- 1Emerging Pathogens Institute, University of Florida, Gainesville, FL, United States
- 2Department of Microbiology and Cell Science, College of Agricultural and Life Sciences, University of Florida, Gainesville, FL, United States
- 3Department of Environmental and Global Health, College of Public Health and Health Professions, University of Florida, Gainesville, FL, United States
Many bacterial pathogens promote biofilms that confer resistance against stressful survival conditions. Likewise Vibrio cholerae O1, the causative agent of cholera, and ubiquitous in aquatic environments, produces vps-dependent biofilm conferring resistance to environmental stressors and predators. Here we show that a 49-bp deletion mutation in the flrA gene of V. cholerae N16961S strain resulted in promotion of vps-independent biofilm in filter sterilized lake water (FSLW), but not in nutrient-rich L-broth. Complementation of flrA mutant with the wild-type flrA gene inhibited vps-independent biofilm formation. Our data demonstrate that mutation in the flrA gene positively contributed to vps-independent biofilm production in FSLW. Furthermore, inactivation of mshA gene, encoding the main pilin of mannose sensitive hemagglutinin (MSHA pilus) in the background of a ΔflrA mutant, inhibited vps-independent biofilm formation. Complementation of ΔflrAΔmshA double mutant with wild-type mshA gene restored biofilm formation, suggesting that mshA mutation inhibited ΔflrA-driven biofilm. Taken together, our data suggest that V. cholerae flrA and mshA act inversely in promoting vps-independent biofilm formation in FSLW. Using a standard chemotactic assay, we demonstrated that vps-independent biofilm of V. cholerae, in contrast to vps-dependent biofilm, promoted bacterial movement toward chitin and phosphate in FSLW. A ΔflrAΔmshA double mutant inhibited the bacterium from moving toward nutrients; this phenomenon was reversed with reverted mutants (complemented with wild-type mshA gene). Movement to nutrients was blocked by mutation in a key chemotaxis gene, cheY-3, although, cheY-3 had no effect on vps-independent biofilm. We propose that in fresh water reservoirs, V. cholerae, on repression of flagella, enhances vps-independent biofilm that aids the bacterium in acquiring nutrients, including chitin and phosphate; by doing so, the microorganism enhances its ability to persist under nutrient-limited conditions.
Introduction
Vibrio cholerae, toxigenic strains of which cause epidemic cholera, is ubiquitous to aquatic environment. Ingestion of water and/or food contaminated with the microorganism leads to cholera (Colwell and Huq, 1994; Kaper et al., 1995; Faruque et al., 1998; Morris, 2011). In aquatic reservoirs, V. cholerae can persist either in a planktonic (free-living) stage or in biofilms (Tamplin et al., 1990; Shukla et al., 1995). In the biofilm mode of persistence, the microorganism can attach to biotic and abiotic surfaces, including phytoplankton, crustaceans, zooplanktons, aquatic plants, sediments, and detritus (Huq et al., 1983; Huq et al., 1990; Islam et al., 1990; Nelson et al., 2009). Despite decades of investigations, the physiological and genetic basis of the persistence of V. cholerae in aquatic reservoirs, particularly during inter-epidemic periods, remains to be fully elucidated. Possible survival mechanisms in nutrient-poor and stressful aquatic reservoirs include: (a) adopting a viable but non-culturable (VBNC) state (Xu et al., 1982; Colwell and Huq, 1994), (b) switching from planktonic to biofilm lifestyle (Wai et al., 1998; Yildiz and Schoolnik, 1999; Ali et al., 2002), and/or (c) assuming “persister” and “growth advantage stationary phase” (“GASP”) phenotypes as described recently (Jubair et al., 2012, 2014).
Vibrio cholerae produces a single polar flagellum that contributes to motility during the planktonic stage of growth, but is inhibited in mature biofilm growth stage. Biogenesis of flagella is encoded by four gene clusters. Gene cluster I harbors a master transcriptional activator, FlrA encoded by the flrA gene. Coupled with δ54, FlrA promotes transcription of other flagellar genes present in clusters I–IV; furthermore, FlrA also contributes to the transcription of a number of genes required for chemotaxis (Klose and Mekalanos, 1998) (see below). During environmental persistence, V. cholerae employs its flagella in two distinct ways: first, flagella promote planktonic bacteria to propel to a growth-permissible environment, while they drive back the bacteria in response to non-permissible and hostile growth conditions. Secondly, flagella play a pivotal role in sensing and attaching to biotic or abiotic surfaces triggering the initial steps of biofilm formation that ultimately lead to mature biofilm (Watnick and Kolter, 1999; Butler and Camilli, 2005). Interestingly, in a mature biofilm, V. cholerae represses flagella synthesis while it promotes exopolysaccharide production, a key ingredient for mature biofilm formation (Watnick and Kolter, 1999; Prouty et al., 2001). However, it is worth noting here that many bacterial species, including Acinetobacter baumannii (Tomaras et al., 2008; Gaddy and Actis, 2009) do not possess flagella, yet they are able to produce biofilm suggesting that flagella may be dispensable in biofilm formation in non-motile bacteria.
