- Department of Biological Sciences, University of Calgary, Calgary, AB, Canada
Control of microbial reduction of sulfate to sulfide in oil reservoirs (a process referred to as souring) with nitrate has been researched extensively. Nitrate is reduced to nitrite, which is a strong inhibitor of sulfate-reducing bacteria (SRB). Perchlorate has been proposed as an alternative souring control agent. It is reduced to chlorate (ClO3-) and chlorite (ClO2-), which is dismutated to chloride and O2. These can react with sulfide to form sulfur. Chlorite is also highly biocidal. Here we compared the effectiveness of perchlorate and nitrate in inhibiting SRB activity in medium containing heavy oil from the Medicine Hat Glauconitic C (MHGC) field, which has a low reservoir temperature and is injected with nitrate to control souring. Using acetate, propionate and butyrate as electron donors, perchlorate-reducing bacteria (PRB) were obtained in enrichment culture and perchlorate-reducing Magnetospirillum spp. were isolated from MHGC produced waters. In batch experiments with MHGC oil as the electron donor, nitrate was reduced to nitrite and inhibited sulfate reduction. However, perchlorate was not reduced and did not inhibit sulfate reduction in these incubations. Bioreactor experiments were conducted with sand-packed glass columns, containing MHGC oil and inoculated with an oil-grown mesophilic SRB enrichment. Once active souring (reduction of 2 mM sulfate to sulfide) was observed, these were treated with nitrate and/or perchlorate. As in the batch experiments, 4 mM nitrate completely inhibited sulfide production, while partial inhibition occurred with 1 and 2 mM nitrate, but injection of 4 mM perchlorate did not inhibit sulfate reduction and perchlorate was not reduced. The enriched and isolated PRB were unable to use heavy oil components, like alkylbenzenes, which were readily used by nitrate-reducing bacteria. Hence perchlorate, injected into a low temperature heavy oil reservoir like the MHGC, may not be reduced to toxic intermediates making nitrate a preferable souring control agent.
Introduction
Waterflooded low temperature reservoirs have a constant temperature from injector to producing wells, allowing growth of different mesophilic bacteria or archaea depending on the electron acceptors present. The presence of sulfate in injection water stimulates the growth of sulfate-reducing bacteria (SRB), which reduce sulfate to sulfide (H2S or HS-) resulting in reservoir souring. H2S is a toxic and corrosive gas, which constitutes a health hazard to oilfield operators. Corrosion of carbon steel infrastructure, contamination of produced oil and reservoir plugging due to precipitation of metal sulfides are other souring-related problems (Vance and Thrasher, 2005). A commonly used control measure for reservoir souring is the injection of nitrate. This is reduced by nitrate-reducing bacteria (NRB) to nitrite and then mostly to N2. Nitrite, a strong metabolic inhibitor of SRB, binds to dissimilatory sulfite reductase which catalyzes the reduction of sulfite to sulfide as the last step in the sulfate reduction pathway. However, many NRB reduce nitrite further to dinitrogen (N2) removing the inhibition. Continuous injection of sulfate-containing, nitrate-amended water into a low temperature reservoir leads, therefore, to microbial zonation where NRB activity is limited to the near-injection wellbore region (NIWR) with SRB activity occurring deeper into the reservoir (Voordouw et al., 2009; Callbeck et al., 2011). Deeper zones of sulfate reduction may be controlled by the batch-wise injection of high concentrations of nitrate to push nitrate deeper into the reservoir (Voordouw et al., 2009; Callbeck et al., 2011) or by the combined injection of nitrate and biocides (Xue and Voordouw, 2015; Fida et al., 2018).
Perchlorate has been proposed as an alternative to nitrate in the control of reservoir souring (Coates, 2014; Liebensteiner et al., 2014). Perchlorate was first reported by Postgate (1952) to directly inhibit sulfate reduction in Desulfovibrio desulfuricans, when added at 10 times the concentration of sulfate in the growth medium. Since then, anaerobic reduction of perchlorate and chlorate has been studied extensively and the mechanism by which perchlorate inhibits sulfate reduction has been elucidated. Perchlorate- and/or chlorate-reducing bacteria (PRB) inhibit SRB via (i) biocompetitive exclusion (Engelbrektson et al., 2014; Liebensteiner et al., 2014), (ii) oxidation of sulfide to sulfur (Coates and Achenbach, 2004; Gregoire et al., 2014; Mehta-Kolte et al., 2017), and/or (iii) direct inhibition of the dissimilatory sulfate respiration pathway (Carlson et al., 2015). In (ii) sulfide is oxidized either biotically through enzyme-catalyzed reactions starting with perchlorate reductase (PcrAB) or through the abiotic reaction of sulfide with chlorite or oxygen formed as intermediate reaction products in microbial perchlorate reduction (Mehta-Kolte et al., 2017). Inhibition of sulfide production by perchlorate in a bioreactor column injected with sulfate and yeast extract has been reported by Engelbrektson et al. (2014). No sulfide was detected in the column effluent and perchlorate injection stimulated the emergence of a diverse microbial community (Engelbrektson et al., 2014).
Studies on the control of sulfide production with nitrate in heavy oil-containing bioreactor columns as well as in the field have shown that alkylbenzenes (toluene, ethylbenzene, and m- and p-xylene) were utilized as electron donors for nitrate reduction (Lambo et al., 2008; Agrawal et al., 2012; Suri et al., 2017), whereas alkylbenzenes and alkanes were used as electron donors for sulfate reduction (Agrawal et al., 2012). Acetate accumulated transiently as an intermediate in the metabolism of these oil components under sulfate-reducing, but not under nitrate-reducing conditions (Callbeck et al., 2013).
It is presently unknown whether perchlorate can prevent souring in oil-containing bioreactors and, by extension, in oil reservoirs. Dechloromonas strain RCB uses benzene as electron donor for perchlorate reduction (Coates et al., 2001; Chakraborty et al., 2005), but oxidized alkylbenzenes only under nitrate-reducing conditions. Pseudomonas chloritidismutans AW-1(T) couples the reduction of chlorate (not perchlorate) to the oxidative metabolism of n-alkanes (Mehboob et al., 2009, 2016). This organism activates hydrocarbon with a mono-oxygenase using oxygen generated by the dismutation of chlorite by the enzyme chlorite dismutase (Wolterink et al., 2002; Mehboob et al., 2009, 2016). Hence, the potential of PRB for controlling souring with perchlorate in bioreactors with oil as the sole electron donor is promising. The objective of the current work was, therefore, to directly compare control of sulfide production in low-temperature, oil-containing bioreactors with nitrate and perchlorate.
Materials and Methods
Source of Field Samples
Produced water samples were obtained from production well 18PW of the Medicine Hat Glauconitic C (MHGC) field near Medicine Hat, Alberta. This is a shallow reservoir (850 m below the surface; reservoir temperature 30°C) from which heavy oil with an American Petroleum Institute (API) gravity of 16° is produced by re-injection of produced water. The water samples were collected in sterile 1 L Nalgene bottles filled to the brim to exclude air during transport. Following arrival at the University of Calgary within half a day from collection, samples were transferred to a Coy anaerobic hood with an atmosphere of 90% N2 and 10% CO2 (N2–CO2).
Batch Culture Enrichments
Anaerobic Coleville synthetic brine medium K (CSBK; Fida et al., 2016), pH 7.5, was dispensed in 20 mL aliquots in 60 mL serum bottles with a head space of N2–CO2, sealed with butyl rubber stoppers and aluminum rings. This medium was amended with electron acceptors (e.g., sulfate, perchlorate, or nitrate) and electron donors like 3 mM volatile fatty acids (VFA, 3 mM each of acetate, propionate and butyrate), 3 mM toluene, 2 mM ethylbenzene, or 1 mL of MHGC oil, as indicated. Batch cultures were inoculated with 5% (v/v) of 18PW unless stated otherwise and were incubated at 30°C.
Serum bottles used to determine the time course of growth with VFA and perchlorate, VFA and nitrate, or VFA and nitrate and perchlorate were inoculated with a PRB enrichment. Samples (1 mL) were taken periodically using N2–CO2 flushed syringes to determine growth (OD600), the concentrations of nitrite (NO2-), nitrate (NO3-), and perchlorate (ClO4-) and to isolate DNA for community analysis.
Inhibition of Oil-Dependent Sulfate Reduction With Perchlorate and/or Nitrate in Batch Cultures
Aliquots (20 mL) of anaerobic CSBK medium in 60 mL serum bottles were amended with 1 mL of MHGC oil (O) and either 5 mM sulfate (S), 5 mM sulfate and 10 mM nitrate (SN), 5 mM sulfate and 10 mM perchlorate (SP), 5 mM sulfate and 10 mM nitrate and 10 mM perchlorate (SNP), or 10 mM perchlorate (P). Bottles were inoculated with 1 mL of 18PW, concentrated 20-fold by centrifugation (Okpala et al., 2017). Sterile controls, containing 20 mL of CSBK medium, 1 mL of MHGC oil and electron acceptors, but no inoculum, were also prepared. Concentrations of sulfate, nitrate, nitrite, perchlorate, and aqueous sulfide were determined as a function of time in duplicate incubations. The concentrations of residual alkylbenzenes and alkanes present in dichloromethane (DCM) extracts of the residual oil were determined using gas chromatography-mass spectrometry (GC-MS).
Control of Souring With Nitrate and/or Perchlorate in Oil-Containing Bioreactors
Glass syringe bioreactors (50 mL) were assembled essentially as described by Callbeck et al. (2011). Bioreactors were sealed at the bottom with a layer of glass wool and polymeric mesh and were tightly packed with silica sand (Sigma-Aldrich, 50–70 mesh). The upper ends were sealed with a layer of polymeric mesh and a rubber stopper perforated with a 1 mL syringe through which effluent flowed into an effluent collector. Sterile anaerobic CSBK from a medium reservoir was injected through the inlet bottom end of the bioreactor using a Minipuls-3 multi-channel peristaltic pump (Gilson Inc.). The medium reservoir was maintained anaerobic through injection of N2–CO2 in the headspace. Once the columns were saturated with medium, their wet weight was determined to calculate their pore volumes (PV, wet weight minus dry weight), which was approximately 25 mL for each column. The columns were then flooded first with 1 PV of MHGC oil and then with multiple PV of anoxic CSBK medium until approximately 0.5 PV of oil was produced, as determined spectrophotometrically (Gassara et al., 2015). At that point little further oil production was observed and approximately 0.5 PV of oil remained in the columns. The columns were then inoculated with SRB culture grown with MHGC oil and sulfate. The three-way valves at the inlet and outlet ends were closed and the columns were incubated for 20 days. Following incubation, injection of anoxic medium containing 2 mM sulfate was resumed at a flow rate of 0.5 PV/day. Concentrations of sulfate, sulfide and other anions tested were monitored for every 1 PV of effluent.
