- 1Jiangsu Key Laboratory of Zoonosis, Jiangsu Co-Innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou University, Yangzhou, China
- 2Key Laboratory of Prevention and Control of Biological Hazard Factors (Animal Origin) for Agrifood Safety and Quality, Ministry of Agriculture of China, Yangzhou, China
- 3Joint International Research Laboratory of Agriculture and Agri-Product Safety, Ministry of Education of China, Yangzhou, China
Although campylobacteriosis is a zoonotic foodborne illness, high-risk isolates from animal sources are rarely characterized, and the pathogenic potential of zoonotic strains remains an obstacle to effective intervention against human infection. HS19 has been acknowledged as a maker serotype represented by Campylobacter jejuni (C. jejuni) isolates from patients with post-infection Guillain-Barré syndrome (GBS), which is circulation in developed countries. However, a previous serotype epidemiological study of C. jejuni isolates in an animal population revealed that HS19 was also prevalent in isolates from cattle in China. In this study, to investigate the hazardous potential of zoonotic strains, 14 HS19 isolates from cattle were systematically characterized both by genotype and phenotype. The results showed that all of these cattle isolates belonged to the ST-22 complex, a high-risk lineage represented by 77.2% HS19 clinical isolates from patients worldwide in the PubMLST database, indicating that the ST-22 complex is the prominent clonal complex of HS19 isolates, as well as the possibility of clonal spread of HS19 isolates across different regions and hosts. Nevertheless, these cattle strains clustered closely with the HS19 isolates from patients, suggesting a remarkable phylogenetic relatedness and genomic similarity. Importantly, both tetracycline genes tet(O) and gyrA (T86I) reached a higher proportional representation among the cattle isolates than among the human clinical isolates. A worrying level of multidrug resistance (MDR) was observed in all the cattle isolates, and two MDR profiles of the cattle isolates also existed in human clinical isolates. Notably, although shared with the same serotype HS19 and sequence type ST-22, 35.7% of cattle isolates induced severe gastrointestinal pathology in the IL-10–/– C57BL/6 mice model, indicating that some bacteria could change due to host adaptation to induce a disease epidemic, thus the associated genetic elements deserve further investigation. In this study, HS19 isolates from cattle were first characterized by a systematic evaluation of bacterial genomics and in vitro virulence, which improved our understanding of the potential zoonotic hazard from food animal isolates with high-risk serotypes, and provided critical information for the development of targeted C. jejuni mitigation strategies.
Introduction
Campylobacter jejuni is the leading cause of bacterial foodborne gastroenteritis in humans, both in developed and developing countries (Ruiz-Palacios, 2007; Kaakoush et al., 2015), making it a great threat to the public’s health. While the majority of cases are self-limiting, they can spread into the bloodstream in immunocompromised individuals and become potentially lethal (Whitehouse et al., 2018). In some instances, affected patients are at risk of Guillain-Barré syndrome (GBS), a severe post-infectious autoimmune disease that occurs weeks or months after the initial infectious gastrointestinal manifestation (Pithadia and Kakadia, 2010), which can also sometimes be life-threatening (Nachamkin, 2002). The high incidence of C. jejuni-associated disease in humans is largely due to its prevalence as a zoonotic agent in animals (Sheppard et al., 2011; Burnham and Hendrixson, 2018). As a part of the commensal microbiota of numerous host species, fecal contamination from carrier animals is considered to be the primary source of C. jejuni (Koenraad et al., 1997; Guirado et al., 2020). Chicken is a common source of C. jejuni in sporadic infection, and the role of cattle is also notable (Hakkinen et al., 2009). Significant associations emerged between certain clonal complexes from human infection and the contact with cattle, the consumption of unpasteurized milk and raw minced meat, raising the question about the pathogenic potential of cattle isolates (Kärenlampi et al., 2007; Wilson et al., 2008; Costard et al., 2017; Hsu et al., 2020). In fact, not all strains or genetic lineages pose equal risks to human health. Although campylobacteriosis is a zoonotic foodborne disease, the majority of the reported high-risk strains were from clinical patients, and isolates from animal sources have rarely been characterized.
The identification and profiling of C. jejuni virulence determinants are crucial for the risk assessment of campylobacteriosis infection (Fiedoruk et al., 2019), whereas gaining insight into the distribution of virulence-associated genes among strains might shed some light on the mechanisms exploited by Campylobacter to trigger infection (Iglesias-Torrens et al., 2018). Notably, C. jejuni does not possess numerous classical virulence factors. Cytolethal distending enterotoxin (Cdt) is the only virulence determinant located on the C. jejuni chromosome, however, its role in pathogenesis is still unclear (Burnham and Hendrixson, 2018). Moreover, the self-limiting feature of most campylobacteriosis cases, as well as the lack of a traceable animal model, hinder the hazard evaluation of Campylobacter species (Crofts et al., 2018). Multiple bacterial factors have been implicated in the pathogenesis of campylobacteriosis supporting Campylobacter to invade the host and evade the host’s defenses (de Oliveira et al., 2019). Capsular polysaccharide (CPS) is the most common virulence determinant, which is the basis of the classical Penner serotyping scheme. Notably, particular serotypes may contribute to disease susceptibility (Heikema et al., 2015), and HS19 has been reported to be over-represented in GBS outbreaks (Kuroki et al., 1993; Nachamkin et al., 1998), indicating that the unique feature of the HS19 isolate might play a causative role in GBS induction. Moreover, lipooligosaccharide (LOS) is another important virulence determinant, and molecular mimicry between the structure of C. jejuni LOS and human gangliosides is thought to be related to the development of GBS in patients previously infected with this pathogen (Perera et al., 2007). Isolates belonging to the LOS classes A, B, or C harbor genes (such as cst-II and wlaN) that enable the incorporation of sialic acid into LOS (Nachamkin, 1997; Parker et al., 2005; Müller et al., 2007). In addition to their association with GBS, strains harboring sialylated LOS are also thought to be related to an increased severity of gastroenteritis (Poly et al., 2011).
In addition to virulence determinants, understanding the status of C. jejuni drug resistance is also essential for isolate hazard characterization, which could be critical in the instruction of antibiotics clinically, as well as to implement efficient control measures to reduce human exposure to the pathogen. Antibiotic treatment is indispensable if severe or immunocompromised cases occur, with macrolides and fluoroquinolones (FQs) being the first choice of drugs (Mourkas et al., 2019). However, as a naturally competent organism, C. jejuni is capable of incorporating exogenous DNA to adapt to antibiotic selective pressure and is spread by the food chain and water (Kashoma et al., 2016). Over the years, increasing rates of Campylobacter strains that are resistant to these two antibiotics. The World Health Organization listed fluoroquinolone-resistant Campylobacter spp. as one of the six high-priority pathogens for research and development of new antibiotics in 2017 (Tacconelli et al., 2018). Gentamicin (GEN) and tetracycline (TET) have been reported as alternative therapies (Koolman et al., 2015). Nevertheless, these resistant strains have also been found in multiple types of food animal facilities. As a result, C. jejuni is increasingly viewed as a reservoir of antibiotic resistance genes in both human medicine and the food supply chain (Mourkas et al., 2019; Hsu et al., 2020), making antimicrobial resistance (AMR) a public health concern. It could be especially dangerous for people with compromised immunity, since drug resistance greatly limits the available therapeutic effects.
Tracking high-risk animal strains will lead to a better understanding of their distribution in the food chain and provides critical information for the development of targeted mitigation strategies to reduce human exposure (Baker et al., 2012; Buchanan et al., 2017). Because campylobacteriosis is often associated with contaminated food products and exposure to animals, whole-genome sequencing (WGS) might become the preferred typing method, and analysis of isolates from various sources should be a major component of studying disease ecology and epidemiology (Park et al., 2020). At present, WGS has been considered the most informative and discriminative typing method to examine the genomic characteristics of Campylobacter isolates with high resolution (Davies et al., 2020), allowing for comprehensive phylogenetic analyses of numerous traits associated with virulence or antibiotic resistance (Fiedoruk et al., 2019). Our previous epidemiological research revealed that the GBS maker serotype HS19 was significantly more prevalent in isolates from cattle than other animal populations circulating in China. Thus, the goal of the current study is to investigate the pathogenic potential of HS19 isolates from cattle. WGS was used for isolate characterization, and the phylogenetic relatedness between HS19 isolates from cattle in China and GBS patients worldwide was analyzed. Additionally, antimicrobial resistance profiles and virulence genes were identified in silico, while antimicrobial susceptibility and in vivo C. jejuni infection assays were conducted to determine the pathogenic phenotypes.
Materials and Methods
HS19 C. jejuni Isolates
A collection of 14 cattle isolates, characterized as serotype HS19, were involved in this study. These isolates were identified from fecal samples of cattle populations circulating in Jiangsu province and Liaoning provinces, in eastern China, between 2005 and 2019. Cattle is a common source of animal protein in this geographical area, and cattle isolates were sampled from three large-scale cattle farms, which were selected as the suppliers for cattle slaughterhouses. The sampling procedure was approved by the Research Ethics Committee of Yangzhou University. Additionally, five control strains with HS19 from other sources (diarrhea patients, n = 2; pet, n = 2; chick, n = 1) but were also isolated from Jiangsu province were chosen for phylogenetic analysis, and the background information of these isolates was shown in Supplementary Table 1.
As previously described (Zang et al., 2016), C. jejuni strains were routinely cultured on Campylobacter selective agar base plates (modified CCDA, Preston; Oxoid, United Kingdom) under microaerophilic conditions (5% O2, 10% CO2, and 85% N2) at 42°C for 48 h. The isolates were identified at the C. jejuni species level by PCR, and then stored at –80°C in 15% glycerol in brain heart infusion broth until use. A capsule genotyping scheme was exploited for isolates serotyping (Poly et al., 2011).
Whole-Genome Sequencing
Genomic DNA of cattle isolates was prepared using the TIANamp Bacterial DNA Kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. DNA was then fragmented to prepare the library and was sequenced using Illumina NovaSeq 6,000 (Illumina, United States) in the Novogene Institution (Tianjing, China). Reads were assembled into contigs and scaffolds using SOAPdenovo v2.04.1 Genomes were annotated using Prokka (Seemann, 2014). WGS data were submitted to the Sequence Read Archive (SRA) database in NCBI with the accession number PRJNA725618 animal isolates,2 and the SRA-BioSample numbers ranged from SAMN18896327-SAMN18896340 (Supplementary Table 1).
