rot, and spot blotch diseases of wheat: a review. Front. Cell. Infect. Microbiol. 11:118. doi: 10.3389/fcimb.2021.584899
Al-Sadi, A. M., and Deadman, M. L. (2010). Influence of seed-borne Cochliobolus sativus (Anamorph Bipolaris sorokiniana) on crown rot and root rot of barley and wheat. J. Phytopathol. 158, 683–690. doi: 10.1111/j.1439-0434.2010.01684.x
Andersen, T. F. (1996). A comparative taxonomic study of Rhizoctonia sensu lato employing morphological, ultrastructural and molecular methods. Mycol. Res. 100, 1117–1128. doi: 10.1016/S0953-7562(96)80224-3
Anderson, J. P., Lichtenzveig, J., Oliver, R. P., and Singh, K. B. (2013). Medicago truncatula as a model host for studying legume infecting Rhizoctonia solani and identification of a locus affecting resistance to root canker. Plant Pathol. 62, 908–921. doi: 10.1111/j.1365-3059.2012.02694.x
Anjanappa, R. B., Mehta, D., Maruthi, M., Kanju, E., Gruissem, W., and Vanderschuren, H. (2016). Characterization of brown streak virus–resistant cassava. Mol. Plant-Microbe Interact. 29, 527–534. doi: 10.1094/MPMI-01-16-0027-R
Assunção, I. P., Nascimento, L. D., Ferreira, M. F., Oliveira, F. J., Michereff, S. J., and Lima, G. S. A. (2011). Reaction of faba bean genotypes to Rhizoctonia solani and resistance stability. Hortic. Bras. 29, 492–497. doi: 10.1590/S0102-05362011000400008
Azam, B., Farhad, N.-F., and Ahmad, I. (2018). Fusion of a chitin-binding domain to an antibacterial peptide to enhance resistance to Fusarium solani in tobacco (Nicotiana tabacum). Biotech 8:331. doi: 10.1007/s13205-018-1416-7
Badis, Y., Bonhomme, M., Lafitte, C., Huguet, S., Balzergue, S., Dumas, B., et al. (2015). Transcriptome analysis highlights preformed defences and signalling pathways controlled by the pr A e1 quantitative trait locus (QTL), conferring partial resistance to Aphanomyces euteiches in Medicago truncatula. Mol. Plant Pathol. 16, 973–986. doi: 10.1111/mpp.12253
Bai, Q. R., Jiang, X. Y., Xie, Y. Y., Sun, H. Y., and Gao, J. (2014). Summer blight of white clover (Trifolium repens) caused by Rhizoctonia solani AG-1-IB in China. Plant Dis. 98:1153. doi: 10.1094/PDIS-09-13-0987-PDN
Balali, G. R., and Kowsari, M. (2004). Pectic zymogram variation and pathogenicity of Rhizoctonia solani AG-4 to bean (Phaseolus vulgaris) isolates in Isfahn, Iran. Mycopathologia 158:377. doi: 10.1007/s11046-004-2227-4
Bao, A. K., Du, B. Q., Touil, L., Kang, P., Wang, Q. L., and Wang, S. M. (2016). Co-expression of tonoplast Cation/H+ antiporter and H+-pyrophosphatase from xerophyte Zygophyllum xanthoxylum improves alfalfa plant growth under salinity, drought and field conditions. Plant Biotechnol. J. 14, 964–975. doi: 10.1111/pbi.12451
Barbetti, M. J. (1983). Relation between plant size and severity of root rot in subterranean clover. Aust. J. Exp. Agric. 23, 126–129. doi: 10.1071/EA9830126
Barbetti, M. J. (1990). Effect of adding lime to potting soil on the pathogenicity of four root pathogens of subterranean clover. Phytophylactica 22, 245–250.
Barbetti, M. J., and MacNish, G. C. (1984). Effects of cultivation and cultural practice on root rot of subterranean clover. Aust. J. Exp. Agric. 24, 550–554. doi: 10.1071/EA9840550
Barbetti, M., You, M., Li, H., Ma, X., and Sivasithamparam, K. (2007). Management of root diseases of annual pasture legumes in Mediterranean ecosystems-a case study of subterranean clover root diseases in the south-west of Western Australia. Phytopathol. Mediterr. 46, 239–258.
Bardin, M., Ajouz, S., Comby, M., Lopez-Ferber, M., Graillot, B., Siegwart, M., et al. (2015). Is the efficacy of biological control against plant diseases likely to be more durable than that of chemical pesticides? Front. Plant Sci. 6:566. doi: 10.3389/fpls.2015.00566
Bartlett, D. W., Clough, J. M., Godwin, J. R., Hall, A. A., Hamer, M., and Parr-Dobrzanski, B. (2002). The strobilurin fungicides. Pest Manag. Sci. 58, 649–662. doi: 10.1002/ps.520
Basbagci, G., and Dolar, F. S. (2020). First report of binucleate Rhizoctonia AG-K causing root rot on chickpea. Arch. Phytopathol. Plant Protect. 53, 13–14. doi: 10.1080/03235408.2020.1789822
Basbagci, G., Unal, F., Uysal, A., and Dolar, F. S. (2019). Identification and pathogenicity of Rhizoctonia solani AG-4 causing root rot on chickpea in Turkey. Span. J. Agric. Res. 17:e1007. doi: 10.5424/sjar/2019172-13789
Bates, G. D., Rothrock, C. S., and Rupe, J. C. (2008). Resistance of the soybean cultivar archer to Pythium damping-off and root rot caused by several Pythium spp. Plant Dis. 92, 763–766. doi: 10.1094/PDIS-92-5-0763
Batnini, M., Fernández Del-Saz, N., Fullana-Pericàs, M., Palma, F., Haddoudi, I., Mrabet, M., et al. (2020). The alternative oxidase pathway is involved in optimizing photosynthesis in Medicago truncatula infected by Fusarium oxysporum and Rhizoctonia solani. Physiol. Plant. 169, 600–611. doi: 10.1111/ppl.13080
Berg, L. E., Miller, S. S., Dornbusch, M. R., and Samac, D. A. (2017). Seed rot and damping-off of alfalfa in Minnesota caused by Pythium and Fusarium species. Plant Dis. 101, 1860–1867. doi: 10.1094/PDIS-02-17-0185-RE
Blanco, A. J. V., Costa, M. O., Silva, R. D. N., Albuquerque, F. S. D., Melo, A. T. D. O., Lopes, F. A. C., et al. (2018). Diversity and pathogenicity of Rhizoctonia species from the Brazilian Cerrado. Plant Dis. 102, 773–781. doi: 10.1094/PDIS-05-17-0721-RE
Bonhomme, M., André, O., Badis, Y., Ronfort, J., Burgarella, C., Chantret, N., et al. (2014). High-density genome-wide association mapping implicates an F-box encoding gene in Medicago truncatula resistance to Aphanomyces euteiches. New Phytol. 201, 1328–1342. doi: 10.1111/nph.12611
Botha, A., Denman, S., Lamprecht, S. C., Mazzola, M., and Crous, P. W. (2003). Characterisation and pathogenicity of Rhizoctonia isolates associated with black root rot of strawberries in the Western Cape Province, South Africa. Australas. Plant Pathol. 32, 195–201. doi: 10.1071/AP02067
Broders, K. D., Parker, M. L., Melzer, M. S., and Boland, G. J. (2014). Phylogenetic Diversity of Rhizoctonia solani Associated with Canola and Wheat in Alberta, Manitoba, and Saskatchewan. Plant Dis. 98, 1695–1701. doi: 10.1094/PDIS-02-14-0146-RE
Brown, L., Harrington, S., Harrington, M., Murdock, M. R., Pizolotto, C. A., and Woodhall, J. W. (2020). Rhizoctonia solani AG 2-2 IIIB Causing Root Rot of Onion in Idaho. Plant Dis. 105:498. doi: 10.1094/PDIS-08-20-1724-PDN
Bruno, L., and Griffiths, P. D. (2004). Evaluation of common bean accessions for resistance to Pythium ultimum. HortScience 39, 1193–1195. doi: 10.21273/HORTSCI.39.6.1193
Bugbee, W. M., and Campbell, L. G. (1990). Combined resistance in sugar beet to Rhizoctonia solani, Phoma betae, and Botrytis cinerea. Plant Dis. 74:353. doi: 10.1094/PD-74-0353
Cai, W., Tian, H., Liu, J., Fang, X., and Nan, Z. (2021). Phytophthora cactorum as a pathogen associated with root rot on Alfalfa (Medicago sativa) in China. Plant Dis. 105, 231. doi: 10.1094/PDIS-04-20-0815-PDN
Cao, S., Liang, Q. W., Nzabanita, C., and Li, Y. Z. (2020). Paraphoma root rot of alfalfa (Medicago sativa) in Inner Mongolia, China. Plant Pathol. 69, 231–239. doi: 10.1111/ppa.13131