Reduction of Perchlorate or Nitrate With Alkylbenzene Electron Donors
Perchlorate and nitrate reduction with toluene or ethylbenzene were compared in serum bottles with 20 mL of CBSK medium, containing 10 mM perchlorate or 10 mM nitrate and 3 mM of aqueous toluene or 2 mM of aqueous ethylbenzene. Alternatively, 1 mL of 60 mM toluene or 40 mM ethylbenzene in the inert carrier 2,2,4,4,6,8,8-heptamethylnonane (HMN) was added. The inoculum used was either 1 mL of 20-fold concentrated 18PW or 2 mL of a chemostat culture derived from 18PW, which was growing on toluene and nitrate or on ethylbenzene and nitrate.
Isolation and Identification of PRB
Perchlorate-reducing bacteria enrichments obtained with VFA and perchlorate at 30°C were diluted 10-fold and plated on CSBK medium with 3 mM VFA and 10 mM perchlorate, solidified with 15 g/L of agar. The plates were incubated at 30°C in anaerobic jars flushed with N2–CO2. Individual colonies were picked and transferred to CSBK medium with 3 mM VFA and 10 mM perchlorate. The isolates were phylogenetically identified by Sanger sequencing of 1500-bp 16S rRNA gene amplicons obtained with universal primers 27F and 1525R (Frank et al., 2008) at the Core DNA Services Laboratory of the University of Calgary. The 16S rRNA gene sequences of the isolates and of reference sequences, retrieved from GenBank, were aligned with Clustal W (Thompson et al., 1994). A phylogenetic tree was constructed using MEGA version 6 (Tamura et al., 2013). The evolutionary history was inferred using the Neighbor-Joining algorithm (Saitou and Nei, 1987). The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al., 2004) in units of the number of base substitutions per site. Confidence estimates of branch clusters were obtained from bootstrap tests of 1000 replicates (Felsenstein, 1985).
Microbial Community Analysis by Illumina Sequencing
DNA was isolated from enrichments using the Fast DNA Spin Kit for Soil and the FastPrep Instrument (MP Biomedicals, Santa Ana, CA, United States) as per the manufacturer’s instructions. The extracted DNAs were quantified, with the Quant-iTTM dsDNA HS assay kit using a Qubit fluorimeter (Invitrogen) and were subjected to a two-step PCR amplification of 16S rRNA. PCR reactions were carried out in duplicate with each replicate containing a 20 μl reaction volume. The amplified PCR products were pooled and cleaned using the QIAquick PCR purification kit (Qiagen) and analyzed on a 1.5% agarose gel. In the first step non-barcoded primers 926Fi5 and 1392Ri7 were used, while the barcoded primers P5-S50X-OHAF and P7-N7XX-OHAF were used in the second step. PCR conditions were 95°C for 5 min, followed by 25 cycles of 95°C for 45 s, 55°C for 2 min, 72°C for 4 min, followed by a final incubation at 72°C for 10 min in the first step and 95°C for 3 min, followed by 10 cycles of 95°C for 45 s, 55°C for 2 min, 72°C for 4 min, followed by a final incubation at 72°C for 10 min in the second step. The final concentrated PCR products were diluted with the Qiagen elution buffer to 4 ng/μl. The 16S amplicons were sequenced using the 300PE (paired-end) MiSeq protocol of the Illumina Miseq system at the Energy Bioengineering and Geomicrobiology Group (EBG) of the University of Calgary.
Analyses of sequencing data involved merging the 300PE reads from both ends with a minimum overlap of 50 bp and a minimum length of 450 bp as cut-offs using the PEAR 0.9.6 software. The merged reads were processed with MetaAmp (Dong et al., 2017b). The sequencing data retrieved were clustered into operational taxonomic units (OTUs) at a taxonomic distance of 3%. Rarefaction curves and alpha diversity indices were calculated including Chao1 (Chao, 1984) and Shannon’s H-index (Shannon, 1948). Relational trees showing the beta diversity of the amplicon sequences were visualized using MEGA 6 (Tamura et al., 2013). All sequences have been submitted to NCBI Sequence Read Archive (SRA) under Bioproject accession number PRJNA181037 with biosample numbers SAMN09519490 to SAMN09519497 and SAMN09521198 to SAMN09521209.
Analytical Techniques
Samples for anion assays were prepared by centrifugation at 14,000 ×g for 5 min, after which 100 μl of supernatant was added to 400 μl of the prepared buffer solution in a vial. Concentrations of NO3-, NO2-, and ClO4- in the samples were analyzed by high-pressure liquid chromatography (HPLC) using a Waters 600E instrument (Waters Corp, Milford, MA, United States), which was fitted with an IC-PAKTM anion HC column (150 mm × 4.6 mm, Waters) and a Waters 2489 UV/Visible detector, set at 220 nm. Nitrate and nitrite were eluted with a sodium borate-gluconate buffer containing 12% (v/v) acetonitrile and 2% (v/v) butanol at a flow rate of 2 mL/min. Perchlorate was eluted using an alkaline buffer mixture of 25 mM NH4HCO3 in 50% (v/v) acetonitrile, pH 10 (adjusted with NH4OH) at a flowrate of 1.5 mL/min. Perchlorate was detected with a Waters 432 conductivity detector using the same column as above. The concentration of VFA components was analyzed with a Prevail organic acid (OA) 5u column (250 mm × 4.6 mm, Alltech, Guelph, ON, Canada) fitted with a Waters 2487 UV detector at 210 nm.
Gas Chromatography-Mass Spectrometry (GC-MS) Analysis of Oil Components
Extraction of oil components with dichloromethane (DCM) was done as described by Agrawal et al. (2012). Prior to DCM extraction of the oil, 50 μL of squalene and mesitylene were added to the 1 mL oil layer of each incubation as internal standards for quantifying the depletion of n-alkanes and alkylbenzenes, respectively. Following this, 9 mL of DCM was added to extract the oil components and one microliter (μL) of the oil-DCM layer was injected by an autoinjector (7683B series, Agilent Technologies) into a GC (7890N series, Agilent Technologies), connected to an MS (5975C inert XL MSD series, Agilent). The GC was equipped with an HP-1 fused silica capillary column (length 50 m, inner diameter 0.32 mm, film thickness 0.52 μm; J&W Scientific) with helium as carrier gas. The amounts of oil components utilized for the reduction of the electron acceptors were determined as the decrease in the ratio of the peak area for a given component to that of the internal standard.
Results
Activity of PRB in the 18PW Field Sample
The results of inoculation of produced water 18PW from the MHGC field into CSBK medium with VFA and perchlorate and incubation at 30°C are shown in Figure 1. Perchlorate was reduced after a 4-day lag phase with complete reduction after 24 days of incubation (Figure 1A). All three VFA (acetate, propionate, and butyrate) were used for perchlorate reduction (Figure 1B), which indicates their suitability as electron donors for reduction of perchlorate in the MHGC field.
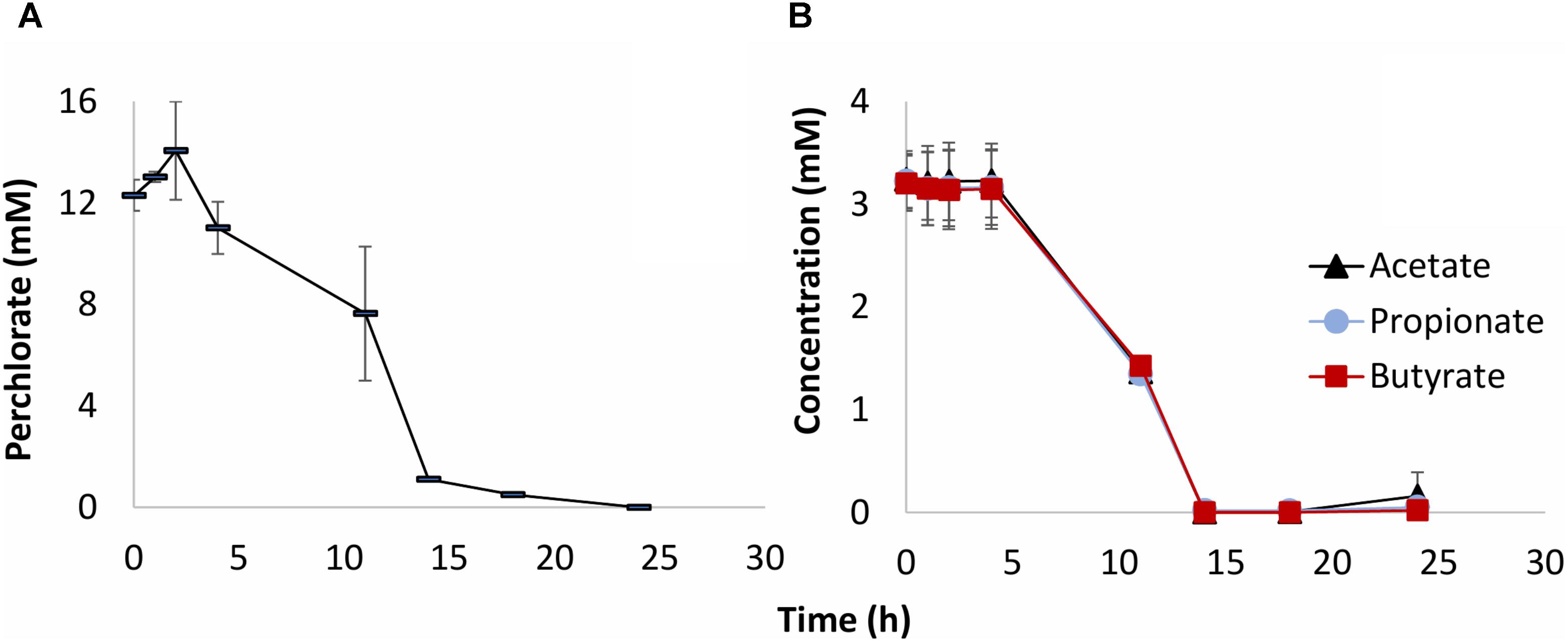
FIGURE 1. Activity of PRB in CSBK medium with 10 mM perchlorate and 3 mM VFA, inoculated with 5% (v/v) of produced water sample 18PW and incubated at 30°C. The concentrations of perchlorate (A) of VFA (B) are shown as a function of time (h). The data presented are averages of duplicate incubations.
Growth and Community Composition of Enrichments With Nitrate and/or Perchlorate
The primary enrichment with VFA and perchlorate (Figure 1) was used to inoculate secondary enrichments in media with VFA and nitrate, VFA and perchlorate, or VFA and nitrate and perchlorate (Figure 2). In cultures with VFA and nitrate growth was indicated as an increase in OD600 to 0.27 at 42 h after which the OD600 was relatively constant up to 336 h (Figure 2A). Nitrate was completely reduced at 48 h with transient formation of 1.2 mM nitrite (Figure 2B). Microbial community compositions at 48 h indicated Pseudomonas (97%) and Thauera (2%), which are genera of known heterotrophic NRB (hNRB). At 336 h the fraction of Pseudomonas had decreased to 74%, whereas that of Thauera had increased to 10.5% (Figure 2C and Supplementary Table S1). Increases were also seen for the genera Petrimonas (0.01–1.74%) and Acholeplasma (0.15–1.7%).