Molecular Typing
WGS data of C. jejuni isolates from cattle were genotyped by in silico multilocus sequence typing (MLST)3 (Jolley et al., 2004). Through searching ‘‘HS19’’ and ‘‘19,’’ a collection of 79 HS19 isolates were accessed from the PubMLST database with the access date 2021-9-1.4 The sources of these isolates from PubMLST database included cattle (3.8%, 3/79), chicken (7.6%, 6/79), GBS patients (17.7%, 14/79), gastroenteritis patients (59.55%, 47/79), human systemic disease (1.3%, 1/79), and human unspecified (10.1%, 8/79). These human isolates were sampled from 11 countries, including the United Kingdom, Netherlands, South Africa, Japan, Thailand, United States, Mexico, Canada, Belgium, Denmark, and China, while the collection year ranged from 1983 to 2018 (Supplementary Table 3).
Housekeeping allelic profiles of cattle isolates were analyzed using the goeBURST algorithm implemented in PHYLOViZ 2.0 (Nascimento et al., 2017), to create a minimum spanning tree (MST), combined with the corresponding data of 79 human isolates. Moreover, two C. jejuni HS19 isolates from GBS patients were downloaded from the NCBI database. The population structures of these 14 cattle isolates were compared with 72 isolates from patients with GBS worldwide, which were accessed from NCBI and PubMLST (Supplementary Table 3).
Homologous Based Phylogenetic Analysis
A total of 62 WGS sequences of C. jejuni isolates were selected for homologous analysis (Supplementary Table 1). Phylogenetic relatedness of 14 HS19 cattle isolates was analyzed, combined with two cattle isolates with HS19 in the United States, 43 control isolates with various serotypes from clinical patients worldwide (including HS19 isolates from GBS patients, n = 10; HS19 isolates from diarrhea patients, n = 9), and three control isolates with HS19 from other animals in China.
To build a phylogenetic tree, homologous genes were screened using OrthoFinder (Levy et al., 2017), 103867 genes (99.1% of total) were assigned to 2306 orthogroups. There were 1390 orthogroups with all species present, and 1132 of these consisted entirely of single-copy genes. Then, a species tree using 1132 orthogroups with a minimum of 100.0% of species having single-copy genes in any orthogroup to construct a phylogenetic tree. The ModelFinder part tested up to 546 protein models, and HIVb+F+R3 was chosen as the best-fit model according to Bayesian statistics criteria (BIC). Finally, a maximum-likelihood-based phylogenetic tree with a bootstrap value of 1,000 iterations was built using the Iq-tree. Table2itol.R was used to generate iTOL annotations from spreadsheet files in R version 3.2.0.
Lipooligosaccharide Typing and Polymorphisms Analysis of cst-II
Cattle isolates of HS19 were characterized using a PCR-based LOS class typing scheme, performed as previously described (Parker et al., 2005). The sequence of gene cst-II was extracted from WGS data by get_homologues-3.3.3 (Contreras-Moreira and Vinuesa, 2013), and the 51st amino acid variations of cst-II from the clustering orthologous sequences were analyzed using the Clustalw program.
In silico Identification of Anti-bacterial Resistance Genes and Virulence Genes
The genomes of C. jejuni isolates (Supplementary Table 1, column 13) were screened for all known resistance and virulence genes using ABRicate v0.8.10 (Park et al., 2020). Anti-bacterial resistance (ABR) genes were identified by a BLASTN comparison against the Resfinder database (Zankari et al., 2012) and Comprehensive Antibiotic Research Database (CARD) database. Point mutations related to antibiotic resistance genes were identified by PointFinder using the pointfinder database (Zankari et al., 2017). Virulence genes were identified by BLASTN comparison against the Virulence Factor Database (VFDB) (Liu et al., 2019). ABRicate classifies the predicted genes based on the proportion of the gene that is covered, and the threshold for identification was taken to be 60% gene identity and 40% sequence coverage.
Antimicrobial Susceptibility
The antimicrobial susceptibilities of HS19 isolates (cattle isolates, n = 14; diarrhea isolates, n = 2; GBS isolate, n = 1) were measured by the agar dilution method recommended by the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2013). C. jejuni ATCC 33560 was used as the quality control strain. Briefly, colonies were subcultured on Campylobacter selective agar base CCDA agar plates for 24 h and then seeded in Mueller Hinton broth supplemented with 5% sheep blood (Oxoid, Basingstoke, United Kingdom), with known scalar concentrations of the following antibiotics: ciprofloxacin (CIP) (0.03–128 μg/ml), erythromycin (ERY) (0.5–256 μg/ml), gentamicin (GEN) (0.25–256 μg/ml), chloramphenicol (CHL) (0.25–128 μg/ml), florfenicol (FFC) (0.25–128 μg/ml), clindamycin (CLI) (0.06–128 μg/ml), and tetracycline (TET) (0.25–256 μg/ml). Strains were classified as resistant (R), intermediate (I) or susceptible (S) according to MIC breakpoints in CLSI (VET01-A4, 2013).
IL-10–/– C57BL/6 Murine Infection Model
IL-10–/– C57BL/6 mice (B6.129P2-IL-10tm1Cgn/J) were obtained from Jackson Laboratories (Bar Harbor, ME, United States). A breeding colony was established under specific pathogen-free conditions. Prior to inoculation, fecal samples were collected to confirm the absence of colitogenic bacteria. Mouse genotypes were identified using a PCR assay from Jackson Laboratories with a one-step mouse genotyping kit (Vazyme, Nanjing, China).
In order to assess the enteritis induction ability of cattle isolates, mice of 6–7 weeks old were orally administrated with fresh suspensions of 14 cattle isolates (0.2 ml of 1 × 1010 CFU per mouse), 14–15 mice were intragastrically administrated with each HS19 C. jejuni isolate. As a positive control, HS19 C. jejuni suspension of one GBS strain was also prepared for mouse infection, whereas sterilized phosphate-buffered saline (PBS) was prepared as the negative control. Fecal pellets were collected at 2 WPI (weeks post-inoculation) to confirm C. jejuni colonization. Diarrhea-associated clinical symptoms were observed weekly. After 5 weeks of observation, the mice were euthanized, and the intestinal gross pathology of each mouse was scored during necropsy as previously reported: Grade 0 = no gross pathology detected; Grade 1 = thickened wall (TW) or enlarged (ENL) colon or cecum; Grade 2 = TW or ENL colon and cecum; Grade 3 = TW or ENL colon and cecum and bloody feces or luminal contents (Brooks et al., 2017). Moreover, intestinal tissue was harvested to assess pathological lesions by staining with hematoxylin and eosin (HE) (Oh et al., 2017).
Results
Multilocus Sequence Typing Profiles of C. jejuni HS19 Isolates From Cattle
MLST analysis revealed restricted genetic diversity for HS19 C. jejuni populations, with the ST-22 complex being the most common clonal complex (CC) for both cattle isolates in China and GBS isolates worldwide. A total of 14 cattle isolates of HS19 belonged to three unique known STs (Figure 1A), and all were grouped into the ST-22 complex. ST-22 was represented of 42.86% (n = 6) cattle isolates, followed by ST-11633 (35.7%, n = 5) and ST-3652 (21.4%, n = 3). In contrast, 15 GBS isolates with HS19 belonged to five unique known STs and were grouped into three CCs. Similarly, the most common ST was ST-22 (73.3%, n = 11), followed by ST-660 (6.6%, n = 1), ST-4051 (6.6%, n = 1), ST-4053 (6.6%, n = 1), and ST-4049 (6.6%, n = 1). CC-22 accounted for 86.6% (n = 13) of GBS isolates, which were mainly sampled from the Netherlands (38.4%, n = 5), followed by Japan (23.0%, n = 3), China (15.3%, n = 2), United States (15.3%, n = 2), and Mexico (7.6%, n = 1). Nevertheless, regarding 71 GBS isolates with various serotypes (Supplementary Table 2), CC-22 was the most common clonal complex (32.3%, n = 23). MLST profiles of four collections of C. jejuni HS19 isolates from different sources were compared (Supplementary Table 3), including animal isolates from China (n = 17), human isolates from China (n = 4), human isolates from other countries (n = 78), and animal isolates from other countries (n = 9). ST-22 (Figure 1B) and ST-22 complex (Figure 1C) were the only sequence type and clonal complex shared by each isolate collection, respectively.