Cao, L. X., Zhao, C. H., Bai, Q. J., and Shao, Z. Z. (2008). Identification of the Pathogens Causing Root Rot of Alfalfa in Inner Mongolia. Acta Agricul. Boreali Sin. 23, 105–107. doi: 10.11686/cyxb2016111
Carisse, O., and Khanizadeh, S. (2006). Relative resistance of newly released apple rootstocks to Phytophthora cactorum. Can. J. Plant Sci. 86, 199–204. doi: 10.4141/P04-176
Carlucci, A., Raimondo, M., Santos, J., and Phillips, A. (2012). Plectosphaerella species associated with root and collar rots of horticultural crops in southern Italy. Persoonia 28, 34. doi: 10.3767/003158512X638251
Chen, Q., Jiang, J. R., Zhang, G. Z., Cai, L., and Crous, P. W. (2015a). Resolving the Phoma enigma. Stud. Mycol. 82, 137–217. doi: 10.1016/j.simyco.2015.10.003
Chen, Y., Yang, X., Gu, C.-Y., Zhang, A.-F., Gao, T.-C., and Zhou, M.-G. (2015b). Genotypes and phenotypic characterization of field Fusarium asiaticum isolates resistant to carbendazim in Anhui Province of China. Plant Dis. 99, 342–346. doi: 10.1094/PDIS-04-14-0381-RE
Coleman, J. J. (2016). The Fusarium solani species complex: ubiquitous pathogens of agricultural importance. Mol. Plant Pathol. 17, 146–158. doi: 10.1111/mpp.12289
Coleman, P. M., Ellerbrock, L. A., and Lorbeer, J. W. (1997). Reaction of selected onion cultigens to pink root under field conditions in New York. Plant Dis. 81, 138–142. doi: 10.1094/PDIS.1997.81.2.138
Cong, L. L., Li, M. N., Sun, Y., Cong, L. L., Yang, Q. C., Long, R. C., et al. (2016b). First report of root rot disease caused by Fusarium tricinctum on Alfalfa in North China. Plant Dis. 100:1503. doi: 10.1094/PDIS-11-15-1293-PDN
Cong, L., Sun, Y., Kang, J., Li, M., Long, R., Zhang, T., et al. (2016a). First report of root rot disease caused by Fusarium proliferatum on alfalfa in China. Plant Dis. 100:2526. doi: 10.1094/PDIS-04-16-0505-PDN
Cong, L. L., Sun, Y., Wang, Z., Kang, J. M., Zhang, T. J., Biligetu, B., et al. (2018). A rapid screening method for evaluating resistance of alfalfa (Medicago sativa L.) to Fusarium root rot. Can. J. Plant Pathol. 40, 61–69. doi: 10.1080/07060661.2017.1402822
Cong, L.-L., Yan, S., Long, R.-c., Kang, J.-m., Zhang, T.-j., LI, M.-n., et al. (2017). Modulation of protein expression in alfalfa (Medicago sativa L.) root and leaf tissues by Fusarium proliferatum. J. Integr. Agric. 16, 2558–2572. doi: 10.1016/S2095-3119(17)61690-3
Couture, L., Dhont, C., Chalifour, F. P., Drapeau, R., Tremblay, G., Castonguay, Y., et al. (2002). Fusarium root and crown rot in alfalfa subjected to autumn harvests. Can. J. Plant Sci. 82, 621–624. doi: 10.4141/P01-132
Curl, E. A. (1963). Control of plant diseases by crop rotation. Bot. Rev. 29, 413–479. doi: 10.1007/BF02860813
da Silva, W. L., and Clark, C. A. (2013). Infection of sweetpotato by Fusarium solani and Macrophomina phaseolina prior to harvest. Plant Dis. 97, 1636–1644. doi: 10.1094/PDIS-05-13-0514-RE
Dale, M., and Irwin, J. (1991). Glasshouse and field screening of chickpea cultivars for resistance to Phytophthora megasperma f.sp. medicaginis. Aust. J. Exp. Agric. 31, 663–667. doi: 10.1071/EA9910663
De Candole, A. P. (1815). Memoire sur les Rhizoctones, nouvean genre de champignons qui attaque les racines, des plantes et en particulier cell de la luzerne cultivee. Mem. Mus. d'Hist. Nat. 2, 209–216.
de Gruyter, J., Aveskamp, M. M., Woudenberg, J. H. C., Verkley, G. J. M., Groenewald, J. Z., and Crous, P. W. (2009). Molecular phylogeny of Phoma and allied anamorph genera: towards a reclassification of the Phoma complex. Mycol. Res. 113, 508–519. doi: 10.1016/j.mycres.2009.01.002
de Gruyter, J., Woudenberg, J. H., Aveskamp, M. M., Verkley, G. J., Groenewald, J. Z., and Crous, P. W. (2010). Systematic reappraisal of species in Phoma section Paraphoma, Pyrenochaeta and Pleurophoma. J. Mycol. 102, 1066–1081. doi: 10.3852/09-240
Deb, D., Khan, A., and Dey, N. (2020). Phoma diseases: Epidemiology and control. Plant Pathol. 69, 1203–1217. doi: 10.1111/ppa.13221
Delang, C. O. (2018). The consequences of soil degradation in China: a review. GeoScape 12. doi: 10.2478/geosc-2018-0010
Dick, M. W., Vick, M. C., Gibbings, J. G., Hedderson, T. A., and Lastra, C. C. L. (1999). 18S rDNA for species of Leptolegnia and other Peronosporomycetes: justification for the subclass taxa Saprolegniomycetidae and Peronosporomycetidae and division of the Saprolegniaceae sensu lato into the Leptolegniaceae and Saprolegniaceae. Mycol. Res. 103, 1119–1125. doi: 10.1017/S0953756299008643
Dijst, G., and Schneider, J. H. M. (1996). “Flowerbulbs diseases incited by Rhizoctonia species,” in Rhizoctonia Species: Taxonomy, Molecular Biology, Ecology, Pathology and Disease Control. eds. S. Baruch, H. Suha Jabaji, N. Stephen, and D. Gerda (Berlin/Heidelberg, Germany: Springer), 279–288.
Ding, S., Li, M., and Hui, Z. (2011). Identification of disease resistance of different varieties of alfalfa to Fusarium spp. Plant Prot. 37, 110–114.
Douhan, G. W., Fuller, E., McKee, B., and Pond, E. (2011). Genetic diversity analysis of avocado (Persea americana Miller) rootstocks selected under greenhouse conditions for tolerance to Phytophthora root rot caused by Phytophthora cinnamomi. Euphytica 182, 209–217. doi: 10.1007/s10681-011-0433-y
Dubey, S. C., Tripathi, A., Upadhyay, B. K., and Deka, U. K. (2014). Diversity of Rhizoctonia solani associated with pulse crops in different agro-ecological regions of India. World J. Microbiol. Biotechnol. 30, 1699–1715. doi: 10.1007/s11274-013-1590-z
Edel-Hermann, V., and Lecomte, C. (2019). Current status of Fusarium oxysporum formae speciales and races. Phytopathology 109, 512–530. doi: 10.1094/PHYTO-08-18-0320-RVW
Eikemo, H., Brurberg, M. B., and Davik, J. (2010). Resistance to Phytophthora cactorum in diploid Fragaria species. HortScience 45, 193–197. doi: 10.21273/HORTSCI.45.2.193
Eken, C., and Demirci, E. (2003). Identification and pathogenicity of Rhizoctonia solani and binucleate Rhizoctonia anastomosis groups isolated from forage legumes in Erzurum. Turkey. Phytoparasitica 31, 76–80. doi: 10.1007/BF02979769
Ellis, M. L., McHale, L. K., Paul, P. A., Martin, S., and Dorrance, A. E. (2013). Soybean germplasm resistant to Pythium irregulare and molecular mapping;of resistance quantitative trait loci derived from the soybean accession. Crop Sci. 53:1008. doi: 10.2135/cropsci2012.08.0461
Eranthodi, A., Schneiderman, D., Harris, L. J., Witte, T. E., Sproule, A., Hermans, A., et al. (2020). Enniatin production influences Fusarium avenaceum virulence on potato tubers, but not on durum wheat or peas. Pathogens 9:75. doi: 10.3390/pathogens9020075
Erginbas-Orakci, G., Morgounov, A., and Dababat, A. A. (2018). Determination of resistance in winter wheat genotypes to the dryland root rots caused by Fusarium culmorum in Turkey. Int. J. Agric. Wildlife Sci. 4, 193–202. doi: 10.24180/ijaws.414501
Erwin, D. C. (1954). Root rot of alfalfa caused by Phytophthora cryptogea. Phytopathology 44, 700–704.
Fahad, S., Hasanuzzaman, M., Alam, M., Ullah, H., Saeed, M., Ali, K. I., et al. (Eds.) (2020). Environment, Climate, Plant and Vegetation Growth. Berlin: Springer Nature Switzerland AG 2020.