In addition to flagella, V. cholerae produces two types of pili, including a mannose-sensitive hemagglutinin (MSHA) pilus (also known as type IVa pilus), and a toxin co-regulated pilus (TCP) (also known as type IVb pilus). The main pilin of MSHA and TCP pili is encoded by mshA and tcpA gene, respectively. While MSHA pilus promotes strong surface attachment facilitating biofilm formation and environmental persistence (Watnick et al., 1999; Hsiao et al., 2009), TCP pilus contributes to virulence by allowing the bacterium to attach to and colonize human intestinal epithelial cells (Herrington et al., 1988; Rhine and Taylor, 1994; Hsiao et al., 2008). Interestingly, biogenesis of the two pili is promoted by opposite conditions. For example, TCP pilus is highly expressed in the human intestine with repression of MSHA pilus. In contrast, MSHA pilus is profusely expressed under in vitro conditions, resulting in biofilm formation with repression of TCP pilus (Hsiao et al., 2008, 2009).
In many bacterial species, chemotaxis promotes the ability of motile bacteria to swim toward or away from specific environmental stimuli in order to provide the microorganism with a survival advantage in a given condition (Wadhams and Armitage, 2004; Butler and Camilli, 2005). Chemotaxis has been extensively investigated in Escherichia coli and Salmonella typhimurium (Bourret and Stock, 2002; Wadhams and Armitage, 2004). In contrast to E. coli or S. typhimurium, multiple paralogs of various chemotaxis genes are present in V. cholerae chromosomes (Heidelberg et al., 2000). With the exception of the mcp gene encoding methyl-accepting chemotaxis protein, which is scattered throughout the genome, most V. cholerae chemotaxis genes are organized into three operons (Heidelberg et al., 2000; Butler and Camilli, 2005). Only one of these three operons, operon 2, is important for chemotaxis in V. cholerae as genes present in this operon promoted chemotaxis (Butler and Camilli, 2005). Previous studies demonstrated that cheA-2 (Gosink et al., 2002) and cheY-3 (Lee et al., 2001) genes of operon 2 were required for chemotaxis, whereas cheA-1 and cheY-1, and cheA-3 and cheY-3 from operons 1 and 3, respectively, were dispensable for chemotaxis (Heidelberg et al., 2000).
Long-term persistence of bacteria in stressful and stationary growth phase can promote a “GASP” phenotype (Finkel and Kolter, 1999; Zinser and Kolter, 2004; Finkel, 2006). We recently reported a V. cholerae GASP phenotype (GASP-700D) after its long-term persistence (700 days) in nutrient-poor lake water microcosms (Jubair et al., 2014). Compared to its wild-type N16961S phenotype, the GASP-700D phenotype was defective in motility and produced significantly increased vps-independent biofilm when grown specifically in filter sterilized lake water (FSLW). Given the non-motile behavior and ∼1000-fold down regulation of flrA gene of GASP-700D, we hypothesized that GASP-700D sustained a mutation in the flrA gene contributing to increased expression of vps-independent biofilm. Here we provide evidence that: (a) flrA gene in GASP-700D sustained a 49-bp internal deletion mutation resulting in an increased production of vps-independent biofilm in FSLW, but not in nutrient-rich L-broth, (b) mutation in the mshA gene inhibited the vps-independent biofilm formation, (c) vps-independent, in contrast to vps-dependent biofilm, promoted biofilm bacterium to move toward nutrients, including chitin and phosphate in FSLW, and (d) mutation in the chemotaxis gene, cheY-3, inhibited the mutant from moving toward nutrients in FSLW.
Materials and Methods
Bacterial Strains and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Table 1. In this study, as needed, we used two variants of V. cholerae N16961 strain, including N16961S (stands for smooth variant) and N16961R (stands for “rugose” variant). The method used to convert smooth variant to rugose variant has been described previously (Ali et al., 2002). As needed, bacterial strains were subcultured from glycerol stock stored at -80°C onto L-agar and the culture was incubated overnight at 37°C. For growth in L-broth, a single colony was transferred to 3 ml L-broth and the culture was grown overnight at 37°C with a shaking speed of 250 × g in an environmental shaker (New Brunswick Scientific, Edison, NJ, United States). Unless otherwise indicated, for culture in FSLW, 1 ml of overnight L-broth grown culture was centrifuged, washed 2× in 1 ml FSLW and finally the pellet was resuspended in 1 ml fresh FSLW. As needed, antibiotics were used at the following concentrations: ampicillin, 100 μg ml-1 and polymyxin B, 50 U ml-1.
Water Source
For this study we used fresh water collected in a sterile 1,000 ml Nalgene bottle (Nalgene, Rochester, NY, United States) from a 30.2-acre natural lake (Wauburg Lake) in Gainesville, FL, United states as described previously (Jubair et al., 2012). For experiment, aliquot (300 ml) of water was filter sterilized using Nalgene 0.22 μm membrane filter unit. Chemical analysis of filter sterilized lake water was performed by Advanced Environmental Laboratories, Inc. (Gainesville, FL, United States). The analysis of lake water revealed following major components: total organic carbon (12.0 mg/l), total nitrogen (1.4 mg/l), ammonia (0.06 mg/l), nitrate plus nitrite (0.014 mg/l), sodium (7.3 mg/l), chloride (14.0 mg/l), calcium (7.0 mg/l), iron (0.03 mg/l), potassium (0.93 mg/l), total phosphate (0.26 mg/l), orthophosphate (0.002 mg/l), and magnesium (1.6 mg/l). Interestingly, low nutrient composition found in Wauburg Lake water was similar to what has been reported by us using the same lake water (Jubair et al., 2012), and in pond water obtained from Bangladesh and other fresh waters obtained from United States (Kierek and Watnick, 2003a; Nelson et al., 2008).