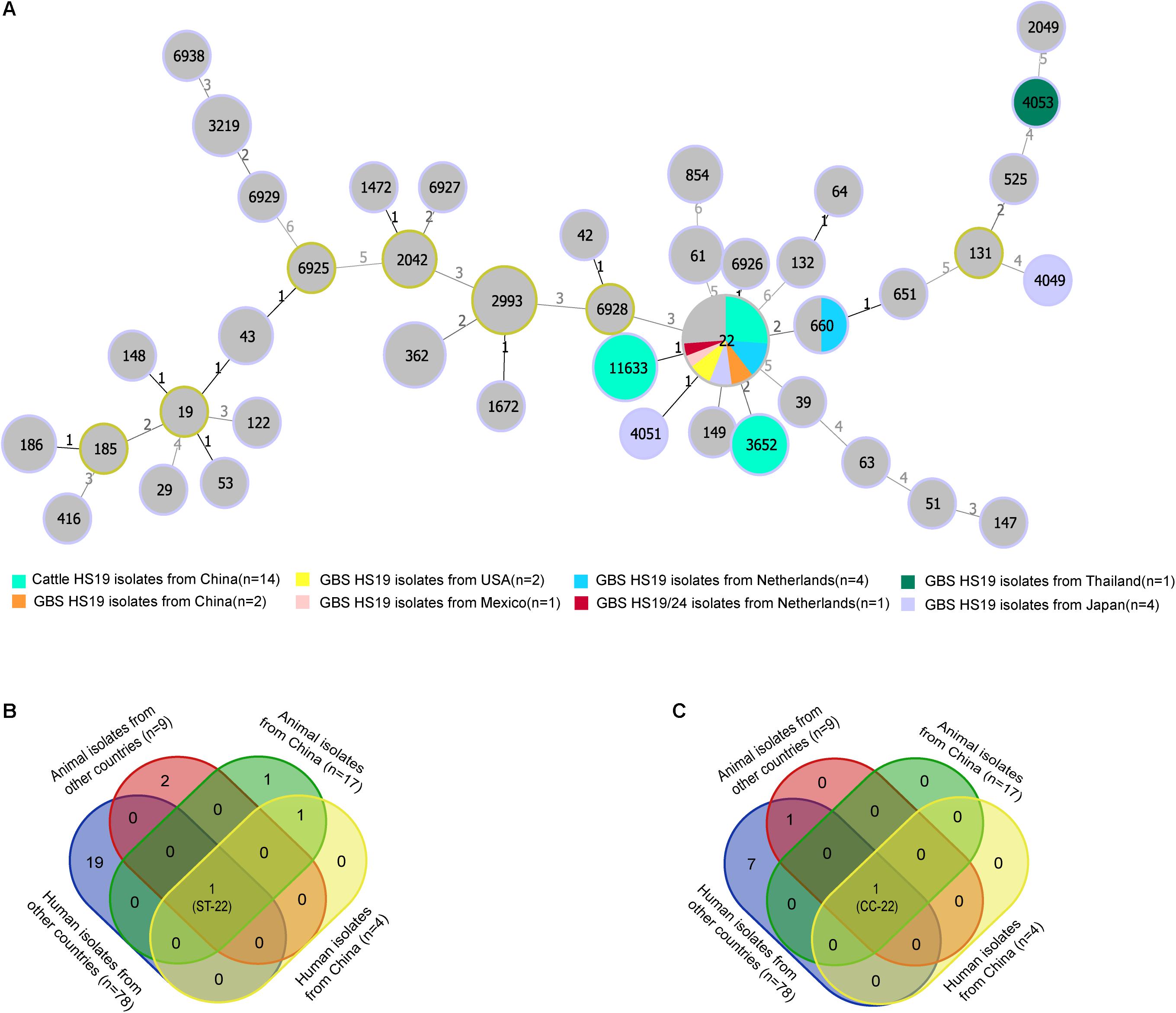
Figure 1. Genotype diversity among Campylobacter jejuni HS19 isolates. (A) A minimum spanning tree generated for C. jejuni HS19 isolates from cattle and Guillain-Barré syndrome (GBS) patients all over the world. The size of the circle proportional to the number of isolates of a particular sequence type. The distance labels correspond to the number of discriminating alleles. GBS isolates with non-HS19 serotypes are represent by gray, whereas HS19 isolates from different geographical locations are represented by other colors. (B) Venn diagram shows the sequence type diversities of isolates from human patients and animals. (C) Venn diagram shows the clonal complex diversities of isolates from human patients and animals.
Phylogenetic Relatedness
C. jejuni HS19 cattle isolates from China showed close phylogenetic relationships with HS19 isolates from GBS patients and diarrhea patients worldwide between 1983 and 2015. Phylogeny was based on orthologous relationships between sixty-two isolates. Overall, the 14 HS19 isolates from cattle in China, together with other 24 HS19 isolates from cattle, poultry, pet, GBS patients, and diarrhea patients worldwide, were grouped into an independent branch marked as “Branch C. jejuni HS19” in Figure 2. In contrast, isolates identified of other serotypes were grouped into other branches. Notably, C. jejuni HS19 strains from clinical patients included 9 GBS strains and 10 diarrhea strains, and the collection dates ranged from 1980 to 2015. The most dominant geographic location was the Netherlands, followed by China, the United States, Mexico, Canada, Japan, and Bangladesh.
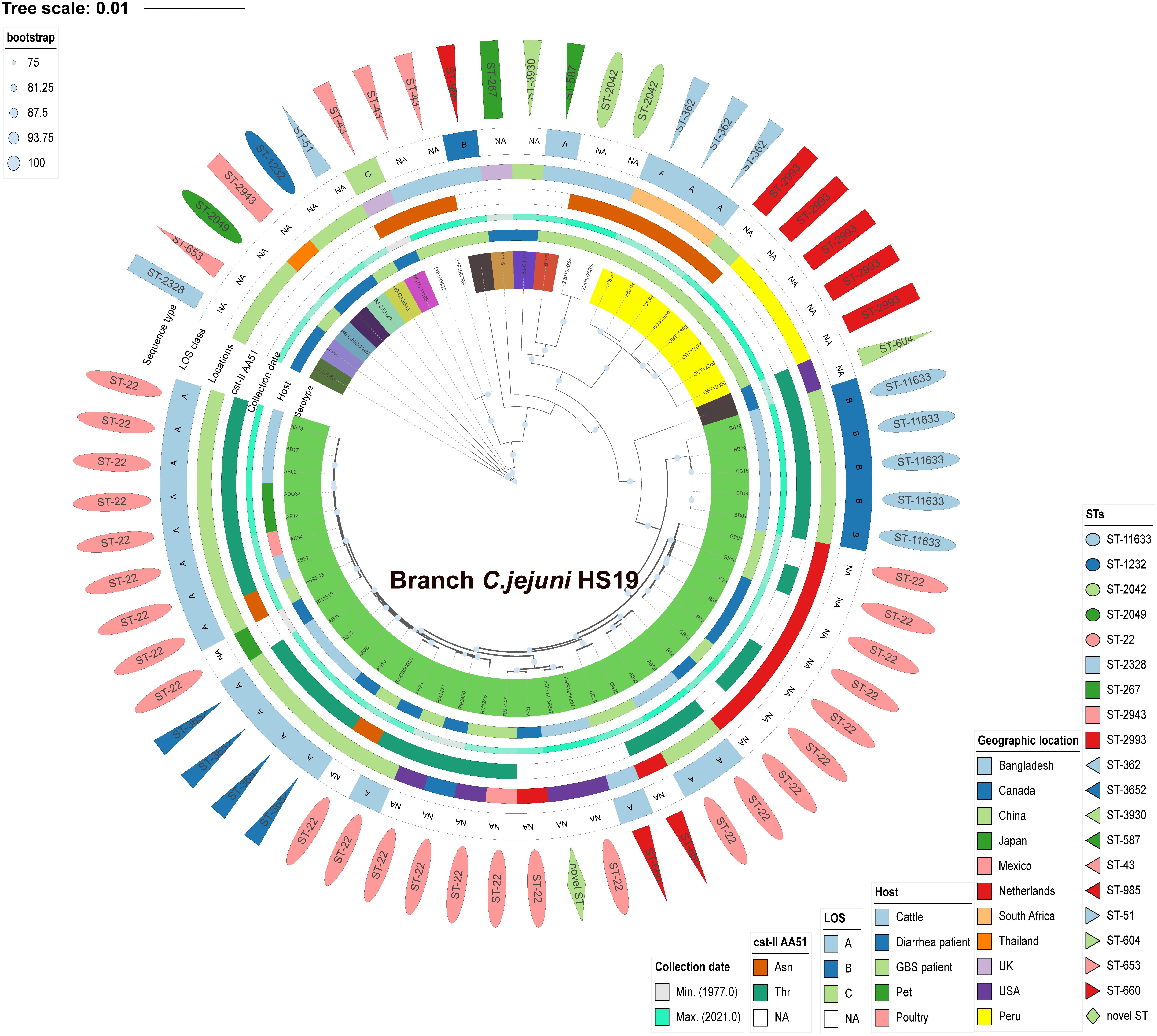
Figure 2. Phylogenetic tree of Campylobacter jejuni isolates according to Bayesian statistics. Serotype and genotype are annotated in this tree. The green tree color refers to the HS19 isolates from cattle and human patients. Other tree colors in the innermost circle (covering strain name) refer to the isolates of other serotypes. Next to the tree color, from inside to outside, five different color strips individually reflect the host, collection year, cst-II AA51, geographical location, and LOS class. The outermost layer of different shapes refers to various sequence types.
Analysis of Sialylated Lipooligosaccharide Class and cst-II Polymorphisms
Sialylated LOS locus classes were detected in all cattle isolates of HS19 (Supplementary Table 1), including LOS A class (64.2%, 9/14) and LOS B class (35.7%, 5/14). Notably, these cattle isolates shared the 51st amino acid Threonine (Thr) in cst-II with six GBS HS19 isolates (the United States, Mexico, and the Netherlands) and four diarrhea isolates (China, the Netherlands, Canada) all over the world, suggesting that these isolates could form a GM1-like ganglioside mimic. Moreover, Asparagine51 (Asn) was detected in the cst-II gene in two GBS isolates of HS19, which could produce ganglioside mimics residues such as GT1a-like, GD3-like, and GD1c-like LOS.
Distribution of Anti-bacterial Resistance Genes
The presence of horizontally acquired genes known to encode resistance to a range of different classes of antibiotics among Chinese cattle isolates (HS19, n = 14) was determined and compared with the corresponding data of GBS isolates (HS19, n = 10), diarrhea isolates (HS19, n = 9), and two cattle isolates from the United States (HS19, n = 2). A total of four unique ABR genes were identified, representing three different major classes of antibiotics (beta-lactams, tetracycline, CmeABC multidrug efflux complex, and CmeR) (Figure 3). All of these isolates (100%, 39/39) carried at least seven horizontally acquired resistance genes, including blaOXA-193, blaOXA-450, cmeA, cmeB, cmeC, and cmeR, with a gene coverage of 99.99–100% and identity percentage of 95.2–99.87%. tet(O) was present in 35.7% (5/14) of cattle isolates and 10.5% of clinical isolates, with a gene coverage of 100% and identity percentage of 99.27–99.74%. The detailed gene identity percentage and sequence coverage of each C. jejuni isolate is shown in Supplementary Table 4. Moreover, gyrA (T86I) was detected in 62.3% of cattle isolates, and 15.8% of clinical isolates, with a gene coverage of 99.99–100% (Table 1).