Fahad, S., Saud, S., Yajun, C., Chao, W., and Depeng, W. (Eds.), (2021f). Abiotic Stress in Plants. London: IntechOpen United Kingdom 2021.
Fahad, S., Sönmez, O., Saud, S., Wang, D., Wu, C., Adnan, M., et al. (eds.), (2021e). Engineering Tolerance in Crop Plants Against Abiotic Stress, First Edition. ed, Footprints of Climate Variability on Plant Diversity. Boca Raton, FL: CRC Press.
Fahad, S., Sönmez, O., Saud, S., Wang, D., Wu, C., Adnan, M., et al. (Eds.), (2021a). Plant Growth Regulators For Climate-Smart Agriculture, First Edition. ed, Footprints of Climate Variability on Plant Diversity. Boca Raton, FL: CRC Press.
Fahad, S., Sonmez, O., Saud, S., Wang, D., Wu, C., Adnan, M., et al. (Eds.), (2021b). Climate Change and Plants: Biodiversity, Growth and Interactions, First Edition. ed, Footprints of Climate Variability on Plant Diversity. Boca Raton, FL: CRC Press.
Fahad, S., Sonmez, O., Saud, S., Wang, D., Wu, C., Adnan, M., et al. (Eds). (2021c). Developing Climate Resilient Crops: Improving Global Food Security and Safety, First Edition. Footprints of Climate variability On Plant Diversity. Boca Raton, FL: CRC Press.
Fahad, S., Sönmez, O., Turan, V., Adnan, M., Saud, S., Wu, C., et al. (Eds.), (2021d). Sustainable Soil and Land Management and Climate Change, First edition. ed, Footprints of Climate Variability On Plant Diversity. Boca Raton, FL: CRC Press.
Fan, Q., Creamer, R., and Li, Y. (2018). Time-course metabolic profiling in alfalfa leaves under Phoma medicaginis infection. PLoS One 13:e0206641. doi: 10.1371/journal.pone.0206641
Fan, Q., and Li, Y.-Z. (2017). The effect of Phoma medicaginis on the photosynthetic physiology of Medicago sativa. Acta Prataculturae Sin. 26:112. doi: 10.11686/cyxb2016275
Fang, M. J., Quan, L. M., Zhang, Z. H., and Chai, Z. X. (2003). Study on relationship between phenylalanine ammonia-lyase (PAL) and resistance to crown and root rot in alfalfa cultivars. Acta Pratacultural Sci. 12, 35–39.
Fang, X. L., Zhang, C. X., and Nan, Z. B. (2019). Research advances in Fusarium root rot of alfalfa (Medicago sativa). Acta Pratacul. Sin. 28:169. doi: 10.11686/cyxb2019105
Fang, X., Zhang, C., Wang, Z., Duan, T., Yu, B., Jia, X., et al. (2021). co-infection by soil-borne fungal pathogens alters disease responses among diverse alfalfa varieties. Frontier Microbiol. 12. doi: 10.3389/fmicb.2021.664385
Faris, M. A., and Sabo, F. E. (1981). Effect of Phytophthora megasperma on yield and survival of resistant and susceptible alfalfa cultivars. Can. J. Plant Sci. 61, 955–960. doi: 10.4141/cjps81-140
Fatemi, J. (1972). Fungi associated with alfalfa root rot in Iran. Phytopathol. Mediterr. 11, 163–165. Available at: http://www.jstor.org/stable/42684143
Fattahi, S. H., Zafari, D., and Mahmoudi, B. (2011). Evaluation of superior sugar beet genotypes for resistance to important root rot pathogens in the greenhouse. J. Sugar Beet. 27, 25–38. doi: 10.22092/jsb.2011.682
Feng, H. E., and Kai, X. I. E. (2014). The role of alfalfa on the maintenance of food security in China. J. Agric. Sci. Technol. 16:7.
Feng, J., Zhao, L., Zhang, Y., Sun, L., Yu, X., and Yu, Y. (2020). Can climate change influence agricultural GTFP in arid and semi-arid regions of Northwest China? J. Arid. Land 12, 837–853. doi: 10.1007/s40333-020-0073-y
Fenille, R. C., Ciampi, M. B., Kuramae, E. E., and Souza, N. L. (2003). Identification of Rhizoctonia solani associated with soybean in Brazil by rDNA-ITS sequences. Fitopatol. Bras. 28, 413–419. doi: 10.1590/S0100-41582003000400011
Fones, H. N., Bebber, D. P., Chaloner, T. M., Kay, W. T., Steinberg, G., and Gurr, S. J. (2020). Threats to global food security from emerging fungal and oomycete crop pathogens. Nat. Food 1, 332–342. doi: 10.1038/s43016-020-0075-0
Fones, H. N., Fisher, M. C., and Gurr, S. J. (2017). Emerging fungal threats to plants and animals challenge agriculture and ecosystem resilience. Microbiol. Spectr. 5:2. doi: 10.1128/microbiolspec.FUNK-0027-2016
Fravel, D. R. (2005). Commercialization and implementation of biocontrol. Annu. Rev. Phytopathol. 43, 337–359. doi: 10.1146/annurev.phyto.43.032904.092924
Fuchs, M. (2017). Pyramiding resistance-conferring gene sequences in crops. Curr. Opin. Virol. 26, 36–42. doi: 10.1016/j.coviro.2017.07.004
Gang, W. X. W. C. Z., Changfu, H., and Dongmei, X. L. W. (1996). Investigation and analysis on lucerne diseases from Altay newly established areas in Northern Xinjiang. J. Xinjiang Agric. Univ. 3.
Gao, P., Guo, Y., Li, Y., and Duan, T. (2018a). Effects of dual inoculation of AMF and rhizobium on alfalfa (Medicago sativa) root rot caused by Microdochium tabacinum. Australas. Plant Pathol. 47, 195–203. doi: 10.1007/s13313-018-0543-2
Gao, Y., Zhang, C., Han, X., Wang, Z. Y., Ma, L., Yuan, D. P., et al. (2018b). Inhibition of OsSWEET11 function in mesophyll cells improves resistance of rice to sheath blight disease. Mol. Plant Pathol. 19, 2149–2161. doi: 10.1111/mpp.12689
Garibaldi, A., Tabone, G., Luongo, I., and Gullino, M. (2021). First Report of Rhizoctonia solani AG 2-1 Causing Crown Rot on Alcea rosea in Italy. Plant Dis. 105:707. doi: 10.1094/PDIS-07-20-1635-PDN
Gaulin, E., Jacquet, C., Bottin, A., and Dumas, B. (2007). Root rot disease of legumes caused by Aphanomyces euteiches. Mol. Plant Pathol. 8, 539–548. doi: 10.1111/j.1364-3703.2007.00413.x
Godoy-Lutz, G., Kuninaga, S., and Powers, J. (2008). Phylogenetic analysis of Rhizoctonia solani subgroups associated with web blight symptoms on common bean based on ITS-5.8S rDNA. J. Gen. Plant Pathol. 74, 32–40. doi: 10.1007/s10327-007-0060-6
Gonzalez, M., Pujol, M., Metraux, J.-P., Gonzalez-Garcia, V., Bolton, M. D., and Borrás-Hidalgo, O. (2011). Tobacco leaf spot and root rot caused by Rhizoctonia solani Kühn. Mol. Plant Pathol. 12, 209–216. doi: 10.1111/j.1364-3703.2010.00664.x
Gónzalez, D., Rodriguez-Carres, M., Boekhout, T., Stalpers, J., Kuramae, E. E., Nakatani, A. K., et al. (2016). Phylogenetic relationships of Rhizoctonia fungi within the Cantharellales. Fungal Biol. 120, 603–619. doi: 10.1016/j.funbio.2016.01.012
Gossen, B. D., Conner, R. L., Chang, K.-F., Pasche, J. S., McLaren, D. L., Henriquez, M. A., et al. (2016). Identifying and managing root rot of pulses on the northern great plains. Plant Dis. 100, 1965–1978. doi: 10.1094/PDIS-02-16-0184-FE
Graham, R. D. (1983). Effects of nutrient stress on susceptibility of plants to disease with particular reference to the trace elements. Adv. Bot. Res. 10, 221–276.
Gray, F. A., Bohl, W. H., and Abernethy, R. H. (1983). Phytophthora root rot of alfalfa in Wyoming. Plant Dis. 67, 291–294. doi: 10.1094/PD-67-291
Gray, F. A., Heald, T. E., Hollingsworth, C. R., and Koch, D. W. (1997). Brown Root Rot Caused by Phoma sclerotioides, a New Disease of Alfalfa in the US. In Proceedings of the 10th Western Alfalfa Improvement Conference, Davis. California, USA: WAIC Executive Committee, 22–24.