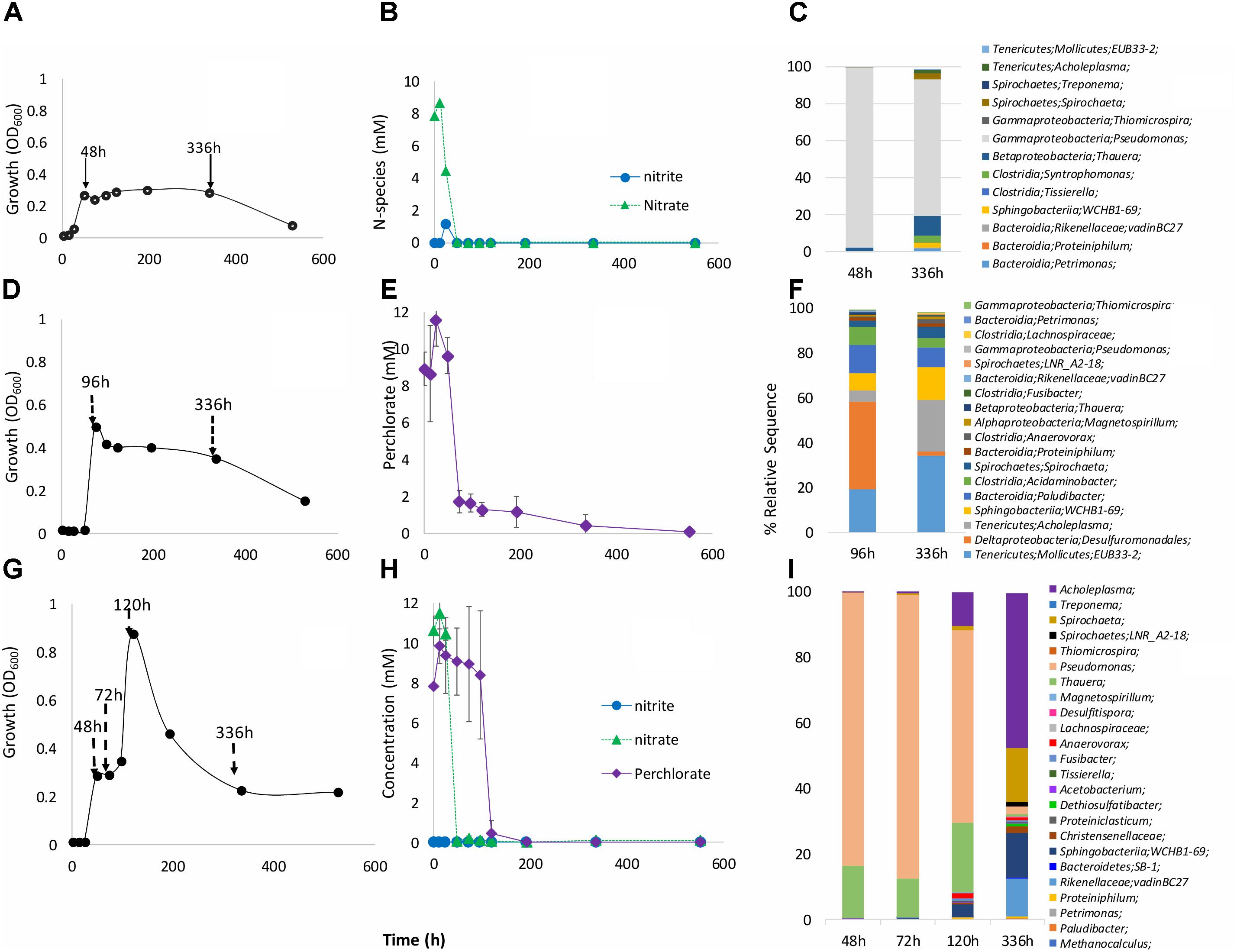
FIGURE 2. Time course of nitrate (A–C), perchlorate (D–F) or nitrate and perchlorate (G–I) reduction in 18PW communities incubated in CSBK medium containing 10 mM each of nitrate and/or perchlorate and VFA. (A,D,G) Growth OD at 600 nm; (B,E,H) concentrations of electron acceptors; (C,F,I) microbial community composition at T = 48, 72, 96, 120, and 336 h, as indicated (Supplementary Table S1).
In enrichments with perchlorate a peak OD600 of 0.50 was observed at 96 h (Figure 2D). Most perchlorate in the medium was reduced during this period with a slower reduction observed from 96 to 336 h (Figure 2E). The microbial community in the perchlorate-reducing enrichment was more diverse, than that in the nitrate-reducing enrichment (Supplementary Table S1: Shannon indices of 1.93 and 2.06 and of 0.18 and 1.13, respectively). The dominant community members at 96 h (Figure 2F and Supplementary Table S1) were Desulfuromonadales (39.2%), Mollicutes EUB33-2 (19.3%), Acholeplasma (5.0%), Acidaminobacter (7.9%), Sphingobacteria WCHB1-69 (7.4%), Paludibacter (12.7%), and Magnetospirillum (0.8%). At 336-h increased fractions were observed for the Mollicutes (34%), Sphingobacteria (14.3%), and Acholeplasma (23.1%).
In medium containing both nitrate and perchlorate, biphasic growth was observed as perchlorate reduction was delayed until nitrate reduction was complete (Figures 2G,H). Initial nitrate-reducing growth occurred from 0 to 48 h, with OD600 = 0.27 at 48 h. This was followed by perchlorate-reducing growth from 72 to 120 h with OD600 = 0.88 at 120 h (Figure 2G). These increases in OD600 (0.27 and 0.51 for nitrate and perchlorate) were very similar to those observed in cultures with only nitrate or perchlorate. Examination of early microbial community compositions showed dominance of Pseudomonas (83% at 48 h and 86% at 72 h) and Thauera (16% at 48 h and 11.8% at 72 h). From 120 to 336 h the taxa Acholeplasma (10.4 to 47.2%), Sphingobacteria (4 to 13.8%), Fusibacter (0.85 to 0.46%), and Rikenellaceae (0.12 to 11.6%) were observed (Figure 2I and Supplementary Table S1).
Effects of Perchlorate and Nitrate on Sulfide Production in Batch Cultures With Oil
Batch incubations were started to determine the effect of nitrate, perchlorate, or nitrate and perchlorate on sulfate reduction in CSBK medium containing 1 mL of MHGC oil and inoculated with concentrated 18PW. Changes in the composition of the microbial community of selected oil components were also monitored.
Sulfate reduction with oil as electron donor proceeded slowly. Following a lag phase of 24 days reduction of 5 mM sulfate to 3.3 mM aqueous sulfide was observed after 77 days of incubation (Figure 3A: SO). In the presence of 10 mM nitrate, sulfate reduction was completely inhibited. Nitrate was completely reduced after 24 days, with peak nitrite (3.5 mM) occurring at day 9. The nitrite concentration was further decreased to 1 mM at day 24 and this concentration persisted preventing the start of sulfate reduction (Figure 3B; SNO). In the presence of 10 mM perchlorate a longer lag phase of 65 days was observed before the start of sulfate reduction, indicating some inhibition of sulfate reduction. However, sulfate reduction was not prevented whereas perchlorate was not reduced (Figure 3C: PSO). No perchlorate reduction was observed in incubations in which only perchlorate and oil were present (Figure 2E: PO). In incubations with all three electron acceptors (nitrate, sulfate, and perchlorate) nitrate was reduced first with transient formation of 4 mM nitrite. Nitrate and nitrite were completely reduced after 34 and 65 days, respectively. Perchlorate reduction was not observed and was unable to prevent reduction of sulfate to sulfide, which started after 100 days (Figure 3D: PSNO).
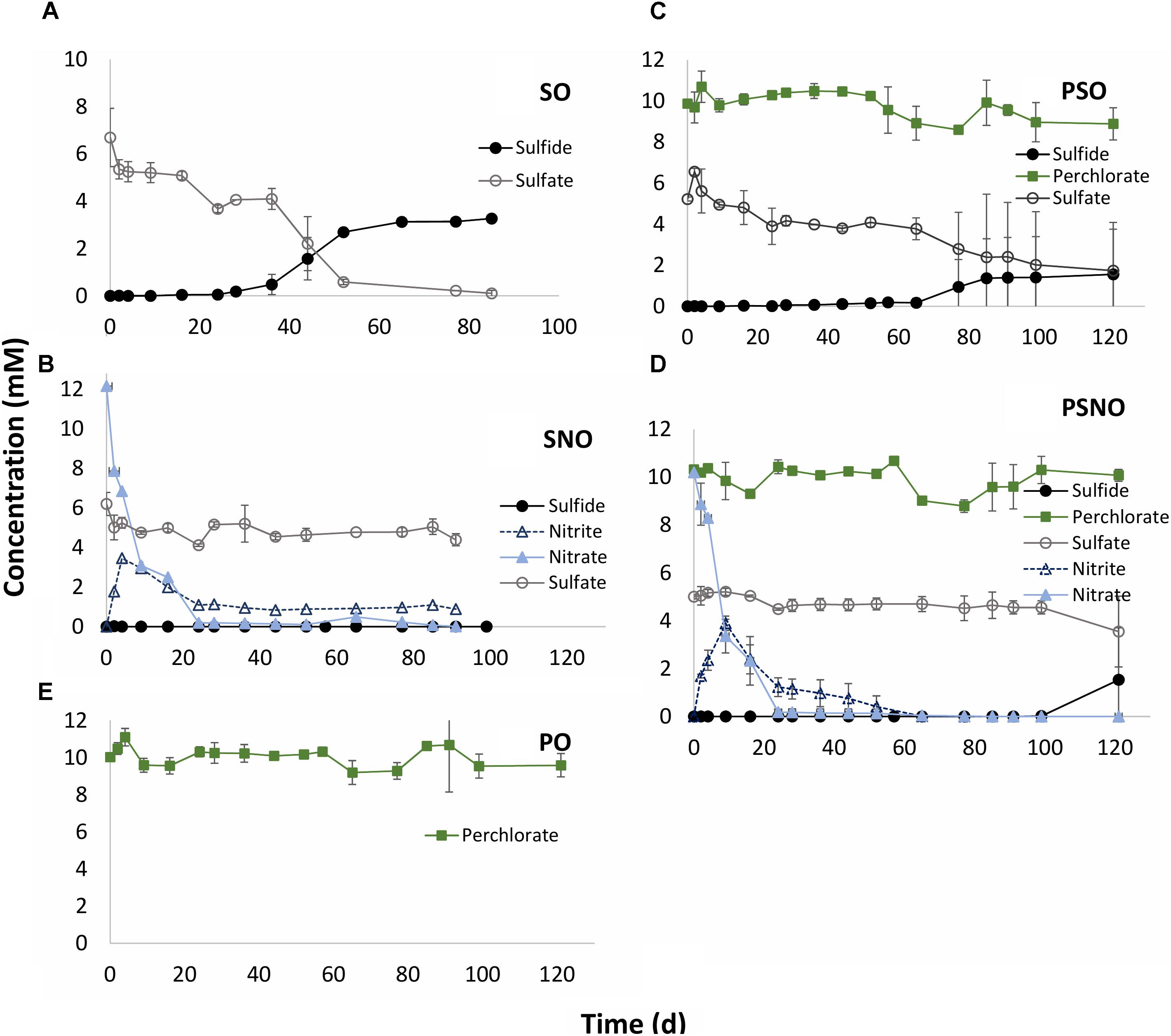
FIGURE 3. Inhibition of sulfide production by oil grown SRB with nitrate, perchlorate or nitrate, and perchlorate. CSBK medium containing 1 mL MHGC oil (represented as O) as electron donor was inoculated with a 20-fold concentrated 18PW sample. Medium was amended with 5 mM sulfate – SO (A); 5 mM sulfate and 10 mM nitrate – SNO (B); 5 mM sulfate and 10 mM perchlorate – PSO (C); 5 mM sulfate and 10 mM each of perchlorate and nitrate – PSNO (D); and 10 mM of perchlorate only – PO (E). Data presented are means of duplicate incubations.