Genetic Manipulations
To determine if GASP-700D (Jubair et al., 2014) sustained a mutation in the flrA gene, we PCR amplified the gene from the GASP-700D genome with two convergent PCR primers [aa778 and aa780 (Supplementary Table 1)] using standard PCR conditions. The amplicon was purified using Qiagen PCR Purification Kit (Qiagen, Valencia, CA, United States) and sequenced with help from the Interdisciplinary Center for Biotechnology Research (ICBR) at University of Florida; sequence analysis revealed that GASP-700D sustained a 49-bp deletion mutation in flrA gene (we hereafter referred this strain as GSΔflrA).
For creating an identical 49-bp deletion mutation in flrA gene in the background of wild-type V. cholerae N16961S strain (Table 1), a one-step PCR cloning strategy was employed using primers, including aa778S and aa780S (Supplementary Table 1) carrying SalI and SacI site at the 5′-end of each primer, respectively. Genomic DNA of GSΔflrA was used as PCR template while primer pairs (aa778S and aa780S) flanking the 49-bp deletion in GSΔflrA were used to amplify the PCR product using standard PCR conditions. The purified PCR product was digested with SalI and SacI, the digested product was purified, and ligated into a similarly digested suicide vector, pCVD442 (Donnenberg and Kaper, 1991); the ligated product was transformed into E. coli SM10 λ pir using standard transformation procedures resulting in plasmid pAAS82. E. coli SM10 λ pir carrying pAAS82 was conjugated to V. cholerae N16961S; selection of transconjugants and counter selection for the flrA mutation resulting from homologous recombination were performed as described previously (Jubair et al., 2014). PCR and DNA sequencing methodologies were used to confirm the 49-bp mutation in flrA gene [AAS84 (N16961SΔflrA), Table 1]. An in-frame deletion in the ΔmshA gene was created in the backgrounds of N16961S and GSΔflrA by using primers and methodologies described previously (Thelin and Taylor, 1996). A two-step PCR cloning strategy, with a set of targeted primers, was used to create a null mutation in cheY-3 gene in the backgrounds of N16961S, N16961SΔflrA, and GSΔflrA strains. Mutation in vpsA gene (VC0917) encoding UDP-N-acetylglucosamine 2-epimerase (Ali et al., 2000b), a key component of vps-dependent biofilm in the background of V. cholerae N16961S, N16961R, and GSΔflrA strains was created as described previously (Jubair et al., 2014). To complement mutants, each wild-type gene of interest was PCR amplified using two convergent PCR primers targeting that gene; chromosomal DNA of N16961S was used as the template for PCR reactions. Following purification of the PCR product, the gene was cloned into pWSK29 vector (Wang and Kushner, 1999). For complementation of ΔmshA, we used plasmid vector pMMB67EH carrying wild-type mshA gene cloned into the vector (complementation plasmid is a kind gift from Andrew Camilli of Tufts University) (Table 1). The complementing plasmid was introduced into the V. cholerae mutant strain of interest using DNA electroporation (Ali et al., 2000a). All PCR primers used either in gene disruptions or in cloning wild-type genes are listed in Supplementary Table 1.
Motility Assay
Swarming behavior of V. cholerae strains on motility agar (0.3% agar) was examined as described previously (Gardel and Mekalanos, 1996). Briefly, each strain of interest was grown overnight in L-broth at 37°C with a shaking speed of 250 × g. A sterile inoculating wire was dipped into the culture and the wire containing the culture was then inoculated onto motility agar. Zone of migration of each bacterial strain around the inoculating site was examined and the image was captured after incubation of the microorganism in motility agar at 37°C for 8 h.
Quantitative Biofilm Assay
Quantification of biofilm production by V. cholerae strains grown in L-broth was measured as described previously (Watnick and Kolter, 1999). For measurement of biofilm production in FSLW microcosm, a single colony of V. cholerae grown on L-agar was transferred into 3 ml L-broth and the culture was incubated overnight at 37°C with a shaking speed of 250 × g. One milliliter of culture was spun down and washed 2× with 1 ml of FSLW. A 24-well polystyrene plastic plate (Corning Incorporated, Corning, NY, United States) was used as the surface for bacterial attachment; 50 μl of culture was added to 450 μl of fresh FSLW (10-fold dilution) to obtain ca. 108 cfu/ml in each well (six replicates). The culture was incubated overnight statically at ambient temperature. Following incubation, the culture was discarded from the wells and washed 2× with water to discard any residual bacterial cells. Crystal violet (CV) dye was added to each well and the plate was incubated at room temperature for 30 min; the CV dye was discarded and the well was washed (2×) with water. Finally, cell-associated CV dye was extracted with 600 μl of dimethyl sulfoxide (DSMO) (Sigma, St. Louis, MO, United States) and the quantitative biofilm production was determined as described previously (Jubair et al., 2014).