Gray, F. A., Hollingsworth, C. R., Koch, D., Groose, R., and Heald, T. (2004). Brown root rot of alfalfa. Plant science timely information No, 1, University of Wyoming, USA.
Gui, Z., and Yuan, Q. (2002). Studies on RAPD polymorphic in alfalfa resistant to brown blot. Acta Agrestia Sin. 10, 274–278.
Guo, T., Xue, B., Bai, J., and Sun, Q. (2019). Discussion of the present situation of China’s forage grass industry development: an example using alfalfa and oats. J. Pratacultural Sci. 36, 1466–1474.
Gupta, S., Kumar, A., Patel, R., and Kumar, V. (2021). Genetically modified crop regulations: scope and opportunity using the CRISPR-Cas9 genome editing approach. Mol. Biol. Rep. 48, 4851–4863. doi: 10.1007/s11033-021-06477-9
Hagerty, C. H., Cuesta-Marcos, A., Cregan, P. B., Song, Q., McClean, P., Noffsinger, S., et al. (2015). Mapping Fusarium solani and Aphanomyces euteiches root rot resistance and root architecture quantitative trait loci in common bean. Crop Sci. 55, 1969–1977. doi: 10.2135/cropsci2014.11.0805
Hancock, J. G. (1983). Seedling diseases of alfalfa in California. Plant Dis. 67, 1203–1208. doi: 10.1094/PD-67-1203
Harper, J. T., Waanders, E., and Keeling, P. J. (2005). On the monophyly of chromalveolates using a six-protein phylogeny of eukaryotes. Int. J. Syst. Evol. 55, 487–496. doi: 10.1099/ijs.0.63216-0
Hollingsworth, C. R., Gray, F. A., Koch, D. W., Groose, R. W., and Heald, T. E. (2003). Distribution of Phoma sclerotioides and incidence of brown root rot of alfalfa in Wyoming, U.S.A. Can. J. Plant Pathol. 25, 215–217. doi: 10.1080/07060660309507071
Huang, N., Sun, X.-B., Wang, T.-M., and Lu, X.-S. (2013). Evaluation of resistance to Fusarium Wilt (Fusarium oxyssorum) and preliminary screening of check varieties of resistant evaluation from alfalfa (Medicago sativa) cultivar. Chin. J. Grassland 1, 1–5.
Hwang, S. F., Chakravarty, P., and Prevost, D. (1993). Effects of rhizobia, metalaxyl, and VA mycorrhizal fungi on growth, nitrogen fixation, and development of Pythium root rot of Sainfoin. Plant Dis. 77, 1093–1098. doi: 10.1094/PD-77-1093
Hwang, S. E., and Flores, G. (1987). Effects of Cylindrocladium gracile, Fusarium roseum and Plenodomus melilotion crown and root rot, forage yield. Can. Plant Dis. Surv. 67:31.
Hwang, S. F., Gossen, B. D., Conner, R. L., Chang, K. F., Turnbull, G. D., Lopetinsky, K., et al. (2007). Management strategies to reduce losses caused by Rhizoctonia seedling blight of field pea. Can. J. Plant Sci. 87, 145–155. doi: 10.4141/P04-172
Hwang, S. F., Gossen, B. D., Turnbull, G. D., Chang, K. F., and Howard, R. J. (2002). Seedbed preparation, timing of seeding, fertility and root pathogens affect establishment and yield of alfalfa. Can. J. Plant Sci. 82, 371–381. doi: 10.4141/P01-121
Hwang, S. F., Howard, R. J., and Moskaluk, E. (1989). Crown and root rot of alfalfa in southern Alberta. Can. Plant Dis. Surv. 69, 9–11.
Jat, M. K., and Ahir, R. R. (2013). Effect of temperature, relative humidity and pH on mycelial growth and sporulation of Fusarium solani causing root rot of Indian Aloe (Aloe barbadensis MILL.). J. Plant Res. 29:181.
Jiang, D., Xu, C., Han, W., Harris-Shultz, K., Ji, P., Li, Y., et al. (2021). Identification of fungal pathogens and analysis of genetic diversity of Fusarium tricinctum causing root rots of alfalfa in north-east China. Plant Pathol. 70, 804–814. doi: 10.1111/ppa.13333
Jun, F., Yu, G., Quanjiu, W., Malhi, S. S., and Yangyang, L. (2014). Mulching effects on water storage in soil and its depletion by alfalfa in the Loess Plateau of northwestern China. Agric. Water Manag. 138, 10–16. doi: 10.1016/j.agwat.2014.02.018
Karppinen, E. M., Payment, J., Chatterton, S., Bainard, J. D., Hubbard, M., Gan, Y., et al. (2020). Distribution and abundance of Aphanomyces euteiches in agricultural soils: Effect of land use type, soil properties, and crop management practices. Appl. Soil Ecol. 150:103470. doi: 10.1016/j.apsoil.2019.103470
Kaur, S., Dhillon, G. S., Brar, S. K., Vallad, G. E., Chand, R., and Chauhan, V. B. (2012). Emerging phytopathogen Macrophomina phaseolina: biology, economic importance and current diagnostic trends. Crit. Rev. Microbiol. 38, 136–151. doi: 10.3109/1040841X.2011.640977
Khoury, C. K., Bjorkman, A. D., Dempewolf, H., Ramirez-Villegas, J., Guarino, L., Jarvis, A., et al. (2014). Increasing homogeneity in global food supplies and the implications for food security. Proc. Natl. Acad. Sci. 111, 4001–4006. doi: 10.1073/pnas.1313490111
Kiewnick, S., Jacobsen, B. J., Braun-Kiewnick, A., Eckhoff, J. L. A., and Bergman, J. W. (2001). Integrated control of Rhizoctonia crown and root rot of sugar beet with fungicides and antagonistic bacteria. Plant Dis. 85, 718–722. doi: 10.1094/PDIS.2001.85.7.718
Kilicoglu, M. C., and Ozkoc, I. (2010). Molecular characterization of Rhizoctonia solani AG4 using PCR-RFLP of the rDNA-ITS region. Turk. J. Biol. 34, 261–269. doi: 10.3906/biy-0812-14
Kim, H.-J., Nahm, S.-H., Lee, H.-R., Yoon, G.-B., Kim, K.-T., Kang, B.-C., et al. (2008). BAC-derived markers converted from RFLP linked to Phytophthora capsici resistance in pepper (Capsicum annuum L.). Theor. Appl. Genet. 118, 15–27. doi: 10.1007/s00122-008-0873-5
Knezevic, M. M., Stajkovic-Srbinovic, O. S., Assel, M., Milic, M. D., Mihajlovski, K. R., Delic, D. I., et al. (2021). The ability of a new strain of Bacillus pseudomycoides to improve the germination of alfalfa seeds in the presence of fungal infection or chromium. Rhizosphere 18:100353. doi: 10.1016/j.rhisph.2021.100353
Kong, Q., Feng, Q., Zhang, Y., Ma, Z., and Wang, H. (2018a). Detection of pathogenicity and mycotoxin chemotypes of Fusarium causing alfalfa root rot. J. China Agric. Univ. 23, 74–85.
Kong, Q., Liu, R., Liu, D., Liu, Z., and Wang, H. (2018b). Biological characteristics of Fusarium causing alfalfa root rot in Hebei Province. J. China Agric. Univ. 23, 59–76.
Kousik, C., Snow, J., Berggren, G., and Harville, B. (1995). Effect of temperature on virulence of Rhizoctonia solani on soybean leaves and seedlings. Plant Pathol. 44, 580–586. doi: 10.1111/j.1365-3059.1995.tb01680.x
Kuan, T. L., and Erwin, D. C. (1980). Predisposition effect of water saturation of soil on Phytophthora root rot of alfalfa. Phytopathology 70, 981–986. doi: 10.1094/Phyto-70-981
Kühn, J. (1858). Die Krankheiten der Kulturgewächse: ihre Ursachen und ihre Verhütung. Berlin: Bosselmann.
Lakhran, L., Ahir, R., Choudhary, M., and Choudhary, S. (2018). Isolation, purification, identification and pathogenicity of Macrophomina phaseolina (Tassi) goid caused dry root rot of chickpea. J. Pharmacogn. Phytochem. 7, 3314–3317.