Analysis of the alkylbenzenes toluene, ethylbenzene, m,p-xylene and o-xylene remaining in the oil after 123 days of incubation showed that these were mostly used in incubations with sulfate and that these were completely used in incubations with sulfate and nitrate with the exception of o-xylene (Supplementary Figure S1: SO and SNO). These alkylbenzenes were not used in incubations with perchlorate only (Supplementary Figure S1: PO). Their use in incubations with sulfate and perchlorate and with sulfate, nitrate, and perchlorate (Supplementary Figure S1: PSO and PSNO) was, therefore, for reduction of sulfate and of sulfate and nitrate, respectively, not for reduction of perchlorate.
Microbial Community Compositions in Batch Cultures With Oil
Illumina sequencing of 16S rRNA amplicons and analysis of the sequences obtained indicated that communities in incubations with nitrate were less diverse than those in incubations without nitrate (Supplementary Table S2: Shannon indices of 0.76–2.04 and 2.82–3.93, respectively). Likewise, comparison of microbial communities derived from these sequenced amplicons in a dendrogram indicated that communities in incubations without nitrate clustered separately from communities in incubations with nitrate (Figure 4: clusters I and II, respectively). In cluster I communities from incubations with sulfate (SO) and sulfate and perchlorate (PSO) were distinct from incubations with perchlorate only (PO), whereas in cluster II communities in short term incubations (9 days) were distinct from those in longer term incubations (65–123 days). At the class level, cluster I had higher fractions of Deltaproteobacteria, whereas cluster II had higher fractions of Beta- and Gammaproteobacteria (Figure 4A). Communities in cluster I also had higher fractions of the phyla Atribacteria, Euryarchaeota, Firmicutes, and Spirochaetae (Figure 4B). Unique to cluster I was the candidate phylum Cloacimonetes, which is mostly found in anaerobic environments, often in close association with Firmicutes (Stolze et al., 2016). Cloacimonetes have been predicted to be syntrophic propionate fermenters (Hamilton et al., 2016) producing CO2 and H2, which is used by hydrogenotrophic methanogens or SRB (Stolze et al., 2016).
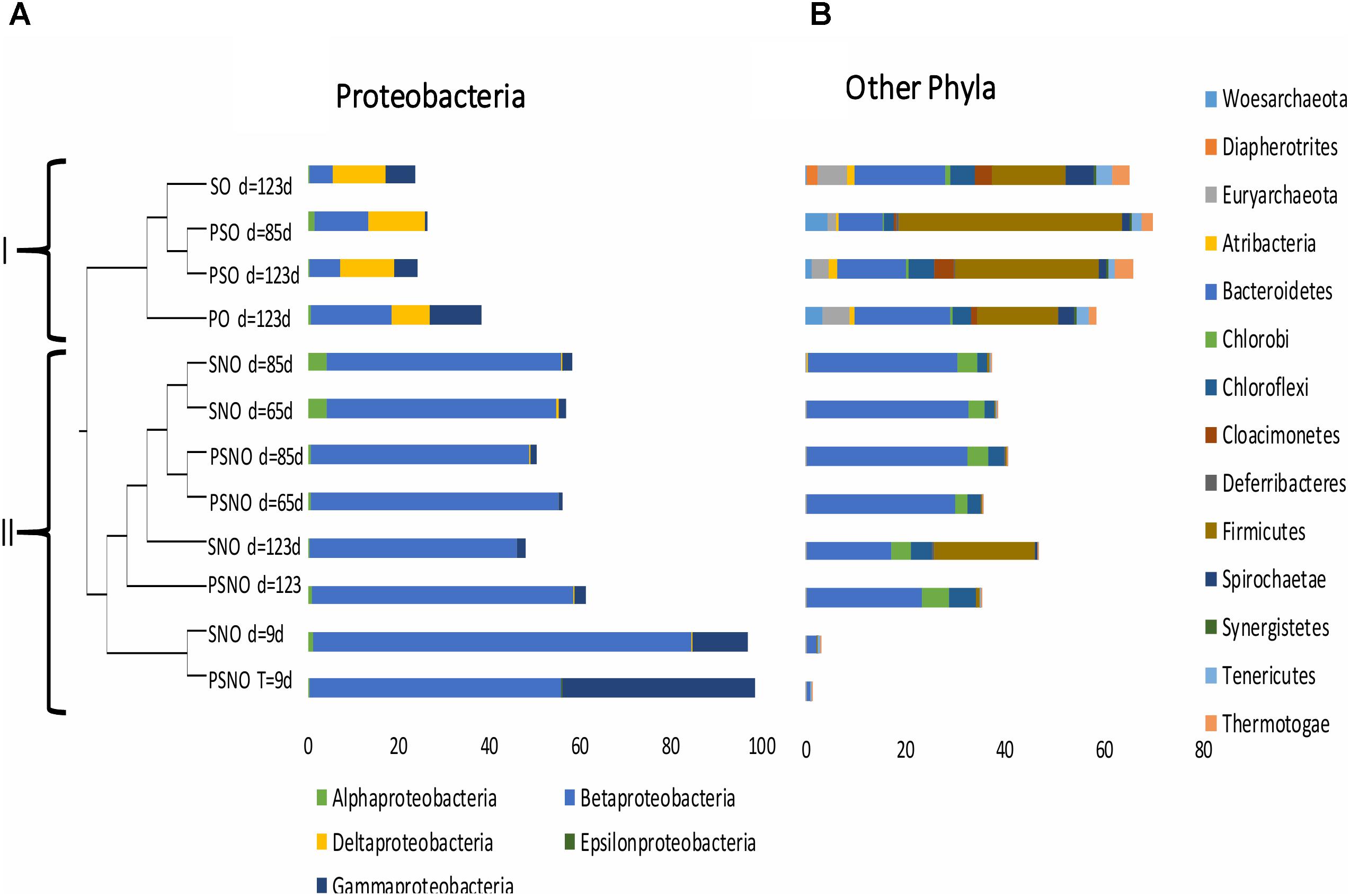
FIGURE 4. Relational tree for 16S rRNA gene libraries from incubations of CSBK medium, inoculated with 18PW and amended with 1 mL MHGC oil and sulfate (SO); sulfate and nitrate (SNO); sulfate and perchlorate (PSO); sulfate, nitrate, and perchlorate (PSNO); or perchlorate only (PO). Community DNA was obtained from cells harvested at day 9, 65, 85, or 123 as indicated. The fractions of reads in classes of Proteobacteria (A) and in other phyla (B) are shown.
At the genus level all nitrate-reducing communities in cluster II had high fractions of Thauera (Supplementary Table S2, entry #1: 44–82%), whereas high fractions of Pseudomonas were observed in early communities (Supplementary Table S2, entry #4: 12–43%). All communities in cluster I had high fractions of Smithella (Supplementary Table S2, entry #9: 2.8–6.5%) and Desulfomicrobium (Supplementary Table S2, entry #18: 0.9–2.4%). However, the putative SRB Desulfarculus, Desulfobacteraceae, and Desulfocurvus were mostly present in the SO and PSO and not in the PO incubations (Supplementary Table S2, entries #16, 20, and 22). Pelotomaculum of the phylum Firmicutes, identified in the SO and PSO incubations (Supplementary Table S2, entry #3: 11–40%), was also lacking in the PO incubations. Dong et al. (2017a) showed that a Pelotomaculum sp., which coupled the reduction of sulfate to the anaerobic oxidation of benzene, possessed genes for dissimilatory sulfate reduction as found in other Gram-positive SRB. In contrast, the microbial community in the PO incubations did not have taxa that were absent in the SO and PSO incubations, which is consistent with the fact that the communities in the PO incubations did not catalyze perchlorate reduction.
Souring Control in Oil-Containing Bioreactors
Eight sand packed columns containing 0.5 PV of residual MHGC oil, were set up to evaluate the control of sulfide production by SRB using nitrate, perchlorate or a combination of these. Duplicate 50 mL glass columns were used for each treatment condition, and two columns remained untreated. To ensure uniformity, all the columns were inoculated with the same SRB culture enriched with MHGC oil and 2 mM sulfate. Following a 20-day incubation period, these columns were flooded continuously with anaerobic CSBK medium, containing 2 mM sulfate and monitored until approximately 2 mM sulfide was produced. Once souring had been established, treatment with either nitrate or perchlorate was started.
In the untreated columns sulfide was observed on day 4 (0.88 ± 0.06 mM), reaching 1.8 ± 0.11 mM on day 45. Production of up to 2 mM sulfide was then maintained for the duration of the experiment up to day 150 (Figure 5A). Injection of soured duplicate columns with 2 mM sulfate and 1 mM nitrate decreased sulfide to 0.1 mM for the first 2 PV after which sulfide production recovered. Increasing the nitrate concentration to 2 mM also caused only transient partial inhibition of sulfide production. However, when the injected nitrate concentration was increased to 4 mM, sulfide production was completely inhibited and the measured effluent sulfate concentrations reached 2 mM. Nitrate (0.6 mM) was detected in the first PV of bioreactor effluent, whereas nitrite was not detected (Figure 5B). When nitrate injection was stopped, the effluent sulfide concentration recovered to 2 mM by day 100.