Confocal Microscopy
In order to determine the three-dimensional architectural structure and thickness of the biofilm, confocal microscopic analysis was performed as described previously (Jubair et al., 2014). V. cholerae strains, including N16961S, N16961SΔflrA, GSΔflrA, GSΔmshA, and GSΔmshA/pAA159 (Table 1) were included in the assay. Briefly, a single colony of each strain was inoculated into 3 ml L-broth and the culture was incubated and processed as described above. An inoculum (ca. 108 cfu/ml) was transferred into a well containing 600 μl of fresh sterile FSLW in a 12-well polystyrene culture plate (Thermo Scientific Nunc, Pittsburgh, PA, United States). To provide a bacterial attachment platform, a 12 mm round glass cover slip (Fischer Scientific, Pittsburgh, PA, United States) was dipped into each culture well, and the culture was incubated statically overnight at room temperature. The following day, attached cells on cover slip were washed twice with Dulbecco’s PBS (DPBS) (HyClone Laboratories, Logan, UT, United States), and fixed in 10% neutral buffered formalin solution (Sigma-Aldrich, St. Louis, MO, United States) for 30 min. Cells were washed again with DPBS and stained with 500 μl/well of 1:1000 SYTO 9 dye (LIVE/DEAD BacLight Bacterial Viability Kit; Invitrogen, Grand Island, NY, United States) as described previously (Jubair et al., 2014). Following two washes with HyPure cell culture grade water (HyClone Laboratories, Logan, UT, United States), the glass cover slip was mounted on a 75 × 25 mm microscopic slide (Corning Inc., Corning, NY, United States). A Zeiss LSM 800 with Axio Observer Z1 inverted microscope (Carl Zeiss Iberia, S.L., Spain) was used to visualize the cover slip with an excitation and emission wavelengths of 484 and 500 nm, respectively. The attached microorganism was examined using Plan-Apochromat 63× objective lens. At least three biological replicates were examined during image analysis. All images and z-stacks were processed using Zen 2 (Blue edition) software and enhanced by adjusting the histogram of the image.
Chemotaxis Assays
A chemotaxis assay was performed as described previously (Butler et al., 2006) with slight modifications. Briefly, a single colony of V. cholerae strain was grown in 3 ml L-broth overnight at 37°C with a shaking speed of 250 × g. One milliliter of culture was centrifuged; after decanting supernatant, the pellet was washed 2× with fresh FSLW and resuspended in 1 ml fresh FSLW. The culture was diluted (10-fold) in FSLW to obtain ca. 108 cfu/ml and incubated overnight statically at room temperature. An aliquot of 150 μl of culture (three replicates) was transferred into 96-well plate. One microliter capillary tube (55 mm) (Drummond Scientific Company, Broomall, PA, United States) filled with either FSLW or FSLW supplemented with chitin (0.05%) or K2HPO4 (1.0 mM at final concentration) was inserted into each culture well. After 6 min of incubation at room temperature, the content of the capillary tube was expelled out into 99 μl of L-broth. The culture was spread onto L-agar plates and the plates were incubated overnight at 37°C; total bacterial number (colony forming units per milliliter) grown on L-agar was determined by standard plate count. The results were expressed as a comparative index (CI) for each strain. CI was calculated by the number of bacterial counts (colony forming units per milliliter) present in FSLW in the capillary tube supplemented with nutrient divided by the number of bacterial counts (colony forming units per milliliter) present in FSLW in the capillary tube not supplemented with nutrient.
Statistical Analysis
One-way ANOVA along with Holm-Sidak’s multiple comparison test was performed using GraphPad Prism version 6.00 for Windows, GraphPad Software, La Jolla, CA, United States, http://www.graphpad.com. A p-value of <0.05 was considered as statistically significant.
Results
GASP-700D Sustained an Internal Deletion Mutation
After PCR amplification and sequencing of the flrA gene of GASP-700D, we found that the flrA gene sustained a 49-bp internal deletion mutation (we hereafter refer this strain as GSΔflrA) (Figure 1) causing a frame shift mutation. To determine if the 49-bp deletion in flrA gene in GSΔflrA inhibited motility, we created an identical 49-bp deletion mutation in the wild-type V. cholerae strain N16961S. Data presented in Figure 2 show that, in contrast to the wild-type V. cholerae N16961S strain (Figure 2A), the deletion mutation in the flrA gene in N16961S strain inhibited motility (Figure 2B). Furthermore, complementation of N16961SΔflrA with the wild-type flrA gene cloned and expressed from pWSK29 (pAA146) restored motility to the level seen with the wild-type strain (Figure 2C). Our data clearly indicate that the potential frame shift mutation (49-bp deletion) in flrA gene had no effect on downstream genes in motility. In contrast, inhibition of motility seen with GSΔflrA (Figure 2D) was only partially complemented by the same complementing vector (pAA146) (Figure 2E) indicating that GSΔflrA might have additional mutation(s) affecting motility. As expected, the N16961SΔflaA mutant was defective in motility (Figure 2F). In summary, our data confirmed that the deletion mutation of the flrA gene in GSΔflrA and N16961S (N16961SΔflrA) inhibited motility.

FIGURE 1. Schematic representation of open reading frame (ORF) of flrA gene. ORFs flanking flrA gene are drawn in the diagram with a black arrow indicating 49-bp deletion site in the flrA gene. ORFs are drawn to a scale using SnapGene software.