Lamichhane, J. R., You, M. P., Laudinot, V., Barbetti, M. J., and Aubertot, J. N. (2020). Revisiting Sustainability of Fungicide Seed Treatments for Field Crops. Plant Dis. 104, 610–623. doi: 10.1094/PDIS-06-19-1157-FE
Larsen, J. E., Hollingsworth, C. R., Flor, J., Dornbusch, M. R., Simpson, N. L., and Samac, D. A. (2007). Distribution of Phoma sclerotioides on alfalfa and winter wheat crops in the North Central United States. Plant Dis. 91, 551–558. doi: 10.1094/PDIS-91-5-0551
Leath, K. T., and Kendall, W. A. (1978). Fusarium root rot of forage species: pathogenicity and host range. Phytopathology 68, 826–831. doi: 10.1094/Phyto-68-826
Lee, S. E., Park, B. S., Kim, M. K., Choi, W. S., Kim, H. T., Cho, K. Y., et al. (2001). Fungicidal activity of pipernonaline, a piperidine alkaloid derived from long pepper, Piper longum L., against phytopathogenic fungi. Crop Protect. 20, 523–528. doi: 10.1016/S0261-2194(00)00172-1
Li, M. (2003). Comparative pathogenicity of isolates of Fusarium spp. and cultivars resistance in alfalfa. J. Grassland of China 25, 39–43.
Li, Y., Duan, T., Nan, Z., and Li, Y. (2021c). Arbuscular mycorrhizal fungus alleviates alfalfa leaf spots caused by Phoma medicaginis revealed by RNA-seq analysis. Appl. Microbiol. 130, 547–560. doi: 10.1111/jam.14387
Li, K.-m., Guo, Q.-y., Zhao, L., and Chen, X.-z. (2009). Study on anastomosis groups and their pathogenicity of Rhizoctonia solani isolated from alfalfa in Xinjiang. J. Pratacultural Sci. 5.
Li, Y., Meng, L., Gong, L., Zhao, T., Ji, P., Zhang, Q., et al. (2019). Occurrence of Bipolaris root rot caused by Bipolaris sorokiniana on Alfalfa in China. Plant Dis. 103:2691. doi: 10.1094/PDIS-03-19-0532-PDN
Li, S., Wang, Z., Tang, B., Zheng, L., Chen, H., Cui, X., et al. (2021b). A Pathogenesis-Related Protein-Like Gene Is Involved in the Panax notoginseng Defense Response to the Root Rot Pathogen. Front. Plant Sci. 11:2217. doi: 10.3389/fpls.2020.610176
Li, Y., Yan, Z., Gao, P., Guo, Y., and Duan, T. (2018). Effect of Glomus mosseae on root rot disease of Medicago sativa caused by Microdochium tabacinum. Chin. J. Biol. 34, 598–605.
Li, B., Zheng, Y., Cai, Y., Liu, J., Wang, R., Cui, G., et al. (2021a). Identification and assessment of a biocontrol agent, Ochrobactrum intermedium I-5, for management of alfalfa root rot caused by Fusarium tricinctum. Phytopathology 111, 1927–1934. doi: 10.1094/PHYTO-12-20-0549-R
Liang, E., Liu, X., Yuan, Y., Qin, N., Fang, X., Huang, L., et al. (2006). The 1920s drought recorded by tree rings and historical documents in the semi-arid and arid areas of northern China. Clim. Chang. 79, 403–432. doi: 10.1007/s10584-006-9082-x
Link, H.F. (1809). Observationes in Ordines plantarum naturales: dissertatio I.ma complectens Anandrarum ordines Epiphytas, Mucedines Gastromycos et Fungos. Aut. Henr. Frid. Link, professore Rostochiensi. S. l.: s. n.
Liu, C.-A., Li, F.-R., Zhou, L.-M., Zhang, R.-H., Lin, S.-L., Wang, L.-J., et al. (2013). Effect of organic manure and fertilizer on soil water and crop yields in newly-built terraces with loess soils in a semi-arid environment. Agric. Water Manag. 117, 123–132. doi: 10.1016/j.agwat.2012.11.002
Liu, Y., Ma, J., Yan, W., Yan, G., Zhou, M., Wei, Y., et al. (2012). Different tolerance in bread wheat, durum wheat and barley to Fusarium crown rot disease caused by Fusarium pseudograminearum. J. Phytopathol. 160, 412–417. doi: 10.1111/j.1439-0434.2012.01920.x
Liu, R., Zhan-Hong, M. A., Liu, Z. Y., Qin, F., and Wang, H. G. (2016). Isolation, identification and toxicity determination of pathogens causing alfalfa root rot. J. China Agric. Univ. 21.
Löffler, M., Kessel, B., Ouzunova, M., and Miedaner, T. (2010). Population parameters for resistance to Fusarium graminearum and Fusarium verticillioides ear rot among large sets of early, mid-late and late maturing European maize (Zea mays L.) inbred lines. Theor. Appl. Genet. 120, 1053–1062. doi: 10.1007/s00122-009-1233-9
Lugtenberg, B. J., Caradus, J. R., and Johnson, L. J. (2016). Fungal endophytes for sustainable crop production. FEMS Microbiol. Ecol. 92. doi: 10.1093/femsec/fiw194
Luo, D., Liu, W., Wang, Y., Zhang, J., and Liu, Z. (2014). Development of a rapid one-step PCR protocol to distinguish between alfalfa (Medicago sativa) and sweet clover (Melilotus spp.) seeds. Seed Sci. Technol. 42, 237–246. doi: 10.15258/sst.2014.42.2.12
Ma, Y., Marzougui, A., Coyne, C. J., Sankaran, S., Main, D., Porter, L. D., et al. (2020). Dissecting the genetic architecture of Aphanomyces root rot resistance in lentil by QTL mapping and genome-wide association study. Int. J. Mol. Sci. 21, 1–24. doi: 10.3390/ijms21062129
Mao, W., Carroll, R. B., and Whittington, D. P. (1998). Association of Phoma terrestris, Pythium irregulare, and Fusarium acuminatum in causing red root rot of corn. Plant Dis. 82, 337–342. doi: 10.1094/PDIS.1998.82.3.337
Marks, G. C., and Mitchell, J. E. (1970). Detection, isolation and pathogenicity of Phytophthora megasperma from soils and estimation of inoculum levels. Phytopathology 60, 1687–1690. doi: 10.1094/Phyto-60-1687
Marquez, N., Giachero, M. L., Declerck, S., and Ducasse, D. A. (2021). Macrophomina phaseolina: general characteristics of pathogenicity and methods of control. Front. Plant Sci. 12, 1–16. doi: 10.3389/fpls.2021.634397
Mårtensson, A. M. (1992). Effects of agrochemicals and heavy metals on fast-growing rhizobia and their symbiosis with small-seeded legumes. Soil Biol. Biochem. 24, 435–445. doi: 10.1016/0038-0717(92)90206-D
Mathew, F. M., Lamppa, R. S., Chittem, K., Chang, Y. W., Botschner, M., Kinzer, K., et al. (2011). Characterization and Pathogenicity of Rhizoctonia solani Isolates Affecting Pisum sativum in North Dakota. Plant Dis. 96, 666–672. doi: 10.1094/PDIS-02-11-0087
McKenzie, J. S., and Davidson, J. (1975). Prevalence of alfalfa crown and root diseases in the Peace River Region of Alberta and British Columbia. Can. Plant Dis. Surv. 55, 121–125.
Miedaner, T., Bolduan, C., and Melchinger, A. E. (2010). Aggressiveness and mycotoxin production of eight isolates each of Fusarium graminearum and Fusarium verticillioides for ear rot on susceptible and resistant early maize inbred lines. Eur. J. Plant Pathol. 127, 113–123. doi: 10.1007/s10658-009-9576-2
Miller-Garvin, J., and Viands, D. (1994). Selection for resistance to Fusarium root rot, and associations among resistances to six diseases in alfalfa. Crop Sci. 34, 1461–1465. doi: 10.2135/cropsci1994.0011183X003400060008x
Min, L. I., Yang, B., Hou, J., and Zhang, Z. (2002). Toxicity measurement of carbendazim on Fusarium spp. Alfalfa. J Acta Agrestia Sin. 10, 271–273.
Min-Quan, L. I., Chai, Z. X., Jin-Hua, L. I., Dong, Y. X., Zhang, L. P., and Zhang, Z. H. (2003). Identification pathogens of crown and root rot of Alfalfa in Dingxi region. Acta Agrestia Sin. 11, 83–86.
Misawa, T., Kurose, D., Shishido, K., Toda, T., and Kuninaga, S. (2020). Characterization of a new subgroup of Rhizoctonia solani anastomosis group 3 (AG-3 TM) associated with tomato leaf blight. J. Gen. Plant Pathol. 86, 457–467. doi: 10.1007/s10327-020-00943-1
Moore, R. T. (1987). The genera of Rhizoctonia-like fungi: Ascorhizoctonia, Ceratorhiza gen. nov., Epulorhiza gen. nov., Moniliopsis, and Rhizoctonia. Mycotaxon 29, 91–99.
Morrison, J. (2004). Transhumant grazing systems in temperate Asia. Plant production and protection series No. 31. Edited by JM Suttie and SG Reynolds. Rome: Food and Agriculture Organization of The United Nations (2003), pp. 331, no price quoted. ISBN 92-5-104977-7. Exp. Agric. 40, 495–495.