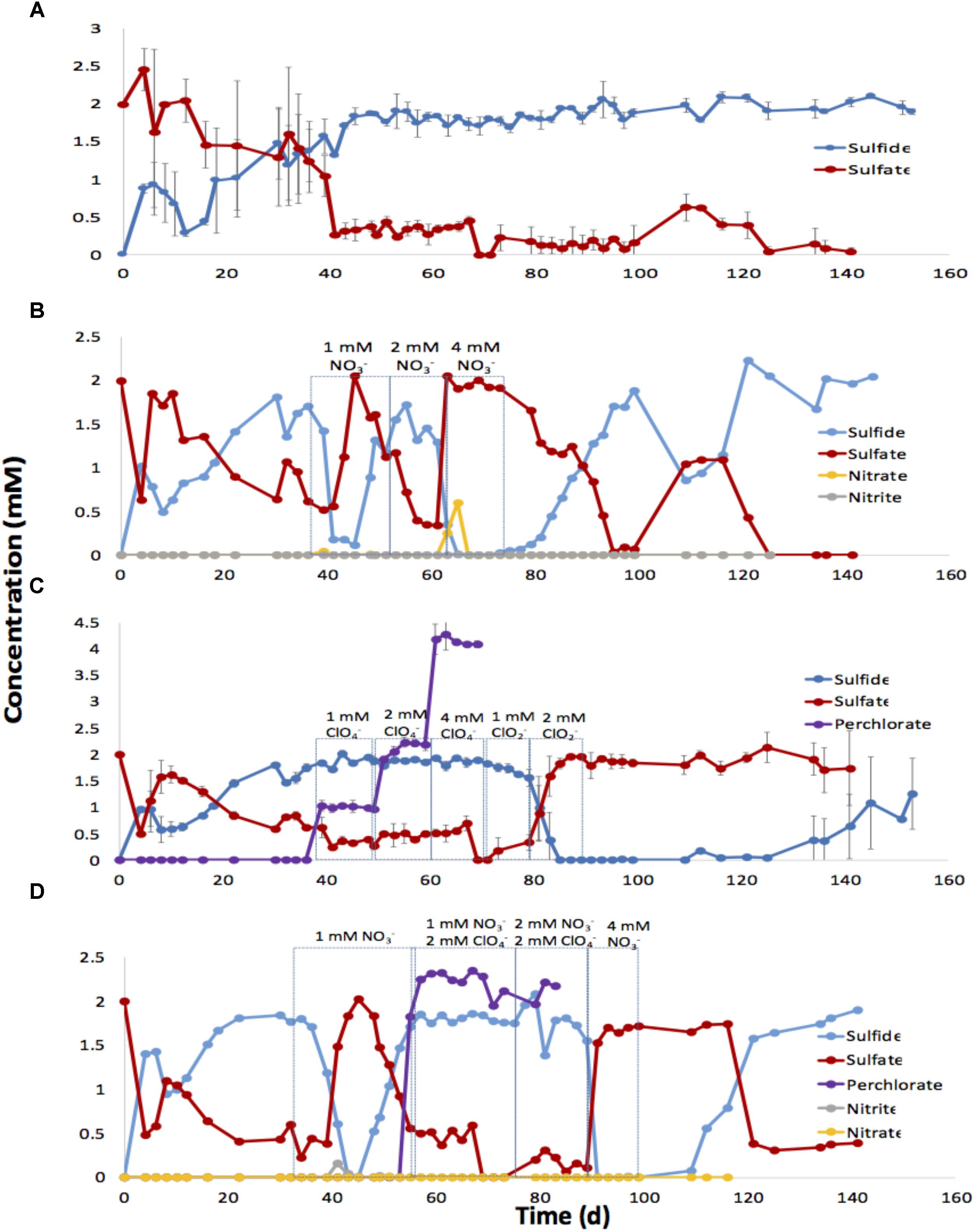
FIGURE 5. Effect of nitrate, perchlorate or chlorite, or nitrate and perchlorate on sulfide production in low temperature, oil-containing bioreactors. Bioreactors were inoculated with SRB enrichments of 18PW grown in CBSK medium with sulfate and MHGC oil. Bioreactors were continuously injected with CBSK medium containing 2 mM sulfate at a flowrate of 0.5 PV/day. The effluent concentrations of sulfate, sulfide, nitrate, nitrite, and perchlorate are shown for untreated columns (A), columns injected with nitrate (B), columns injected with perchlorate or chlorite (C), and columns injected with nitrate and perchlorate (D) as a function of time.
Injection of perchlorate at increasing concentrations of 1, 2, or 4 mM did not control the activity of SRB in the bioreactors as shown in Figure 5C. As in batch cultures incubation with oil, perchlorate was not reduced in the bioreactors and the injected concentrations were fully recovered in the effluent. Since perchlorate showed no inhibition of SRB activity, chlorite was injected instead (Figure 5C). Injection of 1 mM chlorite decreased the effluent sulfide concentration from 1.9 to 1.6 mM, whereas injection of 2 mM chlorite completely inhibited sulfide production by SRB in the bioreactors. Recovery of SRB activity, following the cessation of the chlorite injection, required injection of 28 PV of the CSBK medium with 2 mM sulfate to reach 1 mM sulfide (Figure 5C).
Combined injection of nitrate and perchlorate is shown in Figure 5D. Injection of 1 mM nitrate only caused transient inhibition of sulfide production. Sulfide concentrations decreased from 1.8 to 0 mM following injection of the first 5 PV and then recovered to 1.8 mM. Nitrate was reduced in the bioreactors and only nitrite was detected transiently in the effluent. Combining 2 mM perchlorate with 1 or 2 mM nitrate did not increase inhibition of sulfate reduction, as effluent sulfide remained at 1.8 mM. Switching the injection medium to CSBK with 4 mM nitrate and 2 mM sulfate gave immediate inhibition of sulfide production and increased the effluent sulfate concentration to 1.7 mM. Sulfate reduction resumed 22 days after nitrate injection was stopped (Figure 5D).
Reduction of Nitrite and Chlorite in the Presence of Oilfield Microbes
Nitrite and chlorite are potentially inhibitory products of nitrate and perchlorate reduction. In order to compare their fate directly, batch cultures containing 50 mL of CSBK medium, 1 mL MHGC oil or 3 mM VFA and 4 mM nitrite or 4 mM chlorite, were inoculated with 10% (v/v) 18PW and incubated at 30°C for 30 days. The results indicated that oil field hNRB rapidly reduced nitrite with VFA, while nitrite reduction with oil progressed more slowly over a 26-day period (Figure 6). Chlorite on the other hand was not reduced and remained at a constant concentration in the presence of VFA or oil (Figure 6). Chlorite can be disproportionated to chloride and oxygen through a non-energy yielding reaction, catalyzed by chlorite dismutase (Rikken et al., 1996; van Ginkel et al., 1996; Bender et al., 2005; Bardiya and Bae, 2011; Schaffner et al., 2015). However, this did not occur in the incubations in Figure 6. Chlorite is a strong oxidant, which reacts non-specifically with biomolecules resulting in cell damage and death (McDonnell and Russell, 1999). The results suggest that 4 mM chlorite killed the microbes present in the incubations; chlorite did not react abiotically with either oil or VFA. Hence, nitrite and chlorite behaved very differently with nitrite having less acute toxicity than chlorite.
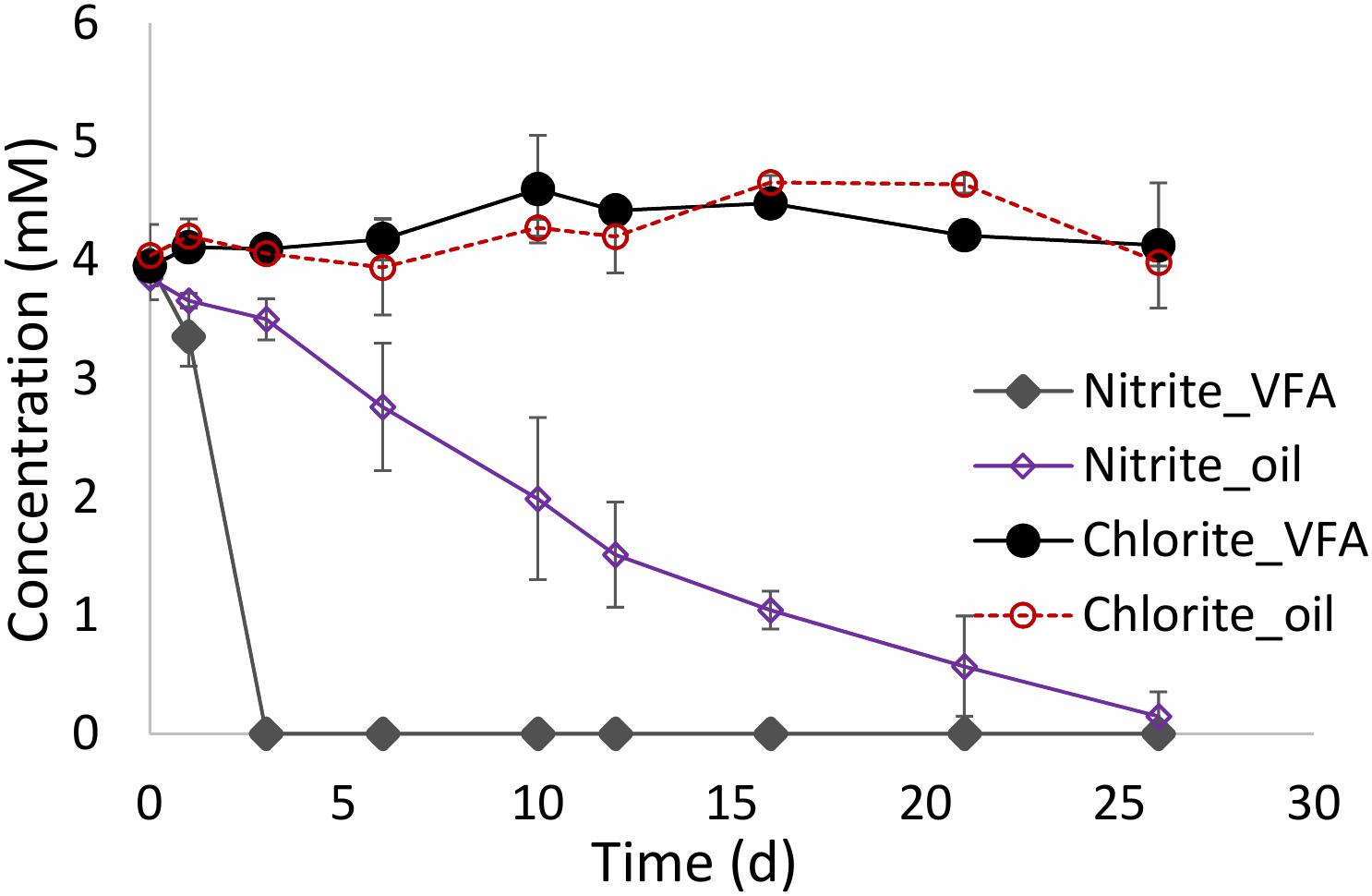
FIGURE 6. Comparison of reduction of nitrite and chlorite in medium containing VFA or MHGC oil and inoculated with 10% (v/v) 18PW.