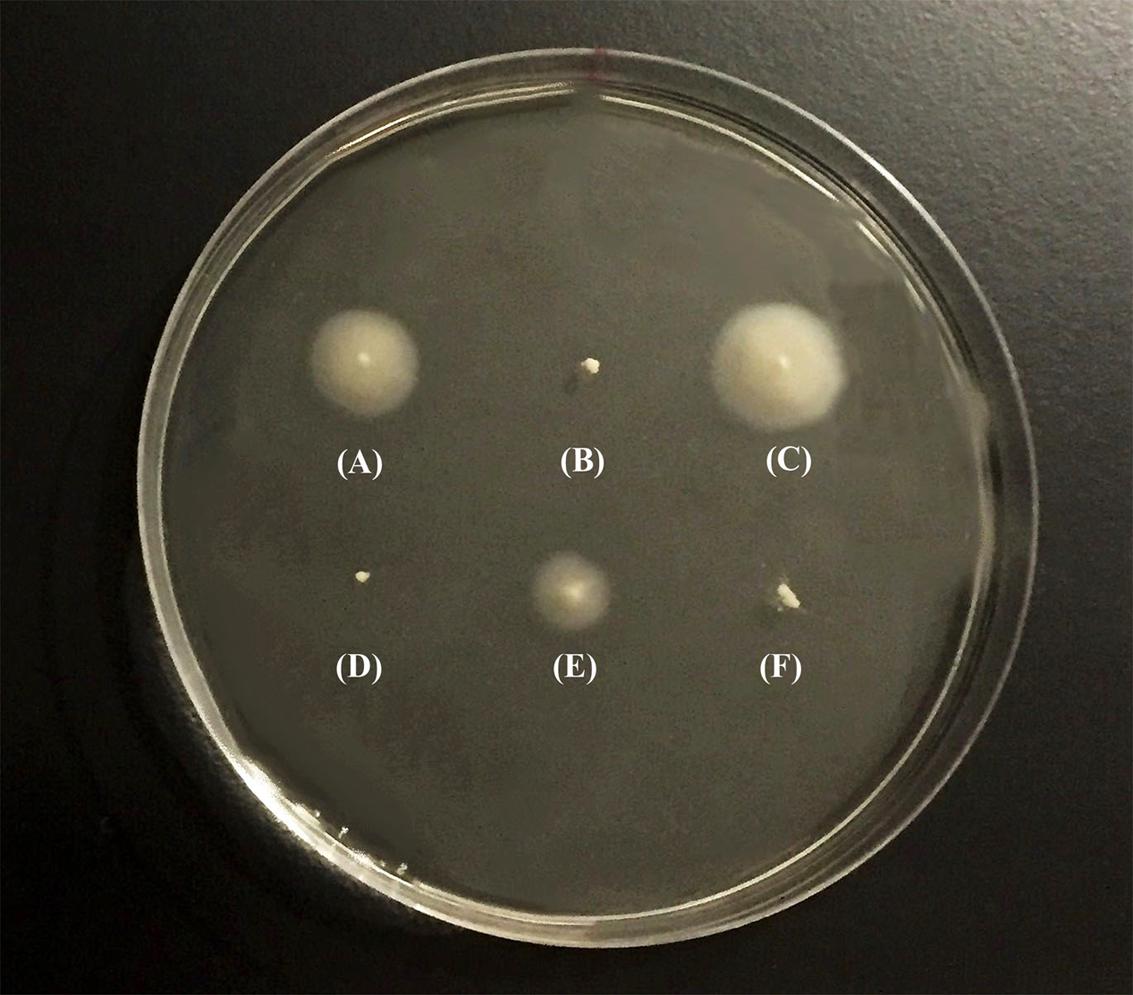
FIGURE 2. Swarming behavior of V. cholerae strains. Swarming behavior of V. cholerae strains was examined on motility agar (0.3% soft agar). Bacterial strains were individually grown overnight in L-broth at 37°C before inoculating them into motility agar. After inoculation, the plates were incubated at 37°C for 8 h and photograph of the culture plate was taken following incubation. (A) V. cholerae N16961S, (B) N16961SΔflrA, (C) N16961SΔflrA/pAA146, (D) GSΔflrA, (E) GSΔflrA /pAA146, and (F) N16961SΔflaA.