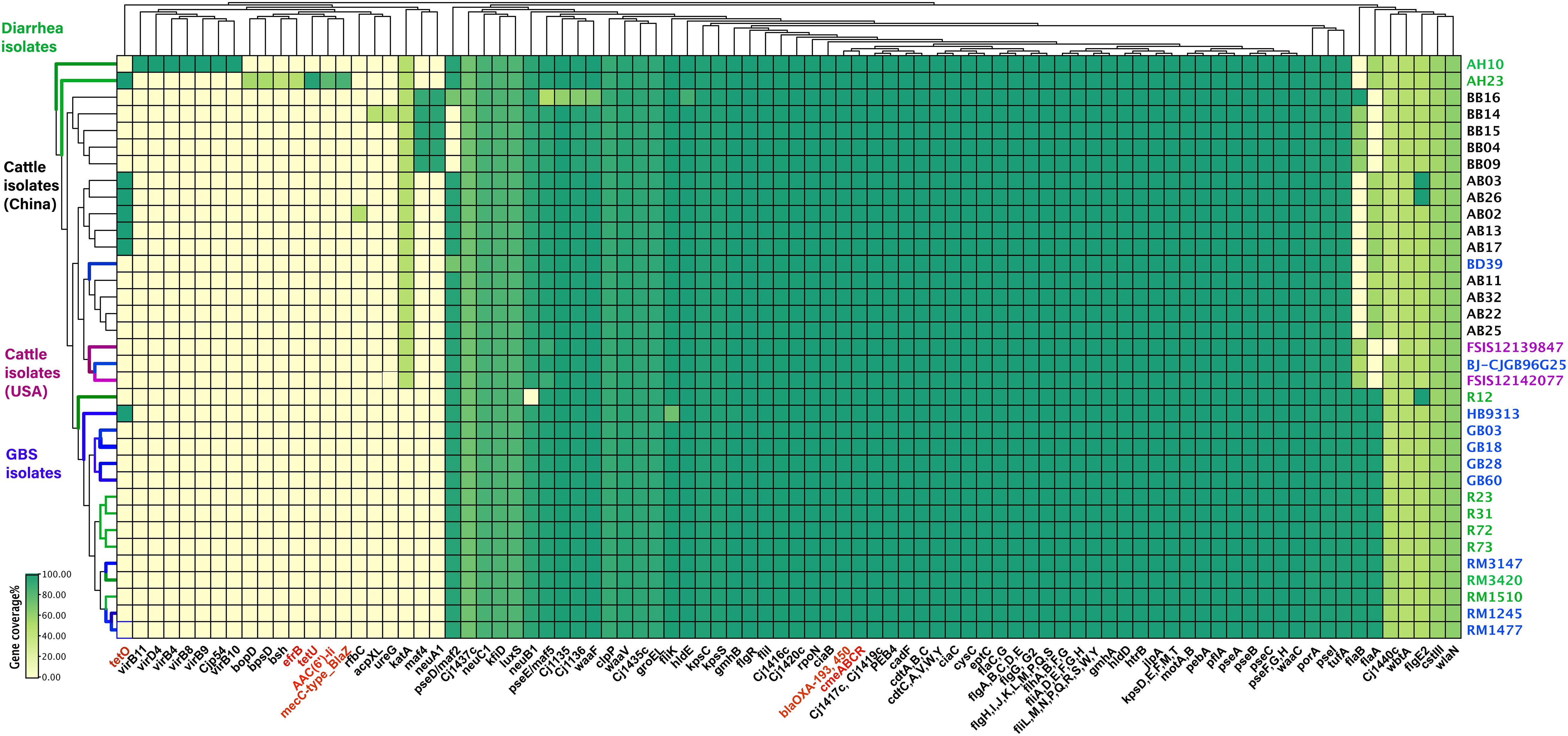
Figure 3. Summary of the antimicrobial resistance genes and virulence profiles of Campylobacter jejuni HS19 isolates. The virulence genes and antimicrobial resistance genes are listed at the bottom part of the figure, and colored in black and red, respectively. Names of isolates are listed on the right, in detail, cattle isolates from China are in black, cattle isolates from United States are in purple, diarrhea isolates worldwide are in green, whereas isolates from patients with Guillain-Barré syndrome are in blue. Further, for the small rectangle, green color indicates the presence of a 100% gene sequence coverage, light green indicates the presence of 60% sequence coverage, and yellow indicates the absence of a gene.
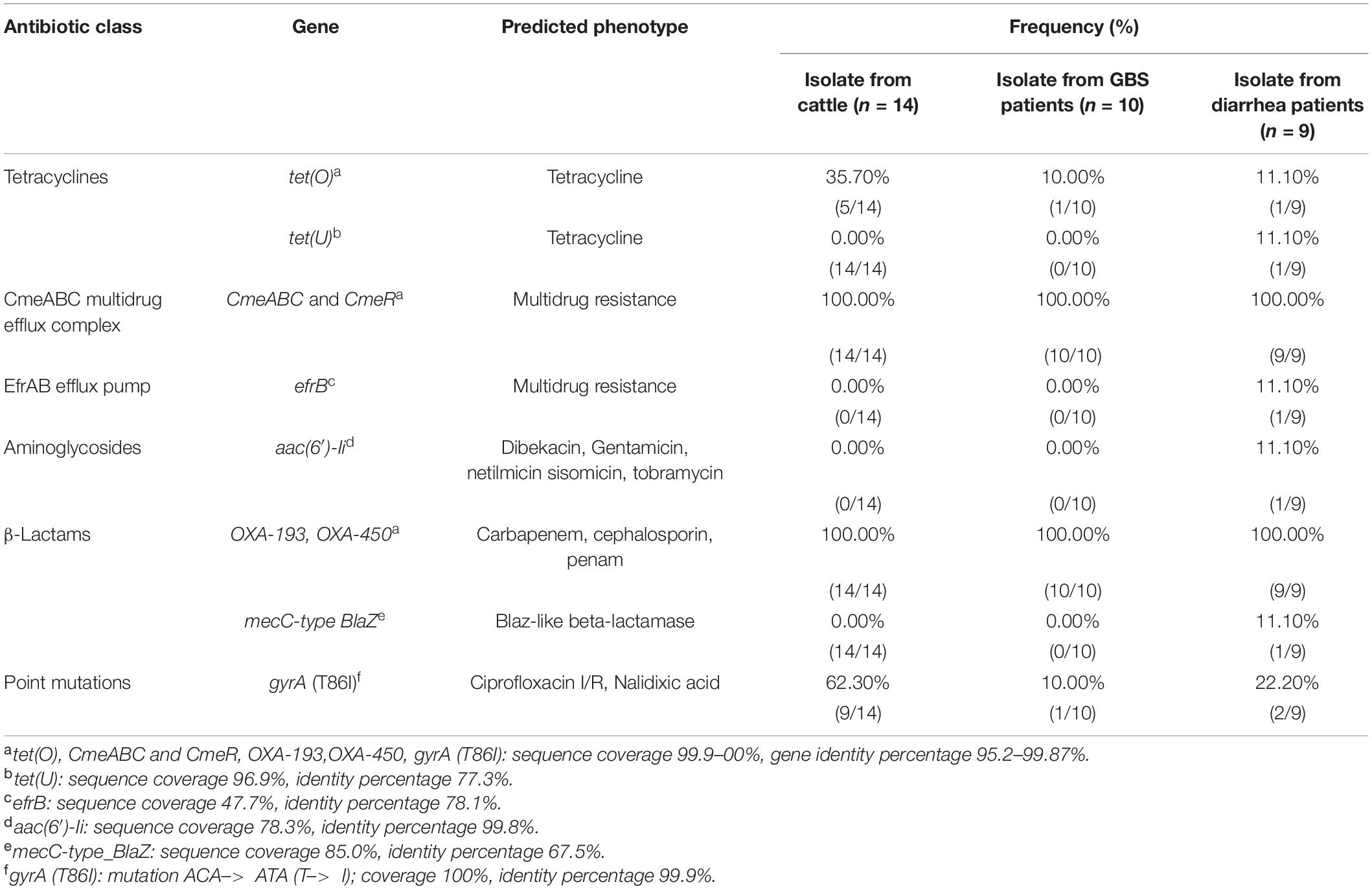
Table 1. Antimicrobial resistance genetic determinants predicted in Campylobacter jejuni HS19 isolates.
Antimicrobial Resistance Phenotypes
Higher levels of AMR in cattle isolates are shown in Figure 4A and Table 2. The most dominant AMR included CIP, CLI, and GEN, all of which were present in all isolates, followed by TET (86%, 12/14), ERY (50%, 7/14), and CHL (36%, 5/14). A lower resistance level was observed for FFC, with a proportional representation of 14% (2/14).
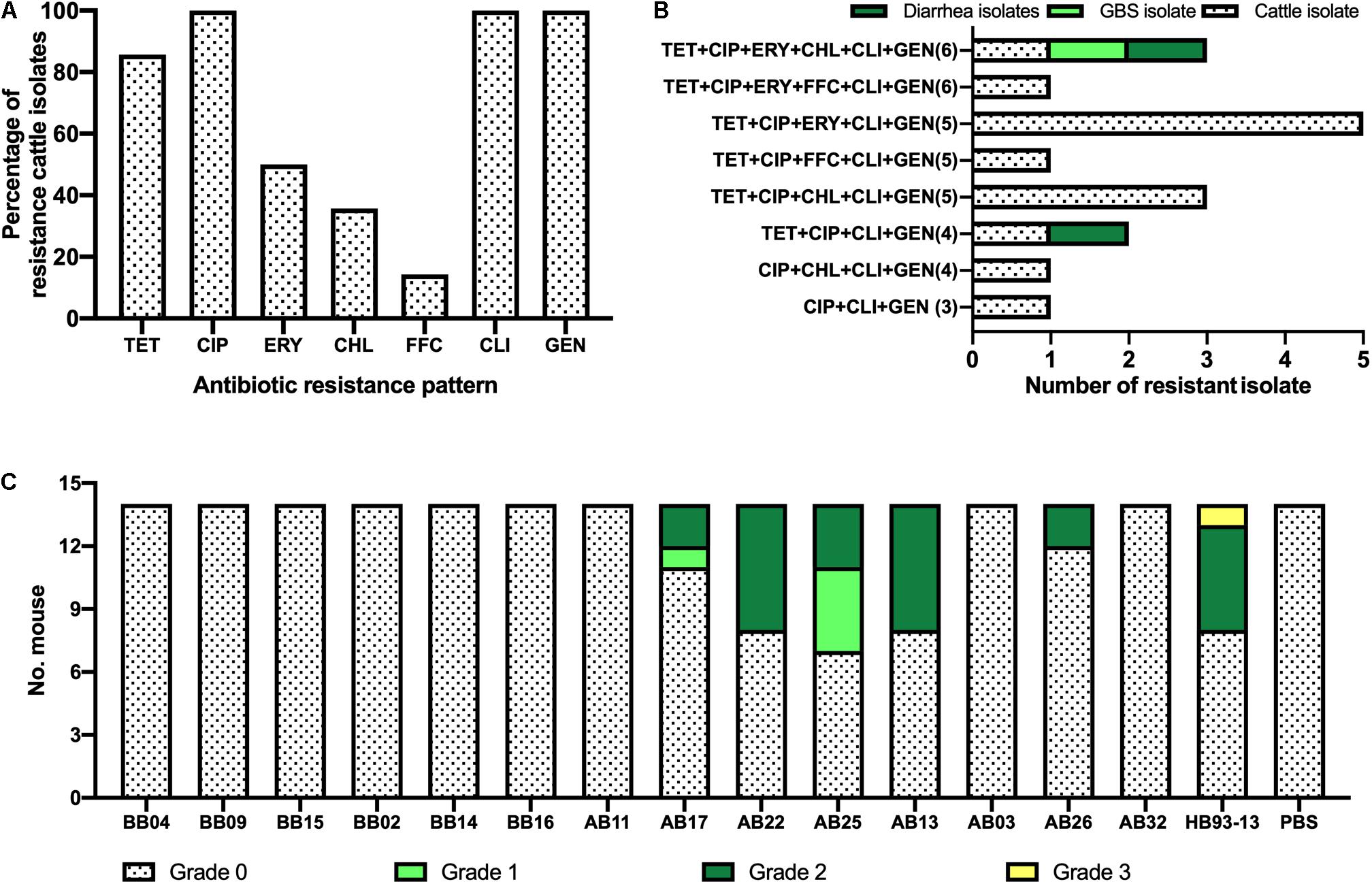
Figure 4. Phenotypic characterization of Campylobacter jejuni HS19 isolates from cattle. (A) Prevalence of C. jejuni antibiotic resistance for seven antibiotics, including ciprofloxacin (CIP), erythromycin (ERY), gentamicin (GEN), chloramphenicol (CHL), florfenicol (FFC), clindamycin (CLI), and tetracycline (TET). (B) Number of resistant C. jejuni isolates from cattle, Guillain-Barré syndrome (GBS) patients, and diarrhea patients showing multi-drug resistance. (C) Gross pathologies of the intestinal tissues induced by the C. jejuni isolates from cattle and GBS patients.