Search for Alkylbenzene-Oxidizing PRB
Although perchlorate concentrations remained unchanged in batch culture and bioreactor experiments with oil, some further attempts were made to enrich microbes using the alkylbenzenes toluene or ethylbenzene as electron donor for perchlorate reduction. Alkylbenzenes were added directly to 50 mL aqueous CSBK medium or to 1 mL of HMN overlaying this medium to minimize toxicity. A 20-fold concentrated 18PW sample (1 mL) or a nitrate-reducing chemostat culture (5 mL), continuously fed with either toluene or ethylbenzene (Suri, 2017), were used as inocula. However, these were unable to use toluene or ethylbenzene for reduction of 10 mM perchlorate within 30 days of incubation. When incubations inoculated with chemostat culture were amended with 5 mM nitrate after day 30, nitrate was reduced with transient formation of nitrite in all four incubations (Supplementary Figures S2A–D), indicating that the alkylbenzene-utilizing hNRB demonstrated in Supplementary Figure S1 were present. When incubations inoculated with concentrated 18PW were amended with 10 mM acetate after day 30, perchlorate was reduced in all four incubations (Supplementary Figures S2E–H), indicating that the acetate-utilizing PRB demonstrated in Figures 1, 2 were present. Hence, the chemostat cultures contained toluene- and ethylbenzene-oxidizing hNRB, but no PRB capable of using these substrates. PRB oxidizing acetate were present.
Characterization of VFA-Oxidizing PRB Isolated From MHGC Produced Water
Plating of perchlorate-reducing enrichments, as in Figure 1, on agar-solidified CSBK medium containing perchlorate and VFA yielded isolates PRB2 and PRB4. 16S rRNA gene sequence analysis indicated that these were closely affiliated with the genus Magnetospirillum. A phylogenetic tree of nearly full length 16S rRNA gene sequences (1367 bp) is presented in Figure 7. Grouping of the 16S rRNA gene sequences of PRB2 and PRB4 with Magnetospirillum moscoviense BB-1 was supported by a bootstrap value of 92%. The 16S rRNA gene sequence similarities of PRB2 and M. moscoviense BB-1, and of PRB4 and M. moscoviense BB-1 were 93.0 and 92.7%, respectively, whereas the pair-wise similarities to other Magnetospirillum strains were between 88.1 and 90.6% (Supplementary Table S3). PRB2 and PRB4 shared 99.2% 16S rRNA gene sequence similarity and clustered together with bootstrap support of 100%.
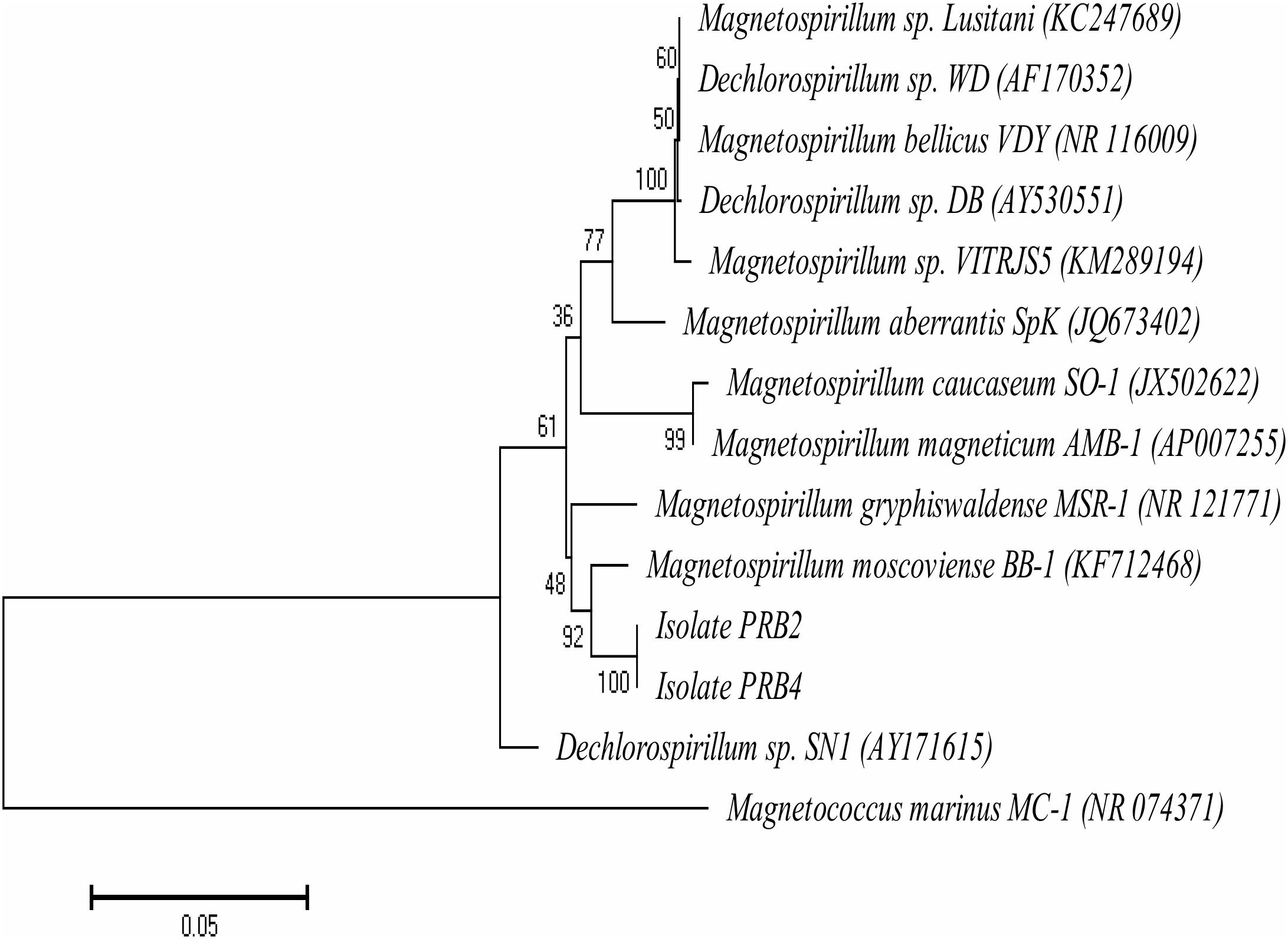
FIGURE 7. Neighbor-joining phylogenetic tree indicating the affiliation of isolates PRB2 and PRB4 with members of the genus Magnetospirillum based on the 16S rRNA gene sequences. Branching points are determined as percentage of bootstrap values based on 1000 replications with those having a cut-off at 50% shown. The scale bar of 0.05 represents the fractional change per nucleotide. The sequence of Magnetococcus marinus MC-1 (NR_074371) was used as an outgroup.
The electron donor and acceptor specificity of PRB2 and PRB4, determined after 7 days of incubation at 30°C, were found to be very similar (Supplementary Table S4). Both coupled the reduction of perchlorate to oxidation of acetate, propionate and butyrate, as well as of lactate, succinate, glucose, glutarate and ethanol but not of methanol. Slow growth and perchlorate reduction occurred in medium with a headspace with 20% (v/v) H2, whereas no growth or perchlorate reduction occurred with sulfide. Benzene, toluene, or ethylbenzene were not used. Chlorate accumulation was not observed during growth with any of the electron donors utilized. In addition to perchlorate, PRB2 and PRB4 used chlorate, nitrate, and nitrite as electron acceptors with acetate as electron donor. Chlorite likely killed the cells, whereas sulfate and sulfite were not used (Supplementary Table S4).
The effect of nitrite or nitrate addition on reduction of perchlorate with lactate as electron donor showed that PRB4 could simultaneously reduce perchlorate and nitrite or nitrate (Supplementary Figure S3). When added singly, perchlorate and nitrate were completely reduced at 18 h (Supplementary Figures S3A,C), whereas nitrite reduction was complete at 48 h (Supplementary Figure S3B). Nitrate addition at 0 or at 6 h did not inhibit perchlorate reduction by PRB4 (Supplementary Figures S3F,G). Reduction of perchlorate and nitrite also occurred simultaneously but the time needed for perchlorate reduction (Supplementary Figures S3D,E; 48 h) increased by 30 h compared to when only perchlorate was present (Supplementary Figure S3A: 18 h).
Discussion
Much has been learned in the past 20 years on the occurrence and microbial reduction of perchlorate in the environment. Different from nitrate, which is an intermediate in the global microbially catalyzed nitrogen cycle, perchlorate has entered the environment predominantly through human industrial activity, although natural sources of perchlorate may yet be identified (Coates and Achenbach, 2004). Nevertheless, perchlorate-reducing bacteria (PRB) have been found in many environments and have been shown to reduce perchlorate and/or chlorate to chlorite, which is then dismutated to chloride and oxygen through a highly conserved periplasmic chlorite dismutase (Cld). The rapid extracellular metabolism of chlorite is crucial as this reactive and strongly oxidative agent (E0’ = +0.78V) is highly biocidal (McDonnell and Russell, 1999). PRB use a variety of organic and inorganic electron donors with acetate being used most frequently (Coates and Achenbach, 2004; Bardiya and Bae, 2011). Use of hydrocarbons, e.g., benzene by Dechloromonas strain RCB (Chakraborty et al., 2005) and n-alkanes by P. chloritidismutans (Mehboob et al., 2009, 2016), is not wide-spread. Sulfide, when present, is oxidized to inorganic sulfur in preference over the use of organic electron donors like acetate (Gregoire et al., 2014). Microbial communities can protect themselves from the negative effects of perchlorate or chlorate metabolism by harboring the cld-gene. Analogs of cld are widespread, although it is unlikely that the gene products of all of these do indeed dismutate chlorite (Liebensteiner et al., 2014).