FlrA and MshA of V. cholerae Inversely Promoted vps-Independent Biofilm in FSLW Microcosm
We previously reported that the GASP-700D phenotype produced vps-independent biofilm in FSLW microcosms with a ∼1,000-fold down regulation of flrA gene (Jubair et al., 2014). However, in that study, we were unable to elucidate which genetic elements, if any, were involved in such biofilm formation. Previous investigations have demonstrated that V. cholerae employs its flagella and mannose sensitive hemagglutinin (MSHA) pilus, in concert for vps-dependent biofilm formation (Watnick et al., 1999; Watnick and Kolter, 1999). To test if flagella and MSHA pilus contribute similarly to the formation of vps-independent biofilm, we compared biofilm-forming ability between a wild-type V. cholerae N16961S strain and its flrA and mshA mutants both in nutrient-rich L-broth and in nutrient-deficient FSLW. As a control, we used a V. cholerae rugose strain (N16961R) that produces copious amounts of exopolysaccharide and robust biofilm both in L-broth and in FSLW (Jubair et al., 2014). As depicted in Figure 3A, except for the V. cholerae rugose strain, none of the tested strains, including wild-type N16961S and flrA mutants (N16961SΔflrA and GSΔflrA) were able to produce significant biofilm in L-broth. In contrast, when grown in FSLW (Figure 3B), both GSΔflrA and N16961SΔflrA produced significantly (p < 0.01) higher amount of biofilm relative to the wild-type N16961S strain. Furthermore, complementation of N16961SΔflrA with the wild-type flrA gene repressed biofilm formation (Figure 3B); however, complementation of GSΔflrA with wild-type flrA gene was unable to significantly repress biofilm formation (Figure 2B) reinforcing our hypothesis that GSΔflrA might have additional mutation(s) affecting motility and vps-independent biofilm formation. In summary, our results demonstrated that mutation in flrA gene in V. cholerae promoted increased biofilm formation specific to FSLW. Furthermore, we did not observe any significant changes in biofilm formation in FSLW among tested strains when biofilm was quantified after 48 h of incubation at room temperature (Supplementary Figure 1).
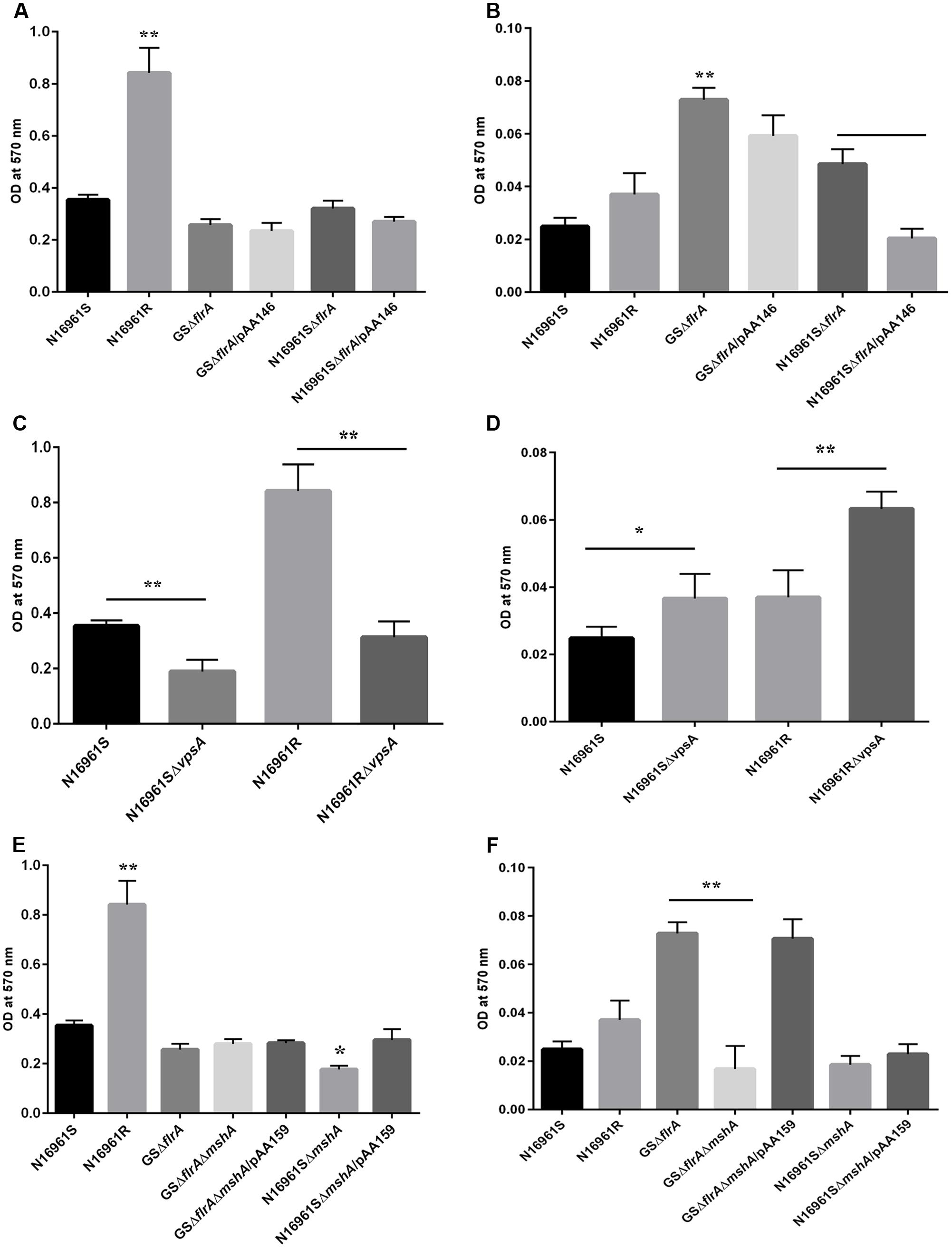
FIGURE 3. Quantitative measurement of biofilm produced by V. cholerae strain. Quantification of biofilm produced by V. cholerae strains grown in: nutrient-rich L-broth (A,C,E), and in nutrient-poor FSLW (B,D,F). The results represent the average of six independent experiments. All the values are expressed as mean ± Δstandard error (SE) calculated from the six readings for each strain. A p-value of <0.05 was considered statistically significant; ∗∗p < 0.01, ∗p < 0.05.