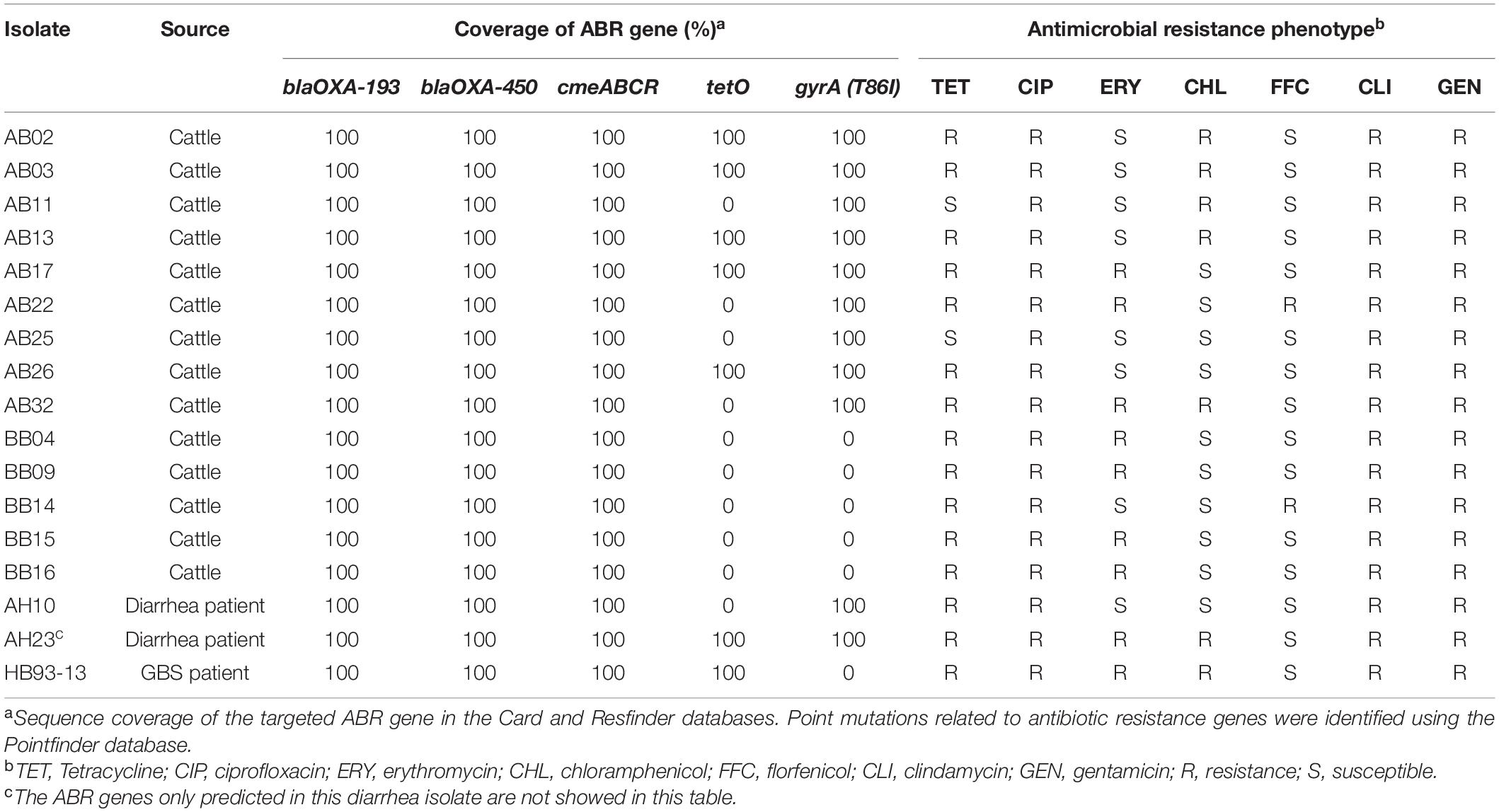
Table 2. ABR gene and antimicrobial resistance phenotype of Campylobacter jejuni HS19 isolates in China.
Notably, worrying levels of multidrug resistance (MDR) were observed in all cattle isolates. TET+CIP+ERY+CLI+GEN was the most common MDR profile with the highest proportion of MDR in cattle isolates (35.7%, n = 5), followed by TET+CIP+CHL+CLI+GEN (21.4%, n = 3), CIP+CHL+CLI+GEN (n = 1, 7.1%), TET+CIP+ ERY+CHL+CLI+GEN (n = 1, 7.1%), TET+CIP+ FFC+CLI+GEN (n = 1, 7.1%), TET+CIP+CLI+GEN(n = 1, 7.1%), TET+CIP+ERY+FFC+CLI+GEN (n = 1, 7.1%), and CIP+CLI+GEN (n = 1, 7.1%). In particular, cattle isolates and human clinical isolates shared two kinds of MDR profiles. In detail, TET+CIP+ERY+CHL+CLI+GEN was present in one diarrheal isolate, one GBS isolate, and one cattle isolate, whereas TET+CIP+CLI+GEN was present in another diarrhea isolate and cattle isolate (Figure 4B).
Distribution of Virulence Determinants
The number of virulence-related genes among the 14 cattle isolates ranged from 106 to 108 (Figure 3 and Supplementary Table 5). Cattle isolates could be divided into two groups based on gene difference; maf4 and neuA1 were the two genes shared by five strains, coding motility accessory factor and bifunctional beta-14-N-acetylgalactosaminyltransferase, respectively, which were absent in both the left cattle isolates and disease control isolates. However, another motility accessory factor associated gene, pseD/maf2, was absent in these cattle isolates, but was present in the remaining isolates.
The GBS-associated genes wlaN and cst-III were prevalent among all the cattle isolates, encoding beta-13 galactosyltransferase involved in the biosynthesis of ganglioside-mimicking LOS and lipooligosaccharide sialyltransferase, respectively. Genes pseA and pseI are required for the biosynthesis and/or transfer of pseudaminic acid to the flagellin, which were also prevalent in all genomes. Other virulence genes shared by each isolate were those encoding traits related to flagella (flgC et al), cytolethal distending toxin (cdtA, cdtB, and cdtC), chemotaxis (cheA, cheV, cheW, cheY), invasion (ciaB, flaC), and adhesin (cadF, jlpA, porA, and pebA).
Genes unique to a certain strain were observed. rfbC was present in a cattle isolate, coding capsule-associated dTDP-4-dehydrorhamnose 35-epimerase, but only blasted with 48.35% coverage. Moreover, ureG and acpXL were detected in another cattle isolate, coding for the LPS-related acyl carrier protein in Brucella melitensis bv. 1 str. 16M and urease accessory protein ureG in Helicobacter pylori 26695, individually, but all blasted with 42.17% coverage. Notably, one cattle isolate carried LOS (Cj1135, Cj1136, hldE, waaF) and motility accessory factor (pseD/maf2, pseE/maf5) associated genes, but showed different gene coverage when compared to other cattle isolates. One cattle isolate from the United States lacked Cj1440c. For human GBS strains, the fliK gene encoding flagellar hook-length control protein FliK was harbored in one GBS isolate with a gene coverage of 72.18%, while the coverage of other GBS isolates was 99.83%, indicating that fliK could not be associated with disease type but could affect disease severity.
Assessment of Enteritis
Although shared with the same serotype HS19 and sequence type ST-22, C. jejuni HB93-13 and 35.7% (5/14) of C. jejuni isolates from cattle induced gross pathology (Score > 0), including bloody feces, inflammation of cecum and colon, pathological, whereas PBS was unable to induce severe gastrointestinal pathology (Figure 4C).
PBS failed to induce inflammation in the mice (Figure 5A). In stark contrast, serious pathological lesions and inflammation were found in the colon cecum ileum junction (ICCJ), colon, and cecum of mice inoculated with GBS patient isolates and part of cattle isolates. In detail, severe inflammation occurred in mice infected with GBS isolate HB93-13, mucosal epithelial cells were necrotic and exfoliated, and lamina propria contained neutrophils and mononuclear cells (Figure 5B). Mice infected with cattle isolates were also observed with submucosal edema, increased monocytes in the mucous layer, individual intestinal gland necrosis, and local inflammatory reaction spread to the muscular layer (Figure 5C).