We have successfully enriched and isolated PRB from produced waters of the MHGC field, a shallow, low-temperature heavy oil reservoir, which has been subject to nitrate injection to prevent souring (Voordouw et al., 2009; Agrawal et al., 2012; Shen et al., 2018). Enrichments with perchlorate and VFA were dominated by Tenericutes, including the genus Acholeplasma (Supplementary Table S1). Some of these are also known to reduce nitrate (Sherry et al., 2013; Engelbrektson et al., 2014; Cheng et al., 2016). Acholeplasma spp. have been enriched from oilfields (Fida et al., 2016) and marine sediments (Masui et al., 2008; Imachi et al., 2011) and are considered scavengers of dead biomass due to their limited biosynthetic capabilities (Hanajima et al., 2015). Microbial communities present during early stages of perchlorate reduction with VFA were also dominated by Desulfuromonadales, as found in other studies where up to a 300-fold increase in this taxon has been reported (Carlström et al., 2016). Desulfuromonadales metabolize VFA and can couple the oxidation of VFA to the reduction of sulfur to sulfide, an activity which may occur in enrichments with perchlorate and sulfide (Gregoire et al., 2014). A small fraction of the microbial community enriched with VFA and perchlorate consisted of Magnetospirillum spp. (Supplementary Table S1: 0.8–0.9%), which was the only taxon obtained by agar plating. Several Magnetospirillum spp., including WD, SN1, VITRJS5, and VDY have been shown to reduce perchlorate (Thrash et al., 2010; Jacob et al., 2017). PRB2 and PBR4, isolated here, reduced perchlorate with short chain organic acids, H2 and ethanol, as do Magnetospirillum spp. WD, SN1, VDY. Other electron donors tested but not utilized for perchlorate reduction were H2S, methanol and alkylbenzenes (Supplementary Table S4). Meyer-Cifuentes et al. (2017) reported that Magnetospirillum sp. 15-1 oxidized toluene with nitrate, but not with perchlorate. Like M. bellicus strain VDY (Thrash et al., 2010), PRB2 and PRB4 used chlorate, nitrate, and nitrite as alternate electron acceptors. PRB2 and PRB4 were phylogenetically most similar to M. moscoviense BB-1 and metabolically most similar to VDY and WD, although they differentiated themselves from these by reducing nitrate or nitrite concurrently with perchlorate (Supplementary Figure S3). Most PRB preferentially reduce nitrate over perchlorate (Chaudhuri et al., 2002; Bardiya and Bae, 2008; Carlström et al., 2013). Sequential reduction of nitrate followed by reduction of perchlorate was also observed in our enrichment cultures (Figure 2H) but not in cultures of the isolated Magnetospirillum spp. PRB2 and PRB4. This indicates that these were not major components of these cultures, which is in agreement with the microbial community data (Supplementary Table S1).
Experiments with batch cultures and bioreactors in which heavy MHGC oil was the only available electron donor indicated absence of PRB capable of coupling the reduction of perchlorate to the oxidation of oil components (Figures 3, 5 and Supplementary Figure S1). Nitrate was reduced to nitrite and dinitrogen under these conditions, which resulted in removal of alkylbenzenes from the oil (Supplementary Figure S1), as found previously (Lambo et al., 2008; Agrawal et al., 2012; Suri et al., 2017). Because perchlorate was not reduced it was inert, i.e., no high-potential electron transport pathways were operating, and sulfide production was observed both in enrichment cultures and bioreactors containing sulfate and perchlorate. Microbial communities in cultures with sulfate and perchlorate were similar to those in cultures with sulfate only (Figure 4: clade I), indicating the lack of impact of perchlorate. In contrast, microbial communities in cultures with nitrate were distinct (Figure 4: clade II) and were dominated by the hNRB Thauera and Pseudomonas (Supplementary Table S2), which couple reduction of nitrate to nitrite and N2 to the oxidation of oil components, especially alkylbenzenes.
The inability of PRB to grow in batch cultures and bioreactors with MHGC oil was due to the absence of usable electron donors, like acetate. Interestingly, acetate has been shown to form in bioreactors with MHGC oil, which were continuously injected with 2 mM sulfate and periodically with 2 mM sulfate and 4 mM nitrate (Callbeck et al., 2013). Transient formation of up to 3 to 4 mM acetate was seen whenever injection of nitrate was stopped. This suggested that upon restoration of sulfate-reducing conditions alkanes were fermented with water to acetate and H2 by syntrophic bacteria (Zengler et al., 1999) with the H2 being used preferentially for the reduction of sulfate and the acetate accumulating. Acetate was then eventually removed by acetotrophic reactions, as discussed by Callbeck et al. (2013). Use of acetate by acetotrophic SRB is inhibited by light oils and may not occur in light oil-containing oil fields (Menon and Voordouw, 2018), which may also cause acetate accumulation. Indeed, high temperature fields, harboring light oil can have very high concentrations of acetate and propionate in produced waters (Menon and Voordouw, 2018).
Thus, even in the absence of direct hydrocarbon oxidation, there is potential activity of PRB in oil fields. It should be pointed out that injection waters with high VFA concentrations are considered poor fits for treatment with nitrate, because a lot of nitrate would be used oxidizing these aqueous VFA and would thus not make it into the reservoir to prevent souring. In the MHGC field VFA concentrations in produced waters and injection waters are low (of the order of 0.1 mM). Hence most of the nitrate amended into injection water is injected into the reservoir, where it is used for oxidation of oil-alkylbenzenes and sulfide. Because heavy MHGC oil has only low concentrations of alkyl benzenes (of the order of 1 mM), long term injection of nitrate can cause the depletion of alkylbenzenes, which then leads to nitrate breakthrough in producing wells, control of souring and the production of toluene-free oil (Agrawal et al., 2012).
Conclusion
In conclusion it appears that perchlorate may not be good souring control agent in the MHGC field, where the pros and cons of using nitrate are well understood. Clearly more research is needed to determine whether PRB using oil components as electron donor are present in other fields. A field test in which perchlorate is injected to determine its effectiveness in decreasing souring would also be valuable.
Author Contributions
GO did the experimental work and wrote a first version of the paper and abstract. GV supervised the work and edited the paper and abstract.
Funding
This work was supported by an NSERC Industrial Research Chair in Petroleum Microbiology Award to GV, as well as by BP America Production Co., Baker Hughes Canada, Computer Modelling Group Ltd., ConocoPhillips Company, Dow Microbial Control, Enbridge, Enerplus Corporation, Intertek, Oil Search Ltd., Shell Global Solutions International, Suncor Energy Inc., Yara Norge AS, and Alberta Innovates.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Yin Shen, Dongshan An, and Tekle Fida for administrative and technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02423/full#supplementary-material
References
Agrawal, A., Park, H. S., Nathoo, S., Gieg, L. M., Jack, T. R., Miner, K., et al. (2012). Toluene depletion in produced oil contributes to souring control in a field subjected to nitrate injection. Environ. Sci. Technol. 46, 1285–1292. doi: 10.1021/es203748b
Bardiya, N., and Bae, J. H. (2008). Isolation and characterization of Dechlorospirillum anomalous strain JB116 from a sewage treatment plant. Microbiol. Res. 163, 182–191. doi: 10.1016/j.micres.2006.05.001
Bardiya, N., and Bae, J. H. (2011). Dissimilatory perchlorate reduction: a review. Microbiol. Res. 166, 237–254. doi: 10.1016/j.micres.2010.11.005
Bender, K. S., Shang, C., Chakraborty, R., Belchik, S. M., Coates, J. D., and Achenbach, L. A. (2005). Identification, characterization, and classification of genes encoding perchlorate reductase. J. Bacteriol. 187, 5090–5096. doi: 10.1128/JB.187.15.5090-5096.2005
Callbeck, C. M., Agrawal, A., and Voordouw, G. (2013). Acetate production from oil under sulfate-reducing conditions in bioreactors injected with sulfate and nitrate. Appl. Environ. Microbiol. 79, 5059–5068. doi: 10.1128/aem.01251-13
Callbeck, C. M., Dong, X., Chatterjee, I., Agrawal, A., Caffrey, S. M., Sensen, C. W., et al. (2011). Microbial community succession in a bioreactor modeling a souring low-temperature oil reservoir subjected to nitrate injection. Appl. Microbiol. Biotechnol. 91, 799–810. doi: 10.1007/s00253-011-3287-2
Carlson, H. K., Kuehl, J. V., Hazra, A. B., Justice, N. B., Stoeva, M. K., Sczesnak, A., et al. (2015). Mechanisms of direct inhibition of the respiratory sulfate-reduction pathway by (per)chlorate and nitrate. ISME J. 9, 1295–1305. doi: 10.1038/ismej.2014.216
Carlström, C. I., Lucas, L. N., Rohde, R. A., Haratian, A., Engelbrektson, A. L., and Coates, J. D. (2016). Characterization of an anaerobic marine microbial community exposed to combined fluxes of perchlorate and salinity. Appl. Microbiol. Biotechnol. 100, 9719–9732. doi: 10.1007/s00253-016-7780-5
Carlström, C. I., Wang, O., Melnyk, R. A., Bauer, S., Lee, J., Engelbrekston, A., et al. (2013). Physiological and genetic description of dissimilatory perchlorate reduction by the novel marine bacterium Arcobacter sp. strain CAB. mBio 4:e00217-13. doi: 10.1128/mBio.00217-13
Chakraborty, R., O’Connor, S. M., Chan, E., and Coates, J. D. (2005). Anaerobic degradation of benzene, toluene, ethylbenzene, and xylene compounds by Dechloromonas strain RCB. Appl. Environ. Microbiol. 71, 8649–8655. doi: 10.1128/AEM.71.12.8649-8655.2005
Chao, A. (1984). Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11, 265–270.
Chaudhuri, S. K., O’Connor, S. M., Gustavson, R. L., Achenbach, L. A., and Coates, J. D. (2002). Environmental factors that control microbial perchlorate reduction. Appl. Environ. Microbiol. 68, 4425–4430. doi: 10.1128/AEM.68.9.4425-4430.2002
Cheng, Y., Hubbard, C. G., Li, L., Bouskill, N., Molins, S., Zheng, L., et al. (2016). Reactive transport model of sulfur cycling as impacted by perchlorate and nitrate treatments. Environ. Sci. Technol. 50, 7010–7018. doi: 10.1021/acs.est.6b00081
Coates, J. D. (2014). Microbial metabolism of chlorine oxyanions as a control of biogenic hydrogen sulfide production. US Patent PCT/US2012/040273. Washington, DC.