Moslemi, A., Ades, P., Crous, P., Groom, T., Scott, J., Nicolas, M., et al. (2018). Paraphoma chlamydocopiosa sp. nov. and Paraphoma pye sp. nov., two new species associated with leaf and crown infection of pyrethrum. Plant Pathol. 67, 124–135. doi: 10.1111/ppa.12719
Nagendran, S., Hammerschmidt, R., and McGrath, J. M. (2009). Identification of sugar beet germplasm EL51 as a source of resistance to post-emergence Rhizoctonia damping-off. Eur. J. Plant Pathol. 123, 461–471. doi: 10.1007/s10658-008-9384-0
Nawrot, R., Barylski, J., Nowicki, G., Broniarczyk, J., Buchwald, W., and Goździcka-Józefiak, A. (2014). Plant antimicrobial peptides. Folia Microbiol. 59, 181–196. doi: 10.1007/s12223-013-0280-4
Neate, S. M. (1989). A comparison of controlled environment and field trials for detection of resistance in cereal cultivars to root rot caused by Rhizoctonia solani. Plant Pathol. 38, 494–501. doi: 10.1111/j.1365-3059.1989.tb01442.x
Nedelnik, J., and Repkova, J. (1997). Phytotoxicity of some Fusarium toxins to red clover and alfalfa. Cereal Res. Commun. 25, 507–509. doi: 10.1007/BF03543767
Nicot, P. C. (2011). Classical and augmentative biological control against diseases and pests: critical status analysis and review of factors. IOBC/WPRS Bull.
Nikraftar, F., Taheri, P., Rastegar, M. F., and Tarighi, S. (2013). Tomato partial resistance to Rhizoctonia solani involves antioxidative defense mechanisms. Physiol. Mol. Plant Pathol. 81, 74–83. doi: 10.1016/j.pmpp.2012.11.004
Ogoshi, A. (1987). Ecology and pathogenicity of anastomosis and intraspecific groups of Rhizoctonia solani Kuhn. Annu. Rev. Phytopathol. 25, 125–143. doi: 10.1146/annurev.py.25.090187.001013
Okubara, P. A., Leston, N., Micknass, U., Kogel, K.-H., and Imani, J. (2016). Rapid quantitative assessment of Rhizoctonia resistance in roots of selected wheat and barley genotypes. Plant Dis. 100, 640–644. doi: 10.1094/PDIS-05-15-0611-SR
Oladzad, A., Zitnick-Anderson, K., Jain, S., Simons, K., Osorno, J. M., McClean, P. E., et al. (2019). Genotypes and genomic regions associated with Rhizoctonia solani resistance in common bean. Front. Plant Sci. 10:956. doi: 10.3389/fpls.2019.00956
Pan, L., Zhang, L., Yang, C., Yuan, Q., Wang, Y., and Miao, L. (2015). Identification and biological characteristics of Fusarium sporotrichioide isolated from Medicago sativa root. Acta Prataculturae Sin. 24, 88–98. doi: 10.11686/cyxb2014466
Park, J., Park, B., Veeraraghavan, N., Jung, K., Lee, Y.-H., Blair, J. E., et al. (2008). Phytophthora database: a forensic database supporting the identification and monitoring of Phytophthora. Plant Dis. 92, 966–972. doi: 10.1094/PDIS-92-6-0966
Pawlowski, M. L., Bowen, C. R., Hill, C. B., and Hartman, G. L. (2016). Responses of soybean genotypes to pathogen infection after the application of elicitors. Crop Prot. 87, 78–84. doi: 10.1016/j.cropro.2016.04.022
Pegg, G. F., and Parry, D. W. (1983). Infection of lucerne (Medicago sativa) by Fusarium species. Ann. Appl. Biol. 103, 45–55. doi: 10.1111/j.1744-7348.1983.tb02739.x
Perez, C. E., Cabral, G. B., and Aragao, F. J. (2021). Host-induced gene silencing for engineering resistance to Fusarium in soybean. Plant Pathol. 70, 417–425. doi: 10.1111/ppa.13299
Pfender, W. F., Hine, R. B., and Stanghellini, M. E. (1976). Production of sporangia and release of zoospores by Phytophthora megasperma in soil. Ecol. Epidemiol. 67, 657–663.
Porter, L. D., Kraft, J. M., and Grünwald, N. J. (2014). Release of Pea Germplasm with Fusarium Resistance Combined with Desirable Yield and Anti-Lodging Traits. J. Plant Regist. 8, 191–194. doi: 10.3198/jpr2013.07.0041crg
Punja, Z. K. (2005). Transgenic carrots expressing a thaumatin-like protein display enhanced resistance to several fungal pathogens. Can. J. Plant Pathol. 27, 291–296. doi: 10.1080/07060660509507227
Qingbin, W., and Yang, Z. O. U. (2020). China’s alfalfa market and imports: development, trends, and potential impacts of the US–China trade dispute and retaliations. J. Integr. Agric. 19, 1149–1158. doi: 10.1016/S2095-3119(19)62832-7
Quan, L. M. (2003). Comparative pathogenicity of isolates of Fusarium spp and cultivars resistance in alfalfa. J. Grassland China 25, 1–5.
Quan, L. M., Zhang, Z. H., Fang, M. J., and Cai, Z. X. (2003). Relationship between POD isozymes and resistance to crown and root rot of alfalfa. J. Grassland China 25, 37–41.
Rahman, M. T., Bhuiyan, M. K. A., Karim, M. A., and Rubayet, M. T. (2018). Screening of soybean resistance genotypes against Fusarium oxysporum, Macrophomina phaseolina, Rhizoctonia solani and Sclerotium rolfsii. Adv. Res. Agric. Vet. Sci. 2, 139–156.
Ramalingam, J., Raveendra, C., Savitha, P., Vidya, V., Chaithra, T. L., Velprabakaran, S., et al. (2020). Gene pyramiding for achieving enhanced resistance to bacterial blight, blast, and sheath blight diseases in rice. Front. Plant Sci. 11:1662. doi: 10.3389/fpls.2020.591457
Rather, I. A., Awasthi, P., Mahajan, V., Bedi, Y. S., Vishwakarma, R. A., and Gandhi, S. G. (2015). Molecular cloning and functional characterization of an antifungal PR-5 protein from Ocimum basilicum. Gene 558, 143–151. doi: 10.1016/j.gene.2014.12.055
Raupach, G. S., and Kloepper, J. W. (1998). Mixtures of plant growth-promoting rhizobacteria enhance biological control of multiple cucumber pathogens. Phytopathology 88, 1158–1164. doi: 10.1094/PHYTO.1998.88.11.1158
Richard, C., and Martin, J. G. (1991). Phytophthora root rot of lucerne in Quebec: geographical distribution of symptoms and study of edaphic factors favouring their expression. Phytoprotection 72, 87–95.
Richard, C., Michaud, R., Frève, A., and Gagnon, C. (1980). Selection for root and crown rot resistance in alfalfa. Crop Sci. 20, 691–695. doi: 10.2135/cropsci1980.0011183X002000060004x
Richard, C., Willemot, C., Michaud, R., Bernier-Cardou, M., and Gagnon, C. (1982). Low-temperature interactions in Fusarium wilt and root rot of alfalfa. Phytopathology 72, 293–297.
Rogers, V. E. (1981). Comparison of lucernes when grown on a heavy clay soil infected by Phytophthora, and their response to attack by the spotted alfalfa aphid (Therioaphis trifolii f. maculata). Aust. J. Exp. Agric. 21, 63–67. doi: 10.1071/EA9810063
Rush, C. M., Carling, D. E., Harveson, R. M., and Mathieson, J. T. (1994). Prevalence and pathogenicity of anastomosis groups of Rhizoctonia solani from wheat and sugar beet in Texas. Plant Dis. 78, 349–352. doi: 10.1094/PD-78-0349
Sakupwanya, M. N., Labuschagne, N., Loots, T., and Apostolides, Z. (2018). Towards developing a metabolic-marker based predictive model for Phytophthora nicotianae tolerance in citrus rootstocks. J. Plant Pathol. 100, 269–277. doi: 10.1007/s42161-018-0080-4
Salter, R., Miller-Garvin, J. E., and Viands, D. R. (1994). Breeding for resistance to alfalfa root rot caused by Fusarium species. Crop Sci. 34, 1213–1217. doi: 10.2135/cropsci1994.0011183X003400050012x
Samac, D. A., Lamb, J. F., Kinkel, L. L., and Hanson, L. (2013). Effect of wheel traffic and green manure treatments on forage yield and crown rot in alfalfa (Medicago sativa). Plant Soil 372, 349–359. doi: 10.1007/s11104-013-1746-5
Sharath, C., Tarafdar, A., Mahesha, H., and Sharma, M. (2021). Temperature and soil moisture stress modulates the host defense response in chickpea during dry root rot Incidence. Front. Plant Sci. 12:932. doi: 10.3389/fpls.2021.653265
Sharon, M., Freeman, S., Kuninaga, S., and Sneh, B. (2007). Genetic diversity, anastomosis groups and virulence of Rhizoctonia spp. from strawberry. Eur. J. Plant Pathol. 117, 247–265. doi: 10.1007/s10658-006-9091-7
Shi, S., Nan, L., and Smith, K. F. (2017). The current status, problems, and prospects of alfalfa (Medicago sativa L.) breeding in China. Agronomy 7:1. doi: 10.3390/agronomy7010001
Shouyan, D., Minquan, L., and Hui, Z. (2011). Identification of disease resistance of different varieties of alfalfa to Fusarium spp. Plant Prot. 37, 110–114.