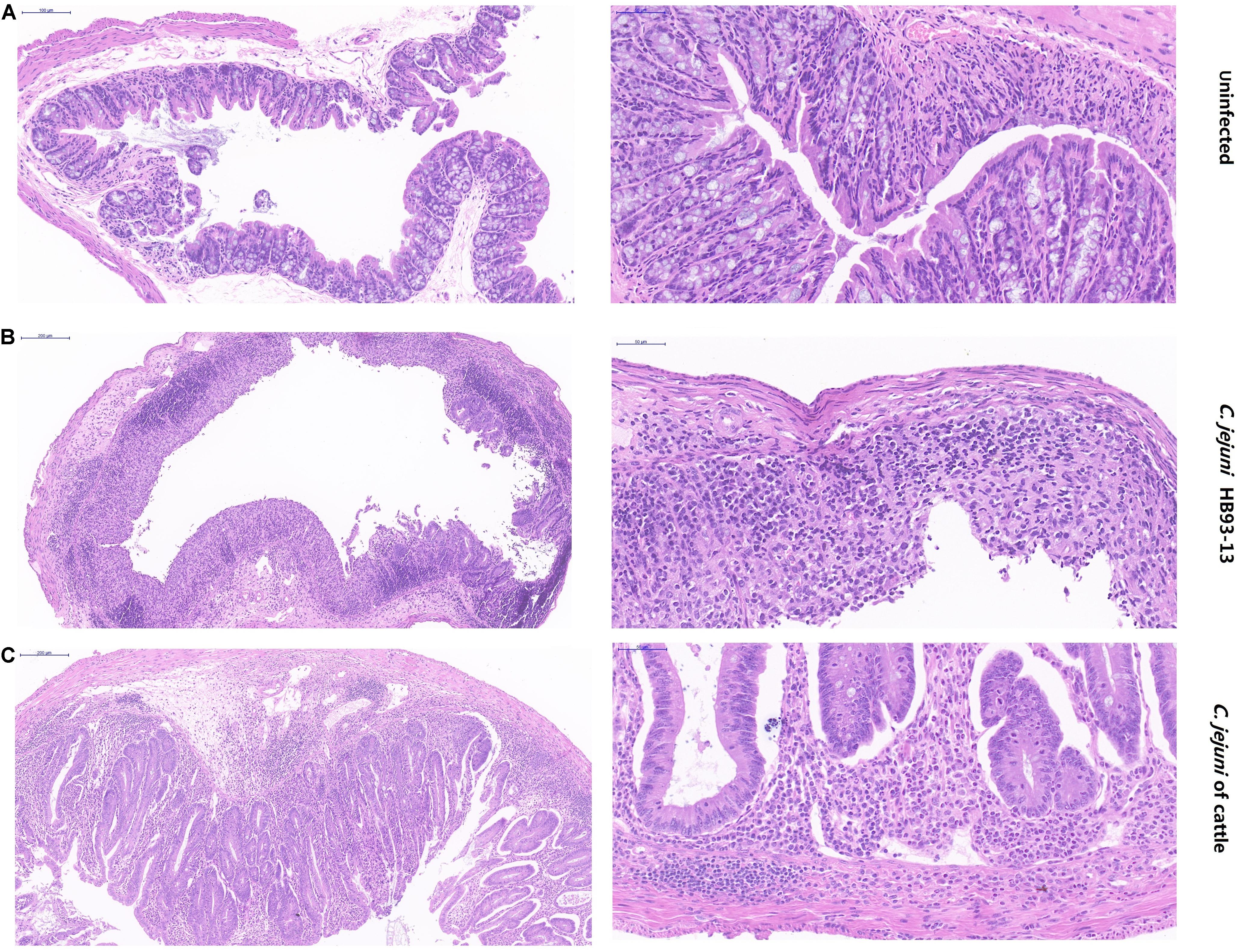
Figure 5. Pathological severity of mouse intestinal tissue. (A) Intestinal lesion is not observed in phosphate-buffered saline sham-inoculated mice. (B) Human Guillain-Barré syndrome Campylobacter jejuni strain HB93-13 inoculated mice showed severe inflammation. (C) Cattle isolates induced local inflammatory reaction. Scale bar, 50 and 200 μm.
Discussion
Identifying high-risk C. jejuni isolates remains an obstacle to effective intervention for campylobacteriosis (Buchanan et al., 2017). Infection of HS19 isolates often increases the risk of developing GBS (Nachamkin et al., 1998), while HS19 isolates are also present in the diarrhea populations circulating in various countries. Since C. jejuni colonizes the intestines of various animals, the source of human infection is thought to be a massive reservoir in animal populations (Parker et al., 2005). Based on the idea of “one health” (Wolfe et al., 2007), campylobacteriosis is a zoonotic foodborne disease, and human infection of C. jejuni needs to be controlled by animal strains. However, 88.61% of the HS19 isolates in PubMLST database were of human origin. In contrast, zoonotic HS19 isolates have rarely been reported, and the pathogenic potential of C. jejuni from animal sources has remained unexplored. In this study, a collection of 14 HS19 cattle isolates from China was systematically characterized both in terms of genotype and phenotype. Remarkably, phylogenetic relatedness and genomic similarity to clinical human isolates were observed among these cattle isolates, as well as a worrying level of multidrug resistance (MDR) and the ability to induce enteritis in a mouse model.
Close phylogenetic relationships between C. jejuni isolates from cattle and patients have been frequently reported, although direct epidemiological evidence linking these zoonotic isolates from a specific source as well as a certain disease type was unavailable (Thépault et al., 2017; Hsu et al., 2020). Our results were consistent with those reported ones; cattle isolates of HS19 were observed to have a close genetic relatedness with clinical isolates. Specifically, all of these HS19 isolates obtained from cattle belonged to the ST-22 complex, which has been significantly overrepresented in the isolates among the patients who developed GBS following campylobacteriosis (Nielsen et al., 2010). In addition to GBS, the ST-22 clonal complex has also been reported as a high-risk lineage represented in HS19 isolates, leading to the development of post-infection irritable bowel syndrome (PI-IBS) (Peters et al., 2021). Compared with cattle isolates collected in east China, the majority of human clinical isolates were recovered from developed countries, such as the United States, Netherlands, Japan, and Australia, suggesting the possibility of clonal spread of capsular genotype HS19 isolates across different regions and hosts. Additionally, the predominance of these clonal complexes could be associated with economic conditions, hygiene conditions, wildlife ecology, population movements, and environmental factors (geography and climate) (Oh et al., 2017; Heimesaat et al., 2021). Multiple introductions and widespread dissemination of C. jejuni lineages between countries may be facilitated by the constant movement of agricultural products, animals, and people, which could also be associated with the risk of emergence and spread of HS19 isolates, potentially causing pandemics.
Animal microbes and human health are intimately coupled, and animals could be transmitters of high-risk C. jejuni isolates to susceptible humans. The sources of HS19 isolates collected in China including cattle, chicken, pets, and human (Supplementary Table 1), while the sources of HS19 isolates from PubMLST database included cattle, chicken, and humans (Supplementary Table 3), indicating HS19 isolates could spread through the food chain “from farm to table.” Thus, in addition to animal types, the prevalence of HS19 isolates could also be influenced by local animal husbandry practices, antibiotic use in farms, transportation of food animals and animal products, and consumption habits. Notably, the collection time also influenced the prevalence of C. jejuni. Epidemiological reports from other countries (Habib et al., 2009) suggest that human campylobacteriosis tends to increase between February and September. Therefore, the isolates in this study were sampled during this period.
Identification of virulence factors is crucial to understand the mechanisms of campylobacteriosis infection and to identify if potentially more virulent strains exist (Wysok et al., 2020). Notably, previous studies have identified that genetic determinants are important for C. jejuni pathogenicity (Dasti et al., 2010), but they are generally conserved across species. Our results are consistent with the reported ones. Except for the maf4 coding motility accessory factor and neuA1 coding bifunctional beta-14-N-acetylgalactosaminyltransferase/CMP-Neu5Ac synthase which were extra represented by five cattle isolates of HS19, other virulence genes harbored in cattle isolates were also represented by human clinical isolates. Notably, even though the cattle isolates from the same collection geography shared the same serotype, ST, AMR genes, virulence genes, as well as similar phylogenetic relatedness with human clinical isolates, only some of the animal isolates could induce the disease phenotype in the mouse model. In fact, although the database of virulence genes is constantly updated, the subject sequence could only be blasted with the reported one, suggesting that unreported pathogenic factors such as accessory genes with a statistically significant difference in carriage rates among animal isolates and human clinical isolates could play a role in C. jejuni pathogenicity (Buchanan et al., 2017). Our results also showed that a few genes present both in human isolates and cattle isolates differed in coverage percentage and copy number, which could be a result of the high frequencies of gene transfer and recombination. Besides host susceptibility, gene mutations and polymorphisms could play roles in C. jejuni infection, which warrants further investigation.
Increasing rates of Campylobacter strains resistant to the drugs of choice and alternative therapies, making AMR Campylobacter a public health concern (Mourkas et al., 2019; Gahamanyi et al., 2021), and MDR is still very common in Campylobacter strains isolated from farmed animals in many European countries (Pascoe et al., 2017). In this study, MDR was common in C. jejuni HS19 isolates from cattle, indicating a potential hazard. TET is often used in the food animal industry because of its low cost and easy administration to animals through drinking water (Jonker and Picard, 2010). Consistent with previous research on a high resistance to TET in China (Zhang et al., 2020), our results showed that tet(O) was predicted in 35.7% of cattle isolates, whereas a high resistance level to TET was observed in 86% of cattle isolates. Except for tet(O), only blaOXA-193, blaOXA-450, cmeABC, and cmeR were predicted in cattle isolates, although an alarming trend toward MDR was also detected among all cattle isolates, while some of these cattle isolates shared the same MDR profiles with clinical isolates. Notably, 62.3% of cattle isolates were predicted to have point mutations in gyrA (T86I), but all of the cattle isolates showed high levels of resistance to CIP. The discrepancies found between the predicted AMR genes and the observed phenotype could be explained by the existence of the efflux pump mechanisms or other unknown resistance mechanisms (Marotta et al., 2020). Moreover, half of the cattle isolates showed resistance to ERY, whereas two clinical isolates showed resistance to ERY. To our knowledge, the use of fluoroquinolones, known to be the first-choice treatment for campylobacteriosis, has recently shifted to erythromycin, against which Campylobacter resistance seemed to develop more slowly with respect to fluoroquinolone resistance (Lapierre et al., 2016).