Coates, J. D., and Achenbach, L. A. (2004). Microbial perchlorate reduction: rocket-fueled metabolism. Nat. Rev. Microbiol. 2, 569–580. doi: 10.1038/nrmicro926
Coates, J. D., Chakraborty, R., Lack, J. G., O’Connor, S. M., Cole, K. A., Bender, K. S., et al. (2001). Anaerobic benzene oxidation coupled to nitrate reduction in pure culture by two strains of Dechloromonas. Nature 411, 1039–1043. doi: 10.1038/35082545
Dong, X., Droge, J., von Toerne, C., Marozava, S., McHardy, A. C., and Meckenstock, R. U. (2017a). Reconstructing metabolic pathways of a member of the genus Pelotomaculum suggesting its potential to oxidize benzene to carbon dioxide with direct reduction of sulfate. FEMS Microbiol. Ecol. 93:fiw254. doi: 10.1093/femsec/fiw254
Dong, X., Kleiner, M., Sharp, C. E., Thorson, E., Li, C., Liu, D., et al. (2017b). Fast and simple analysis of miseq amplicon sequencing data with meta Amp. Front. Microbiol. 8:1461. doi: 10.3389/fmicb.2017.01461
Engelbrektson, A., Hubbard, C. G., Tom, L. M., Boussina, A., Jin, Y. T., Wong, H., et al. (2014). Inhibition of microbial sulfate reduction in a flow-through column system by (per)chlorate treatment. Front. Microbiol. 5:315. doi: 10.3389/fmicb.2014.00315
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Fida, T. T., Chen, C., Okpala, G., and Voordouw, G. (2016). Implications of limited thermophilicity of nitrite reduction for control of sulfide production in oil reservoirs. Appl. Environ. Microbiol. 82, 4190–4199. doi: 10.1128/AEM.00599-16
Fida, T. T., Voordouw, J., Ataeian, M., Kleiner, M., Okpala, G., Mand, J., et al. (2018). Synergy of sodium nitroprusside and nitrate in inhibiting the activity of sulfate reducing bacteria in oil-containing bioreactors. Front. Microbiol. 9:981. doi: 10.3389/fmicb.2018.00981
Frank, J. A., Reich, C. I., Sharma, S., Weisbaum, J. S., Wilson, B. A., and Olsen, G. J. (2008). Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 74, 2461–2470. doi: 10.1128/AEM.02272-07
Gassara, F., Suri, N., Stanislav, P., and Voordouw, G. (2015). Microbially enhanced oil recovery by sequential injection of light hydrocarbon and nitrate in low- and high-pressure bioreactors. Environ. Sci. Technol. 49, 12594–12601. doi: 10.1021/acs.est.5b03879
Gregoire, P., Engelbrektson, A., Hubbard, C. G., Metlagel, Z., Csencsits, R., Aue, R. M., et al. (2014). Control of sulfidogenesis through bio-oxidation of H2S coupled to (per)chlorate reduction. Environ. Microbiol. Rep. 6, 558–564. doi: 10.1111/1758-2229.12156
Hamilton, T. L., Bovee, R. J., Sattin, S. R., Mohr, W., Gilhooly, W. P., Lyons, T. W., et al. (2016). Carbon and sulfur cycling below the chemocline in a meromictic lake and the identification of a novel taxonomic lineage in the FCB superphylum, Candidatus Aegiribacteria. Front. Microbiol. 7:598. doi: 10.3389/fmicb.2016.00598
Hanajima, D., Aoyagi, T., and Hori, T. (2015). Survival of free-living Acholeplasma in aerated pig manure slurry revealed by 13C-labeled bacterial biomass probing. Front. Microbiol. 6:1206. doi: 10.3389/fmicb.2015.01206
Imachi, H., Aoi, K., Tasumi, E., Saito, Y., Yamanaka, Y., Saito, Y., et al. (2011). Cultivation of methanogenic community from subseafloor sediments using a continuous-flow bioreactor. ISME J. 5, 1913–1925. doi: 10.1038/ismej.2011.64
Jacob, J. J., Sumana, S., Jayasri, M. A., and Suthindhiran, K. (2017). Isolation, characterization and kinetics of perchlorate reducing Magnetospirillum species. Geomicrobiol. J. 35, 120–126. doi: 10.1080/01490451.2017.1338796
Lambo, A. J., Noke, K., Larter, S. R., and Voordouw, G. (2008). Competitive, microbially-mediated reduction of nitrate with sulfide and aromatic oil components in a low-temperature, western Canadian oil reservoir. Environ. Sci. Technol. 2008, 8941–8946. doi: 10.1021/es801832s
Liebensteiner, M. G., Tsesmetzis, N., Stams, A. J. M., and Lomans, B. P. (2014). Microbial redox processes in deep subsurface environments and the potential application of (per)chlorate in oil reservoirs. Front. Microbiol. 5:428. doi: 10.3389/fmicb.2014.00428
Masui, N., Morono, Y., and Inagaki, F. (2008). Microbiological assessment of circulation mud fluids during the first operation of riser drilling by the deep-earth research vessel Chikyu. Geomicrobiology J. 25, 274–282. doi: 10.1080/01490450802258154
McDonnell, G., and Russell, A. D. (1999). Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12, 147–179.
Mehboob, F., Junca, H., Schraa, G., and Stams, A. J. M. (2009). Growth of Pseudomonas chloritidismutans AW-1(T) on n-alkanes with chlorate as electron acceptor. Appl. Microbiol. Biotechnol. 83, 739–747. doi: 10.1007/s00253-009-1985-9
Mehboob, F., Oosterkamp, M. J., Koehorst, J. J., Farrakh, S., Veuskens, T., Plugge, C. M., et al. (2016). Genome and proteome analysis of Pseudomonas chloritidismutans AW-1T that grows on n-decane with chlorate or oxygen as electron acceptor. Environ. Microbiol. 18, 3247–3257. doi: 10.1111/1462-2920.12880
Mehta-Kolte, M. G., Loutey, D., Wang, O., Youngblut, M. D., Hubbard, C. G., Wetmore, K. M., et al. (2017). Mechanism of H2S oxidation by the dissimilatory perchlorate reducing microorganism Azospira suillum PS. mBio 8:02023-16. doi: 10.1128/mBio.02023-16
Menon, P., and Voordouw, G. (2018). Impact of light oil toxicity on sulfide production by acetate-oxidizing, sulfate-reducing bacteria. Int. Biodeterior. Biodegr. 126, 208–215. doi: 10.1016/j.ibiod.2016.11.021
Meyer-Cifuentes, I., Martinez-Lavanchy, P. M., Marin-Cevada, V., Böhnke, S., Harms, H., Müller, J. A., et al. (2017). Isolation and characterization of Magnetospirillum sp. strain 15-1 as a representative anaerobic toluene-degrader from a constructed wetland model. PLoS One 12:e0174750. doi: 10.1371/journal.pone.0174750
Okpala, G. N., Chuan, C., Fida, T., and Voordouw, G. (2017). Effect of thermophilic nitrate reduction on sulfide production in high temperature oil reservoir samples. Front. Microbiol. 8:1573. doi: 10.3389/fmicb.2017.01573
Postgate, J. R. (1952). Competitive and noncompetitive inhibitors of bacterial sulphate reduction. J. Gen. Microbiol. 6, 128–142. doi: 10.1099/00221287-6-1-2-128
Rikken, G. B., Kroon, A. G. M., and van Ginkel, C. G. (1996). Transformation of (per)chlorate into chloride by a newly isolated bacterium: reduction and dismutation. Appl. Microbiol. Biotechnol. 45, 420–426. doi: 10.1007/s002530050707
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Schaffner, I., Hofbauer, S., Krutzler, M., Pirker, K. F., Furtmüller, P. G., and Obinger, C. (2015). Mechanism of chlorite degradation to chloride and dioxygen by the enzyme chlorite dismutase. Arch. Biochem. Biophys. 574, 18–26. doi: 10.1016/j.abb.2015.02.031
Shannon, C. E. (1948). A mathematical theory of communication. Bell Syst. Tech. J. 27, 379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x
Shen, Y., Agrawal, A., Suri, N. K., An, D., Voordouw, J. K., Clark, R. G., et al. (2018). Control of microbial sulfide production by limiting sulfate dispersal in a water-injected oil field. J. Biotechnol. 266, 14–19. doi: 10.1016/j.jbiotec.2017.11.016
Sherry, A., Gray, N. D., Ditchfield, A. K., Aitken, C. M., Jones, D. M., Röling, W. F. M., et al. (2013). Anaerobic biodegradation of crude oil under sulphate-reducing conditions leads to only modest enrichment of recognized sulphate-reducing taxa. Int. Biodeterior. Biodegr. 81, 105–113. doi: 10.1016/j.ibiod.2012.04.009
Stolze, Y., Bremges, A., Rumming, M., Henke, C., Maus, I., Puhler, A., et al. (2016). Identification and genome reconstruction of abundant distinct taxa in microbiomes from one thermophilic and three mesophilic production-scale biogas plants. Biotechnol. Biofuels 9:156. doi: 10.1186/s13068-016-0565-3
Suri, N. (2017). Microbially-Enhanced Oil Recovery Through Activity of Alkylbenzene-Oxidizing Nitrate-Reducing Bacteria. Ph.D. thesis, University of Calgary, ıCalgary, AB.
Suri, N., Voordouw, J., and Voordouw, G. (2017). The effectiveness of nitrate-mediated control of the oil field sulfur cycle depends on the toluene content of the oil. Front. Microbiol. 8:956. doi: 10.3389/fmicb.2017.00956
Tamura, K., Nei, M., and Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. PNAS 101, 11030–11035. doi: 10.1073/pnas.0404206101
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Thrash, J. C., Ahmadi, S., Torok, T., and Coates, J. D. (2010). Magnetospirillum bellicus sp. nov., a novel dissimilatory perchlorate-reducing alphaproteobacterium isolated from a bioelectrical reactor. Appl. Environ. Microbiol. 76, 4730–4737. doi: 10.1128/AEM.00015-10
van Ginkel, C. G., Rikken, G. B., Kroon, A. G. M., and Kengen, S. W. M. (1996). Purification and characterization of chlorite dismutase: a novel oxygen-generating enzyme. Arch. Microbiol. 166, 321–326. doi: 10.1007/s002030050390
Vance, I., and Thrasher, D. R. (2005). “Reservoir souring: mechanisms and prevention,” in Petroleum Microbiology (American Society of Microbiology), eds B. Ollivier and M. Magot (Boston, MA: ASM Press), 123–142.
Voordouw, G., Grigoryan, A. A., Lambo, A., Lin, S., Park, H. S., Jack, T. R., et al. (2009). Sulfide remediation by pulsed injection of nitrate into a low temperature Canadian heavy oil reservoir. Environ. Sci. Technol. 43, 9512–9518. doi: 10.1021/es902211j
Wolterink, A. F., Jonker, A. B., Kengen, S. W., and Stams, A. J. (2002). Pseudomonas chloritidismutans sp. nov., a non-denitrifying, chlorate-reducing bacterium. Int. J. Syst. Evol. Microbiol. 52, 2183–2190. doi: 10.1099/00207713-52-6-2183
Xue, Y., and Voordouw, G. (2015). Control of microbial sulfide production with biocides and nitrate in oil reservoir simulating bioreactors. Front. Microbiol. 6:1387. doi: 10.3389/fmicb.2015.01387
Keywords: sulfate reduction, nitrate, perchlorate, souring, hydrogen sulfide, oil field
Citation: Okpala GN and Voordouw G (2018) Comparison of Nitrate and Perchlorate in Controlling Sulfidogenesis in Heavy Oil-Containing Bioreactors. Front. Microbiol. 9:2423. doi: 10.3389/fmicb.2018.02423
Received: 30 June 2018; Accepted: 21 September 2018;
Published: 09 October 2018.
Edited by:
Marc Mussmann, Universität Wien, AustriaReviewed by:
John Senko, The University of Akron, United StatesAmy Michele Grunden, North Carolina State University, United States
Copyright © 2018 Okpala and Voordouw. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gerrit Voordouw, voordouw@ucalgary.ca
 Gloria Ngozi Okpala
Gloria Ngozi Okpala Gerrit Voordouw
Gerrit Voordouw