Sivasithamparam, K. (1993). Ecology of root infecting pathogenic fungi in mediterranean environments. Adv. Plant Pathol. 10, 245–280.
Smiley, R. W., and Yan, H. (2009). Variability of Fusarium crown rot tolerances among cultivars of spring and winter wheat. Plant Dis. 93, 954–961. doi: 10.1094/PDIS-93-9-0954
Sneh, B., Jabaji-Hare, S., Neate, S., and Dijst, G. (2013). Rhizoctonia Species: Taxonomy, Molecular Biology, Ecology, Pathology and Disease Control. eds. S. Baruch, J.-H. Suha, N. Stephen, and D. Gerda. Berlin/Heidelberg, Germany: Springer Science & Business Media.
Snyder, C., Hans, H., and sambucinum Fuckel, F. (1991). Fusarium species associated with crown rot of alfalfa in Nevada. Plant Dis. 75:51.
Song, L., Jiang, L., Chen, Y., Shu, Y., Bai, Y., and Guo, C. (2016). Deep-sequencing transcriptome analysis of field-grown Medicago sativa L. crown buds acclimated to freezing stress. Funct. Integr. Genom. 16, 495–511. doi: 10.1007/s10142-016-0500-5
Spedaletti, Y., Aparicio, M., Mercado Cárdenas, G., Rodriguero, M., Taboada, G., Aban, C., et al. (2016). Genetic characterization and pathogenicity of Rhizoctonia solani associated with common bean web blight in the main bean growing area of Argentina. J. Phytopathol. 164, 1054–1063. doi: 10.1111/jph.12526
Stalpers, J. A., and Andersen, T. F. (1996). “A synopsis of the taxonomy of teleomorphs connected with Rhizoctonia S.L,” in Rhizoctonia Species: Taxonomy, Molecular Biology, Ecology, Pathology and Disease Control. eds. B. Sneh, S. Jabaji-Hare, S. Neate, and G. Dijst (Dordrecht: Springer Netherlands), 49–63.
Stanton-Geddes, J., Paape, T., Epstein, B., Briskine, R., Yoder, J., Mudge, J., et al. (2013). Candidate genes and genetic architecture of symbiotic and agronomic traits revealed by whole-genome, sequence-based association genetics in Medicago truncatula. PLoS One 8:e65688. doi: 10.1371/journal.pone.0065688
Steinmetz, O. J., Huset, D. E., Rouse, D. I., Raasch, J. A., Gutiérrez, L., and Riday, H. (2020). Synthetic cultivar parent number impacts on genetic drift and disease resistance in alfalfa. J. Agron. Crop Sci. 60, 2304–2316. doi: 10.1002/csc2.20219
Stone, A., Vallad, G., Cooperband, L., Rotenberg, D., Darby, H., James, R., et al. (2003). Effect of organic amendments on soilborne and foliar diseases in field-grown snap bean and cucumber. Plant Dis. 87, 1037–1042. doi: 10.1094/PDIS.2003.87.9.1037
Su, Y.-Z., Zhao, H.-L., Zhang, T.-H., and Zhao, X.-Y. (2004). Soil properties following cultivation and non-grazing of a semi-arid sandy grassland in northern China. Soil Tillage Res. 75, 27–36. doi: 10.1016/S0167-1987(03)00157-0
Summerell, B. A., Laurence, M. H., Liew, E. C., and Leslie, J. F. (2010). Biogeography and phylogeography of Fusarium: a review. Fungal Divers. 44, 3–13. doi: 10.1007/s13225-010-0060-2
Summerell, B. A., Leslie, J. F., Liew, E. C. Y., Laurence, M. H., Bullock, S., Petrovic, T., et al. (2011). Fusarium species associated with plants in Australia. Fungal Divers. 46, 1–27. doi: 10.1007/s13225-010-0075-8
Summers, C. G. (1998). Integrated pest management in forage alfalfa. J. Integr. Pest Manag. 3, 127–154.
Sumner, D. (1994). “Cultural management,” in Epidemiology and Management of Root Diseases. eds. C. Lee Campbell and D. Michael Benson (Berlin: Springer), 309–333.
Sunpapao, A., and Pornsuriya, C. (2016). Overexpression of β-1, 3-glucanase gene in response to Phytophthora palmivora infection in leaves of Hevea brasiliensis clones. Walailak J. Sci. Technol. 13, 35–43. doi: 10.14456/WJST.2016.4
Tabares-da Rosa, S., Signorelli, S., Del Papa, M., Sabatini, O., Reyno, R., Lattanzi, F., et al. (2019). Rhizobia inoculants for alfalfa in acid soils: a proposal for Uruguay. Agrociencia Uruguay 23, 4–16. doi: 10.31285/agro.23.120
Taguchi, K., Okazaki, K., Takahashi, H., Kubo, T., and Mikami, T. (2010). Molecular mapping of a gene conferring resistance to Aphanomyces root rot (black root) in sugar beet (Beta vulgaris L.). Euphytica 173, 409–418. doi: 10.1007/s10681-010-0153-8
Tang, B., Li, X., Pu, L., Zhao, Q., Cui, X., Ge, F., et al. (2019). A pathogenesis-related protein 10 gene PnPR10–3 was involved in molecular interaction between Panax notoginseng and Fusarium solani. Australas. Plant Pathol. 48, 447–456. doi: 10.1007/s13313-019-00644-0
Van West, P., Appiah, A. A., and Gow, N. A. (2003). Advances in research on oomycete root pathogens. Physiol. Mol. Plant Pathol. 62, 99–113. doi: 10.1016/S0885-5765(03)00044-4
Vandemark, G. J., and Barker, B. M. (2010). Quantifying Phytophthora medicaginis in Susceptible and Resistant Alfalfa with a Real-Time Fluorescent PCR Assay. J. Phytopathol. 151, 577–583. doi: 10.1046/j.0931-1785.2003.00768.x
Venancio, W. S., Rodrigues, M. A. T., Begliomini, E., and de Souza, N. L. (2003). Physiological effects of strobilurin fungicides on plants. Publicatio UEPG: Ciências Exatas e da Terra, Agrárias e Engenharias 9. doi: 10.5212/publicatio.v9i03.814
Verma, P. R. (1996). Biology and control of Rhizoctonia solani on rapeseed: a review. Phytoprotection 77, 99–111.
Vilgalys, R., and Cubeta, M. A. (1994). Molecular systematics and population biology of Rhizoctonia. Annu. Rev. Phytopathol. 32, 135–155. doi: 10.1146/annurev.py.32.090194.001031
Vincelli, P. C., and Herr, L. J. (1992). Two diseases of alfalfa caused by Rhizoctonia solani AG-1 and AG-4. Plant Dis. 76:1283B. doi: 10.1094/PD-76-1283B
Wang, X., Ding, T., Li, Y., Guo, Y., Li, Y., and Duan, T. (2020a). Dual inoculation of alfalfa (Medicago sativa L.) with Funnelliformis mosseae and Sinorhizobium medicae can reduce Fusarium wilt. J. Appl. Microbiol. 129, 665–679. doi: 10.1111/jam.14645
Wang, Q., Ma, Y., Yang, H., and Chang, Z. (2014). Effect of biofumigation and chemical fumigation on soil microbial community structure and control of pepper Phytophthora blight. World J. Microbiol. Biotechnol. 30, 507–518. doi: 10.1007/s11274-013-1462-6
Wang, D., Meng, Y., Li, W., and Li, W. (2005). Isolation and identification of pathogens causing root rot in lucerne. J. Pratacultural Sci. 22, 78–81.
Wang, Z., Tian, H., Zhang, C., Fang, X., and Nan, Z. (2020b). Occurrence of Macrophomina phaseolina causing root and crown rot on Alfalfa (Medicago sativa) in China. Plant Dis. 104:2521. doi: 10.1094/PDIS-03-20-0555-PDN
Wang, X., Wang, C., Zhou, G., Huang, C., Xu, L., and Wang, D. (1996). Investigation and analysis on Lucerne diseases from altay newly-established areas in northern Xinjiang. J. Xinjiang Agric. University 19, 40–43.