In particular, this study first predicted the tet(U) gene in a C. jejuni isolate from a patient, with a sequence coverage of 96.86%, and an identity percentage of 77.27%. However, a previous bioinformatic analysis provided compelling evidence that “tet(U)” was not a tetracycline resistance determinant, but the misannotated 3′ end of a gene encoding a rolling-circle replication initiator (Rep) protein (Caryl et al., 2012). The potential function of this gene in C. jejuni will be investigated in future studies. A few limitations of this study need to be acknowledged. First, the limited sample size of cattle isolates suggested that we have merely touched on the existing genomic diversity of this pathogen. In fact, 14 HS19 cattle isolates and 3 control isolates from other animal sources involved in this study were selected from 1146 animal isolates in a comprehensive genomic epidemiological study, regarding the serotype diversity of C. jejuni isolates from various animals in China, within a long sampling time span, since cattle and chicken are the common sources of animal protein, while pets are commonly raised by local citizens. In total, 14 cattle strains of HS19 were identified from 277 cattle isolates. In the future, more cattle isolates will be identified. Another limitation was the underrepresentation of C. jejuni isolates from GBS patients and HS19 isolates in the PubMLST database, which needs to be replenished through international cooperation.
Herein, HS19 isolates of animal origin were firstly characterized by a systematic evaluation of bacterial genomics and in vitro virulence. Surprisingly, all of the zoonotic isolates belonged to the clinical high-risk lineage with a worrying level of MDR, while the ability to induce enteritis in vitro varied among different isolates. Our research is not only a supplement to the HS19 animal isolates in the Public database, but also provides new insight into the pathogenic potential of C. jejuni isolates from a putative cattle host. Genetic elements associated with the differences in susceptibility of species as well as the interspecies transmission of these zoonotic isolates to humans should be investigated to advance a better understanding of C. jejuni-associated zoonosis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
Animal experimental procedures were approved by the Laboratory Animal Center of Yangzhou University, according to the experimental animal 3R principle. Animals were housed in the experimental animal center at Yangzhou University, where the environmental conditions meet China’s national standards for environment and facilities for laboratory animals (GB14925-2001). The experimental animal permit license was issued by the Science and Technology Department of Jiangsu Province in China [SYXK (Su) 2017-0044]. Animal experiments were supervised and inspected by the Animal Welfare and Ethics Committee, in accordance with the guidelines of the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH publication number 80-23). All mice were euthanized before the autopsy. Animal carcasses, tissues, or body fluids were centralized for pollution-free treatment.
Author Contributions
XZ: conceptualization, draft preparation, methodology, formal analysis, and visualization. XZ, PH, and JL: investigation. XZ and JH: review. JH and XJ: project administration. JH: funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (31872493), National Key Research and Development Program of China (2018YFD0500500), and six talent peaks project in Jiangsu Province (2015-SWYY-02).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank J. Ruan, Technical Information Scientist at the Jackson Laboratory (United States), for their mouse breeding and husbandry support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.783750/full#supplementary-material
Footnotes
- ^ https://github.com/aquaskyline/SOAPdenovo2
- ^ https://www.ncbi.nlm.nih.gov/sra/
- ^ https://github.com/tseemann/mlst
- ^ https://pubmlst.org/bigsdb?db=pubmlst_campylobacter_isolates&page=query
References
Baker, M. G., Kvalsvig, A., Zhang, J., Lake, R., Sears, A., and Wilson, N. (2012). Declining Guillain-Barré syndrome. after campylobacteriosis control, New Zealand, 1988–2010. Emerg. Infect. Dis. 18, 226–233. doi: 10.3201/eid1802.111126
Brooks, P. T., Brakel, K. A., Bell, J. A., Bejcek, C. E., Gilpin, T., Brudvig, J. M., et al. (2017). Transplanted human fecal microbiota enhanced Guillain-Barré syndrome auto-antibody responses after Campylobacter jejuni infection in C57BL/6 mice. Microbiome. 5, 92. doi: 10.1186/s40168-017-0284-4
Buchanan, C. J., Webb, A. L., Mutschall, S. K., Kruczkiewicz, P., Barker, D. O. R., Hetman, B. M., et al. (2017). A genome-wide association study to identify diagnostic markers for human pathogenic Campylobacter jejuni strains. Front. Microbiol. 8:1224. doi: 10.3389/fmicb.2017.01224
Burnham, P. M., and Hendrixson, D. R. (2018). Campylobacter jejuni: collective components promoting a successful enteric lifestyle. Nat. Rev. Microbiol. 16, 551–565. doi: 10.1038/s41579-018-0037-9
Caryl, J. A., Cox, G., Trimble, S., and O’ Neill, A. J. (2012). “tet(U)” is not a tetracycline resistance determinant. Antimicrob. Agents Chemother. 56, 3378–3379. doi: 10.1128/AAC.05957-11
CLSI (2013). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals; Approved Standard CLSI Document VET01-A4, 4th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Contreras-Moreira, B., and Vinuesa, P. (2013). GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl. Environ. Microbiol. 79, 7696–7701. doi: 10.1128/AEM.02411-13
Costard, S., Espejo, L., Groenendaal, H., and Zagmutt, F. J. (2017). Outbreak-related disease burden associated with consumption of unpasteurized cow’s milk and cheese, United States, 2009-2014. Emerg. Infect. Dis. 23, 957–964. doi: 10.3201/eid2306.151603
Crofts, A. A., Poly, F. M., Ewing, C. P., Kuroiwa, J. M., Rimmer, J. E., Harro, C., et al. (2018). Campylobacter jejuni transcriptional and genetic adaptation during human infection. Nat. Microbiol. 3, 494–502. doi: 10.1038/s41564-018-0133-7
Dasti, J. I., Tareen, A. M., Lugert, R., Zautner, A. E., and Gross, U. (2010). Campylobacter jejuni: a brief overview on pathogenicity-associated factors and disease-mediating mechanisms. Int. J. Med. Microbiol. 300, 205–211. doi: 10.1016/j.ijmm.2009.07.002
Davies, E., Ebbesen, M., Johansson, C., Kaden, R., and Rautelin, H. (2020). Genomic and phenotypic characterisation of Campylobacter jejuni isolates from a waterborne outbreak. Front. Cell. Infect. Microbiol. 10:594856. doi: 10.3389/fcimb.2020.594856
de Oliveira, M. G., Rizzi, C., Galli, V., Lopes, G. V., Haubert, L., Dellagostin, O. A., et al. (2019). Presence of genes associated with adhesion, invasion, and toxin production in Campylobacter jejuni isolates and effect of temperature on their expression. Can. J. Microbiol. 65, 253–260. doi: 10.1139/cjm-2018-0539
Fiedoruk, K., Daniluk, T., Rozkiewicz, D., Oldak, E., Prasad, S., and Swiecicka, I. (2019). Whole-genome comparative analysis of Campylobacter jejuni strains isolated from patients with diarrhea in northeastern Poland. Gut Pathog. 11:32. doi: 10.1186/s13099-019-0313-x
Gahamanyi, N., Song, D. G., Yoon, K. Y., Mboera, L. E. G., Matee, M. I., Mutangana, D., et al. (2021). Antimicrobial resistance profiles, virulence genes, and genetic diversity of thermophilic Campylobacter species isolated from a layer poultry farm in Korea. Front. Microbiol. 12:622275. doi: 10.3389/fmicb.2021.622275
Guirado, P., Paytubi, S., Miró, E., Iglesias-Torrens, Y., Navarro, F., Cerdà-Cuéllar, M., et al. (2020). Differential distribution of the wlaN and cgtB genes, associated with Guillain-Barré Syndrome, in Campylobacter jejuni isolates from humans, broiler chickens, and wild birds. Microorganisms 8:325. doi: 10.3390/microorganisms8030325
Habib, I., Louwen, R., Uyttendaele, M., Houf, K., Vandenberg, O., Nieuwenhuis, E. E., et al. (2009). Correlation between genotypic diversity, lipooligosaccharide gene locus class variation, and caco-2 cell invasion potential of Campylobacter jejuni isolates from chicken meat and humans: contribution to virulotyping. Appl. Environ. Microbiol. 75, 4277–4288. doi: 10.1128/AEM.02269-08
Hakkinen, M., Nakari, U. M., and Siitonen, A. (2009). Chickens and cattle as sources of sporadic domestically acquired Campylobacter jejuni infections in Finland. Appl. Environ. Microbiol. 75, 5244–5249. doi: 10.1128/AEM.00374-09
Heikema, A. P., Islam, Z., Horst-Kreft, D., Huizinga, R., Jacobs, B. C., Wagenaar, J. A., et al. (2015). Campylobacter jejuni capsular genotypes are related to Guillain-Barré syndrome. Clin. Microbiol. Infect. 21, 852.e1–9. doi: 10.1016/j.cmi.2015.05.031
Heimesaat, M. M., Backert, S., Alter, T., and Bereswill, S. (2021). Human campylobacteriosis-a serious infectious threat in a one health perspective. Curr. Top. Microbiol. Immunol. 431, 1–23. doi: 10.1007/978-3-030-65481-8_1
Hsu, C. H., Harrison, L., Mukherjee, S., Strain, E., McDermott, P., Zhang, Q., et al. (2020). Core genome multilocus sequence typing for food animal source attribution of human Campylobacter jejuni infections. Pathogens 9:532. doi: 10.3390/pathogens9070532
Iglesias-Torrens, Y., Miró, E., Guirado, P., Llovet, T., Muñoz, C., Cerdà-Cuéllar, M., et al. (2018). Population structure, antimicrobial resistance, and virulence–associated genes in Campylobacter jejuni isolated from three ecological niches: gastroenteritis patients, broilers, and wild birds. Front. Microbiol. 9:1676. doi: 10.3389/fmicb.2018.01676
Jolley, K. A., Chan, M. S., and Maiden, M. C. (2004). MLSTdbNet-distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5:86. doi: 10.1186/1471-2105-5-86