To determine if gene(s) required for vps-dependent biofilm formation enhances vps-independent biofilm formation both in L-broth and in FSLW, we compared biofilm-forming ability of vpsA mutants to that of corresponding wild-type strains both in L-broth and in FSLW. Data presented in Figure 3C exhibited that, as expected, in L-broth, vpsA mutants significantly inhibited vps-dependent biofilm formation compared to their respective wild-type variants; in contrast, vpsA mutants significantly increased biofilm production compared to their wild-type strains in FSLW (Figure 3D). In summary, consistent with our previous report (Jubair et al., 2014) we demonstrated that mutation in vpsA gene positively contributed to vps-independent biofilm in FSLW, but not in L-broth.
To determine the role for MSHA pilus, if any, for vps-independent biofilm production in FSLW, we created an in-frame deletion within the mshA gene both in GSΔflrA and in the N16961S background resulting in GSΔflrAΔmshA and N16961SΔmshA, respectively. Mutation in mshA gene in GSΔflrA (GSΔflrAΔmshA) had no effect on biofilm production in L-broth compared to its wild-type strain (Figure 3E), while N16961SΔmshA significantly reduced (p < 0.05) biofilm formation relative to its wild-type strain N16961S (Figure 3E). In contrast to L-broth, mutation in the mshA gene in GSΔflrA significantly (p < 0.01) repressed biofilm formation in FSLW (Figure 3F). Moreover, complementation of GSΔflrAΔmshA with the wild-type mshA gene cloned and expressed in trans from plasmid pAA159 completely restored biofilm formation as seen with GSΔflrA in FSLW (Figure 3F). Wild-type V. cholerae strain N16961S and its isogenic mshA mutant (N16961SΔmshA) exhibited no significant difference in biofilm formation in FSLW (Figure 3F). In summary, our results confirmed that, while FlrA negatively affects the expression of vps-independent biofilm production in FSLW, MSHA pilus positively contributes to vps-independent biofilm production elicited by GSΔflrA in FSLW. Unlike ΔflrA, a strain carrying mshA mutation in N16961S was unable to promote vps-independent biofilm formation in FSLW (Figure 3F).
To further examine the role for mshA gene in the expression of ΔflrA-driven biofilm in FSLW, we compared three-dimensional architectural structures and thickness of biofilm among wild-type N16961S, N16961SΔflrA, N16961SΔflrA/pAA146, GSΔflrA, GSΔflrAΔmshA, and GSΔflrAΔmshA/pAA159 using confocal microscopic analysis. As expected, N16961ΔflrA produced more biofilm (Figure 4B) compared to wild-type N16961S (Figure 4A). Complementation of N16961ΔflrA with wild-type flrA gene (N16961SΔflrA/pAA146) repressed biofilm formation (Figure 4C). In contrast to N16961ΔflrA, GSΔflrA produced much more biofilm when grown in FSLW (Figure 4D). GSΔflrAΔmshA completely inhibited biofilm formation (Figure 4E); complementation of GSΔflrAΔmshA strain with the wild-type mshA gene fully restored biofilm production at the level seen with GSΔflrA (Figure 4F). To rule out the possibility that the ΔmshA mutant may not have survived in FSLW, we enumerated microorganisms’ counts for each culture before making the slides used for confocal microscopic analysis using standard plate counts. Each culture yielded ca. 108 cfu/ml suggesting that spotty occurrence of GSΔflrAΔmshA (Figure 4E) cells on slide resulted from the inability of the cells to form biofilm, and, not due to low cell counts present in the culture.