Wang, L., Xie, J., Luo, Z., Niu, Y., Coulter, J. A., Zhang, R., et al. (2021). Forage yield, water use efficiency, and soil fertility response to alfalfa growing age in the semiarid Loess Plateau of China. Agric. Water Manag. 243:106415. doi: 10.1016/j.agwat.2020.106415
Wang, L., Xu, L., He, Y., and Wu, L. (2010). Separation and screening of biocontrol actinomyces against alfalfa root rot. J. Plant Dis. Pests 1, 10–37.
Wang, Y., Yuan, Q. H., Miao, L. H., Zhang, L., and Pan, L. Q. (2016). The major types and epidemic trends of alfalfa diseases in Northeast and North China. Acta Prataculturae Sin. 25, 52–59. doi: 10.11686/cyxb2015260
Wang, S.-R., Zhang, L., and Yuan, Q.-H. (2015a). Identification of the pathogen of the alfalfa Phoma leaf spot and chemical control. Proc. Annu. Meeting Chin. Soc. Plant Pathol. Haikou, Hainan, China.
Wei, R., Guo, Q. Y., Bai, J. Y., and Zhang, S. H., University, X.A (2016). Molecular detection of various pathogens of alfalfa root rot. J. Xinjiang Agric. Sci. 53, 1268–1275.
Wei, X., Xu, Z., Zhang, N., Yang, W., Liu, D., and Ma, L. (2020). Synergistic action of commercially available fungicides for protecting wheat from common root rot caused by Bipolaris sorokiniana in China. Plant Dis. 105, 667–674. doi: 10.1094/PDIS-03-20-0627-RE
Wen, Z., Duan, T., Christensen, M. J., and Nan, Z. (2015a). Bacillus subtilis subsp. spizizenii MB29 controls alfalfa root rot caused by Fusarium semitectum. Biocontrol Sci. Technol. 25, 898–910. doi: 10.1080/09583157.2015.1020759
Wen, Z., Duan, T., Christensen, M. J., and Nan, Z. (2015b). Microdochium tabacinum, confirmed as a pathogen of alfalfa in Gansu Province, China. Plant Dis. 99, 87–92. doi: 10.1094/PDIS-10-13-1048-RE
White, J. F., Kingsley, K. L., Zhang, Q., Verma, R., Obi, N., Dvinskikh, S., et al. (2019). Review: endophytic microbes and their potential applications in crop management. Pest Manag. Sci. 75, 2558–2565. doi: 10.1002/ps.5527
Wiggins, B. E., and Kinkel, L. L. (2005). Green manures and crop sequences influence alfalfa root rot and pathogen inhibitory activity among soil-borne Streptomycetes. Plant Soil 268, 271–283. doi: 10.1007/s11104-004-0300-x
Williamson-Benavides, B. A., and Dhingra, A. (2021). Understanding root rot disease in agricultural crops. Horticulturae 7:33. doi: 10.3390/horticulturae7020033
Wong, D. H., Barbetti, M. J., and Sivasithamparam, K. (1984). Effects of soil temperature and moisture on the pathogenicity of fungi associated with root rot of subterranean clover. Aust. J. Agric. Res. 35, 675–684. doi: 10.1071/AR9840675
Woodhall, J. W., Wharton, P. S., and Peters, J. C. (2012). First Report of Rhizoctonia solani AG4 HG-II Infecting Potato Stems in Idaho. Plant Dis. 96:1701. doi: 10.1094/PDIS-06-12-0568-PDN
Xiao, K., Kinkel, L. L., and Samac, D. A. (2002). Biological Control of Phytophthora Root Rots on Alfalfa and Soybean with Streptomyces. Biol. Control 23, 285–295. doi: 10.1006/bcon.2001.1015
Xu, Y., Liu, J., Liu, X., Li, H., Yang, Z., Wang, H., et al. (2021). Continuous cropping of alfalfa (Medicago sativa L.) reduces bacterial diversity and simplifies cooccurrence networks in aeolian sandy soil. Soil Ecol. Lett. 4, 131–143. doi: 10.1007/s42832-021-0083-5
Yang, X., Tyler, B. M., and Hong, C. (2017). An expanded phylogeny for the genus Phytophthora. JIMA Fungus 8, 355–384. doi: 10.5598/imafungus.2017.08.02.09
Yao, C. (1989). The preliminary study on root rot diseases of alfalfa in xingjiang uygur autonomous region. J. Grassland China 2.
Yinghua, L., Chenghao, J., Meihua, X., and Xiangping, L. (2019). Research advances of root rot in Medicago sativa L. J. Plant Dis. Pests 10, 15–27. doi: 10.19579/j.cnki.plant-d.p.2019.01.004
You, M. P., Barbett, M. J., and Nichols, P. (2005). New sources of resistance identified in Trifolium subterraneum breeding lines and cultivars to root rot caused by Fusarium avenaceum and Pythium irregulare and their relationship to seedling survival. Australas. Plant Pathol. 34, 237–244. doi: 10.1071/AP04092
Yuan, Q. H., Zhang, W. S., Zhou, W. X., and Bai, S. (2003). Preliminary study on Scleroitinia trifoliorum in alfalfa. J. Plant Protect. Beijing 29, 22–24.
Zaccardelli, M., Balmas, V., Altomare, C., Corazza, L., and Scotti, C. (2006). Characterization of Italian isolates of Fusarium semitectum from alfalfa (Medicago sativa L.) by AFLP analysis, morphology, pathogenicity and toxin production. J. Phytopathol. 154, 454–460. doi: 10.1111/j.1439-0434.2006.01128.x
Zeng, L., Chai, J. K., Zhao, G. Q., Chun-Jie, L. I., Liu, W. H., and Yao, T. (2019). Screening, Identification and Control Effect of Biocontrol Fungi Against Alfalfa Root Rot. Acta Agrestia Sin. 27, 1–7. doi: 10.11733/j.issn.1007-0435.2019.04.005
Zhang, C., Wang, Z., Fang, X., and Nan, Z. (2020). Occurrence of Root Rot on Alfalfa (Medicago sativa) Caused by Pythium coloratum in China. Plant Dis. 104:3269. doi: 10.1094/PDIS-06-20-1211-PDN
Zhang, J. X., Xue, A. G., Zhang, H. J., Nagasawa, A. E., and Tambong, J. T. (2010). Response of soybean cultivars to root rot caused by Fusarium species. Can. J. Plant Sci. 90, 767–776. doi: 10.4141/CJPS09133
Zhang, C., Yu, S., Tian, H., Wang, Z., Yu, B., Ma, L., et al. (2021). Varieties with a high level of resistance provide an opportunity to manage root rot caused by Rhizoctonia solani in alfalfa. Eur. J. Plant Pathol. 160, 983–989. doi: 10.1007/s10658-021-02287-8
Zhao, Y. Q., Shi, K., Yu, X., and Zhang, L. J. (2021). First report of alfalfa root rot caused by Plectosphaerella cucumerina in inner Mongolia autonomous region of China. Plant Dis 105, 1–4. doi: 10.1094/PDIS-03-21-0515-PDN
Zhimin, D. (1996). Studies on the species and pathogenicity of root disease of peas in Qinghai province. J. Microbiol. 1, 31–34.
Zhou, Q. X., Chang, K. F., Hwang, S. F., Strelkov, S. E., and Chen, Y. Y. (2009). Pathogenicity and genetic diversity of Rhizoctonia solani isolates from lupin and other crops in Alberta, Canada. Can. J. Plant Pathol. 31, 340–347. doi: 10.1080/07060660909507608
Keywords: alfalfa, root rot, China, distribution, climatic factors, management
Citation: Abbas A, Mubeen M, Sohail MA, Solanki MK, Hussain B, Nosheen S, Kashyap BK, Zhou L and Fang X (2022) Root rot a silent alfalfa killer in China: Distribution, fungal, and oomycete pathogens, impact of climatic factors and its management. Front. Microbiol. 13:961794. doi: 10.3389/fmicb.2022.961794
Edited by:
Shekhar Jain, Mandsaur University, IndiaReviewed by:
Aradhana Mishra, National Botanical Research Institute (CSIR), IndiaShah Fahad, The University of Haripur, Pakistan
Niu Yanbing, Shanxi Agricultural University, China
Copyright © 2022 Abbas, Mubeen, Sohail, Solanki, Hussain, Nosheen, Kashyap, Zhou and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangling Fang, xlf@lzu.edu.cn; Lei Zhou, zhoul@zaas.ac.cn
 Aqleem Abbas
Aqleem Abbas Mustansar Mubeen
Mustansar Mubeen Muhammad Aamir Sohail
Muhammad Aamir Sohail Manoj Kumar Solanki
Manoj Kumar Solanki Babar Hussain
Babar Hussain Shaista Nosheen7
Shaista Nosheen7 Brijendra Kumar Kashyap
Brijendra Kumar Kashyap Lei Zhou
Lei Zhou Xiangling Fang
Xiangling Fang