Jonker, A., and Picard, J. A. (2010). Antimicrobial susceptibility in thermophilic Campylobacter species isolated from pigs and chickens in South Africa. J. S. Afr. Vet. Assoc. 81, 228–236. doi: 10.4102/jsava.v81i4.153
Kaakoush, N. O., Castaño-Rodríguez, N., Mitchell, H. M., and Man, S. M. (2015). Global epidemiology of Campylobacter infection. Clin. Microbiol. Rev. 28, 687–720. doi: 10.1128/CMR.00006-15
Kärenlampi, R., Rautelin, H., Schönberg-Norio, D., Paulin, L., and Hänninen, M. L. (2007). Longitudinal study of Finnish Campylobacter jejuni and C. coli isolates from humans, using multilocus sequence typing, including comparison with epidemiological data and isolates from poultry and cattle. Appl. Environ. Microbiol. 73, 148–155. doi: 10.1128/AEM.01488-06
Kashoma, I. P., Kassem, I. I., John, J., Kessy, B. M., Gebreyes, W., Kazwala, R. R., et al. (2016). Prevalence and antimicrobial resistance of Campylobacter isolated from dressed beef carcasses and raw milk in Tanzania. Microb. Drug Resist 22, 40–52. doi: 10.1089/mdr.2015.0079
Koenraad, P. M. F. J., Rombouts, F. M., and Notermans, S. H. W. (1997). Epidemiological aspects of thermophilic Campylobacter in water-related environments: a review. Water Environ. Res. 69, 52–63. doi: 10.2175/106143097X125182
Koolman, L., Whyte, P., Burgess, C., and Bolton, D. (2015). Distribution of virulence-associated genes in a selection of Campylobacter isolates. Foodborne Pathog. Dis. 12, 424–432. doi: 10.1089/fpd.2014.1883
Kuroki, S., Saida, T., Nukina, M., Haruta, T., Yoshioka, M., Kobayashi, Y., et al. (1993). Campylobacter jejuni strains from patients with Guillain-Barré syndrome belong mostly to Penner serogroup 19 and contain beta-N-acetylglucosamine residues. Ann. Neurol. 33, 243–247. doi: 10.1002/ana.410330304
Lapierre, L., Gatica, M. A., Riquelme, V., Vergara, C., Yañez, J. M., San Martín, B., et al. (2016). Characterization of antimicrobial susceptibility and its association with virulence genes related to adherence, invasion, and cytotoxicity in Campylobacter jejuni and Campylobacter coli isolates from animals, meat, and humans. Microbial. Drug Resist. 22, 432–444. doi: 10.1089/mdr.2015.0055
Levy, A., Salas Gonzalez, I., Mittelviefhaus, M., Clingenpeel, S., Herrera Paredes, S., Miao, J., et al. (2017). Genomic features of bacterial adaptation to plants. Nat. Genet. 50, 138–150. doi: 10.1038/s41588-017-0012-9
Liu, B., Zheng, D., Jin, Q., Chen, L., and Yang, J. (2019). VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 47, 687–692. doi: 10.1093/nar/gky1080
Marotta, F., Di Marcantonio, L., Janowicz, A., Pedonese, F., Di Donato, G., Ardelean, A., et al. (2020). Genotyping and antibiotic resistance traits in Campylobacter jejuni and coli from pigs and wild boars in Italy. Front. Cell. Infect. Microbiol. 10:592512. doi: 10.3389/fcimb.2020.592512
Mourkas, E., Florez-Cuadrado, D., Pascoe, B., Calland, J. K., Bayliss, S. C., Mageiros, L., et al. (2019). Gene pool transmission of multidrug resistance among Campylobacter from livestock, sewage and human disease. Environ. Microbiol. 21, 4597–4613. doi: 10.1111/1462-2920.14760
Müller, J., Meyer, B., Hänel, I., and Hotzel, H. (2007). Comparison of lipooligosaccharide biosynthesis genes of Campylobacter jejuni strains with varying abilities to colonize the chicken gut and to invade Caco-2 cells. J. Med. Microbiol. 56, 1589–1594. doi: 10.1099/jmm.0.47305-0
Nachamkin, I. (1997). Microbiologic approaches for studying Campylobacter species in patients with Guillain-Barré syndrome. J. Infect. Dis. 176(Suppl. 2), S106–S114. doi: 10.1086/513789
Nachamkin, I. (2002). Chronic effects of Campylobacter infection. Microbes. Infect. 4, 399–403. doi: 10.1016/S1286-4579(02)01553-8
Nachamkin, I., Allos, B. M., and Ho, T. (1998). Campylobacter species and Guillain-Barré syndrome. Clin. Microbiol. Rev. 11, 555–567. doi: 10.1128/CMR.11.3.555
Nascimento, M., Sousa, A., Ramirez, M., Francisco, A. P., Carriço, J. A., and Vaz, C. (2017). PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics 33, 128–129. doi: 10.1093/bioinformatics/btw582
Nielsen, L. N., Sheppard, S. K., McCarthy, N. D., Maiden, M. C., Ingmer, H., and Krogfelt, K. A. (2010). MLST clustering of Campylobacter jejuni isolates from patients with gastroenteritis, reactive arthritis and Guillain-Barré syndrome. J. Appl. Microbiol. 108, 591–599. doi: 10.1111/j.1365-2672.2009.04444.x
Oh, J. Y., Kwon, Y. K., Wei, B., Jang, H. K., Lim, S. K., Kim, C. H., et al. (2017). Epidemiological relationships of Campylobacter jejuni strains isolated from humans and chickens in South Korea. J. Microbiol. 55, 13–20. doi: 10.1007/s12275-017-6308-8
Park, C. J., Li, J., Zhang, X., Gao, F., Benton, C. S., and Andam, C. P. (2020). Genomic epidemiology and evolution of diverse lineages of clinical Campylobacter jejuni cocirculating in New Hampshire, USA, 2017. J. Clin. Microbiol. 58, e02070-19. doi: 10.1128/JCM.02070-19
Parker, C. T., Horn, S. T., Gilbert, M., Miller, W. G., Woodward, D. L., and Mandrell, R. E. (2005). Comparison of Campylobacter jejuni lipooligosaccharide biosynthesis loci from a variety of sources. J. Clin. Microbiol. 43, 2771–2781. doi: 10.1128/JCM.43.6.2771-2781.2005
Pascoe, B., Meric, G., Yahara, K., Wimalarathna, H., Murray, S., Hitchings, M. D., et al. (2017). Local genes for local bacteria: evidence of allopatry in the genomes of transatlantic Campylobacter populations. Mol. Ecol. 26, 4497–4508. doi: 10.1111/mec.14176
Perera, V. N., Nachamkin, I., Ung, H., Patterson, J. H., McConville, M. J., Coloe, P. J., et al. (2007). Molecular mimicry in Campylobacter jejuni: role of the lipo-oligosaccharide core oligosaccharide in inducing anti-ganglioside antibodies. FEMS Immunol. Med. Microbiol. 50, 27–36. doi: 10.1111/j.1574-695X.2007.00225.x
Peters, S., Pascoe, B., Wu, Z., Bayliss, S. C., Zeng, X., Edwinson, A., et al. (2021). Campylobacter jejuni genotypes are associated with post-infection irritable bowel syndrome in humans. Commun. Biol. 4:1015. doi: 10.1038/s42003-021-02554-8
Pithadia, A. B., and Kakadia, N. (2010). Guillain-Barré syndrome (GBS). Pharmacol. Rep. 62, 220–232. doi: 10.1016/s1734-1140(10)70261-9
Poly, F., Serichatalergs, O., Schulman, M., Ju, J., Cates, C. N., Kanipes, M., et al. (2011). Discrimination of major capsular types of Campylobacter jejuni by multiplex PCR. J. Clin. Microbiol. 49, 1750–1757. doi: 10.1128/JCM.02348-10
Ruiz-Palacios, G. M. (2007). The health burden of Campylobacter infection and the impact of antimicrobial resistance: playing chicken. Clin. Infect. Dis. 44, 701–703. doi: 10.1086/509936
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics. 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sheppard, S. K., Colles, F. M., McCarthy, N. D., Strachan, N. J. C., Ogden, I. D., Forbes, K. J., et al. (2011). Niche segregation and genetic structure of Campylobacter jejuni populations from wild and agricultural host species. Mol. Ecol. 20, 3484–3490. doi: 10.1111/j.1365-294X.2011.05179.x
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). WHO pathogens priority list working group. discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3
Thépault, A., Méric, G., Rivoal, K., Pascoe, B., Mageiros, L., Touzain, F., et al. (2017). Genome-wide identification of host-segregating epidemiological markers for source attribution in Campylobacter jejuni. Appl. Environ. Microbiol. 83, e03085–16. doi: 10.1128/AEM.03085-16
Whitehouse, C. A., Zhao, S., and Tate, H. (2018). Antimicrobial resistance in Campylobacter species: mechanisms and genomic epidemiology. Adv. Appl. Microbiol. 103, 1–47. doi: 10.1016/bs.aambs.2018.01.001
Wilson, D. J., Gabriel, E., Leatherbarrow, A. J., Cheesbrough, J., Gee, S., Bolton, E., et al. (2008). Tracing the source of campylobacteriosis. PLoS Genet. 4:e1000203. doi: 10.1371/journal.pgen.1000203
Wolfe, N. D., Dunavan, C. P., and Diamond, J. (2007). Origins of major human infectious diseases. Nature 447, 279–283. doi: 10.1038/nature05775
Wysok, B., Wojtacka, J., Hänninen, M. L., and Kivistö, R. (2020). Antimicrobial resistance and virulence-associated markers in campylobacter strains from diarrheic and non-diarrheic humans in Poland. Front. Microbiol. 11:1799. doi: 10.3389/fmicb.2020.01799
Zang, X., Tang, H., Jiao, X., and Huang, J. (2016). Can a visual loop-mediated isothermal amplification assay stand out in different detection methods when monitoring Campylobacter jejuni from diverse sources of samples? Food Control. 75, 220–227. doi: 10.1016/j.foodcont.2016.12.010
Zankari, E., Allesøe, R., Joensen, K. G., Cavaco, L. M., Lund, O., and Aarestrup, F. M. (2017). PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 72, 2764–2768. doi: 10.1093/jac/dkx217
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: Campylobacter jejuni isolates from cattle, serotype HS19, zoonotic hazard, whole-genome sequencing, phylogenetic relatedness, IL-10–/– C57BL/6 mice
Citation: Zang X, Huang P, Li J, Jiao X and Huang J (2021) Genomic Relatedness, Antibiotic Resistance and Virulence Traits of Campylobacter jejuni HS19 Isolates From Cattle in China Indicate Pathogenic Potential. Front. Microbiol. 12:783750. doi: 10.3389/fmicb.2021.783750
Received: 26 September 2021; Accepted: 08 November 2021;
Published: 30 November 2021.
Edited by:
Xiaonan Lu, McGill University, CanadaReviewed by:
Luyao Ma, University of California, Davis, United StatesBen Pascoe, University of Bath, United Kingdom
Copyright © 2021 Zang, Huang, Li, Jiao and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinlin Huang, jinlin@yzu.edu.cn
 Xiaoqi Zang
Xiaoqi Zang Pingyu Huang2
Pingyu Huang2 Xinan Jiao
Xinan Jiao Jinlin Huang
Jinlin Huang