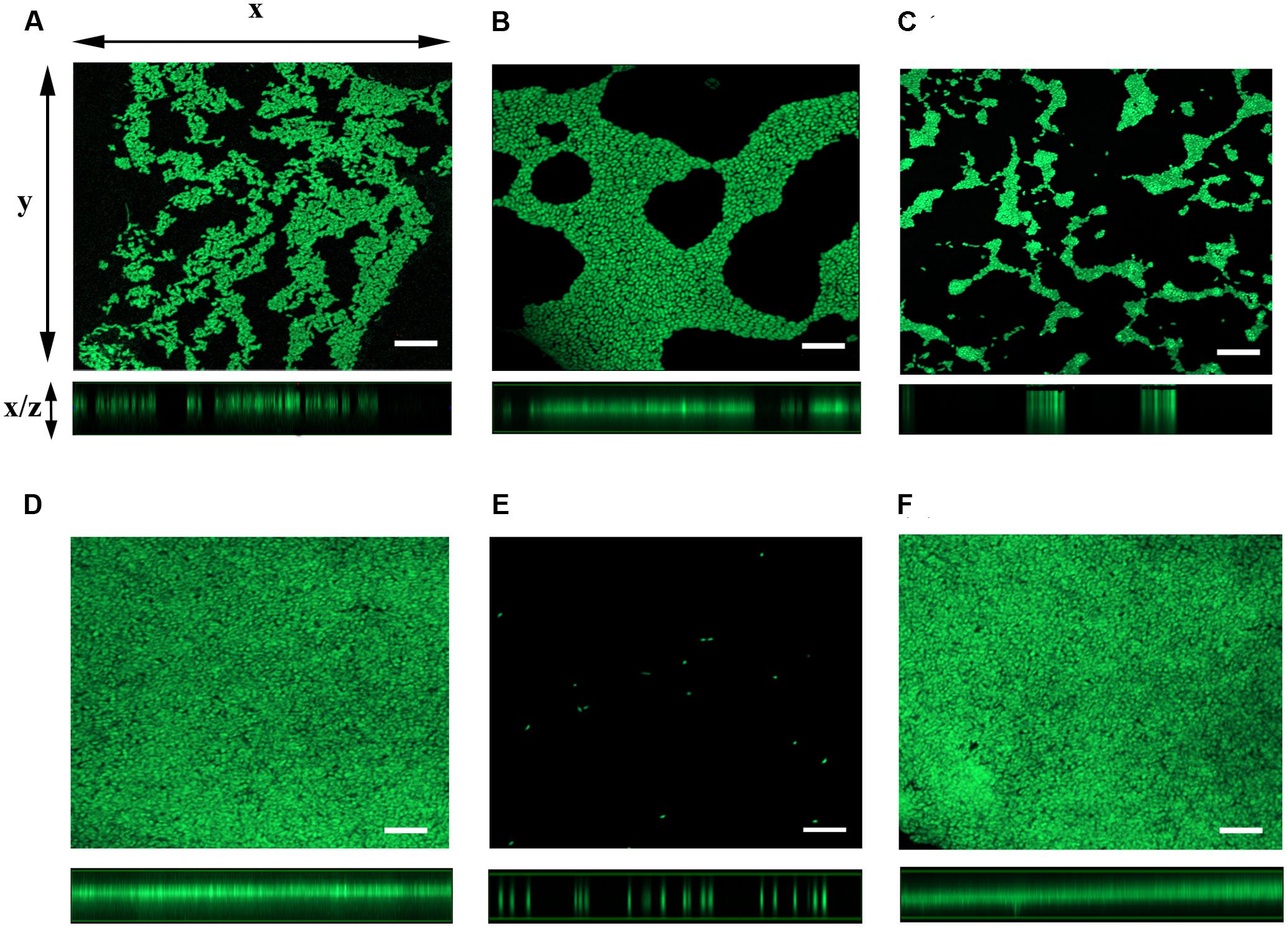
FIGURE 4. Analysis of three-dimensional architecture of V. cholerae biofilm using confocal microscope. Each strain of interest was grown in a 12-well culture plate containing 600 μl FSLW. A glass cover slip was dipped into each culture well and the cultures were incubated overnight statically at room temperature. SYTO 9 dye was added to the culture wells to stain the cover slip and the image was obtained using a laser scanning confocal microscope with an excitation and emission wavelengths of 484 and 500 nm, respectively. Images of x–y sections (top panels) of biofilm and x–z projections (bottom panels) of the same biofilm were captured and analyzed using ZEN 2 imaging software (Blue edition). (A) V. cholerae N16961S, (B) N16961SΔflrA, (C) N16961SΔflrA/pAA146, (D) GSΔflrA, (E) GSΔflrAΔmshA, and (F) GSΔflrAΔmshA/pAA159; bar-10 μm.
vps-Independent Biofilm Promoted V. cholerae to Move toward Nutrients in FSLW
In aquatic environments, many planktonic bacterial species use a combination of chemotaxis and flagellar motility to sense and acquire nutrients (Butler and Camilli, 2005). V. cholerae is no exception to this phenomenon. However, it is not known how non-motile bacterial phenotypes or phenotypes lacking productive motility (commonly found in biofilm communities) acquire nutrients from their aquatic reservoir. Specifically, we questioned whether non-motile V. cholerae, promoting ΔflrA-driven vps-independent biofilm in FSLW, can sense and move toward nutrients and, if so, does chemotaxis play any role in such mechanism? In our previous study (Jubair et al., 2012), we reported that nutrients, including chitin and phosphate (when added to FSLW microcosm) promoted the growth of highly starved (180 and 700 days surviving microorganisms) V. cholerae persisting in nutrient-poor FSLW microcosms. Based on that observation we hypothesized that chitin and phosphate serve as chemo-attractants in chemotaxis assays in FSLW. As illustrated in Figure 5, GSΔflrA and N1696S1ΔflrA mutants moved toward chitin (Figure 5A) and phosphate (Figure 5B) compared to their wild-type phenotype (N16961S). Strikingly, a V. cholerae rugose strain (N16961R), capable of producing robust vps-dependent biofilm (Ali et al., 2002), was unable to move toward nutrients in FSLW (Figures 5A,B). Complementation of GSΔflrA and N1696S1ΔflrA with a wild-type flrA gene inhibited movement to nutrients (Figures 5A,B). In contrast and, as expected, GSΔflrAΔmshA, that repressed vps-independent biofilm (described above), was unable to move toward nutrients (Figures 5A,B). Not surprisingly, unlike the N16961SΔflrA mutant, the N16961SΔmshA mutant, that was unable to induce vps-independent biofilm, failed to move to nutrients (Figures 5A,B).
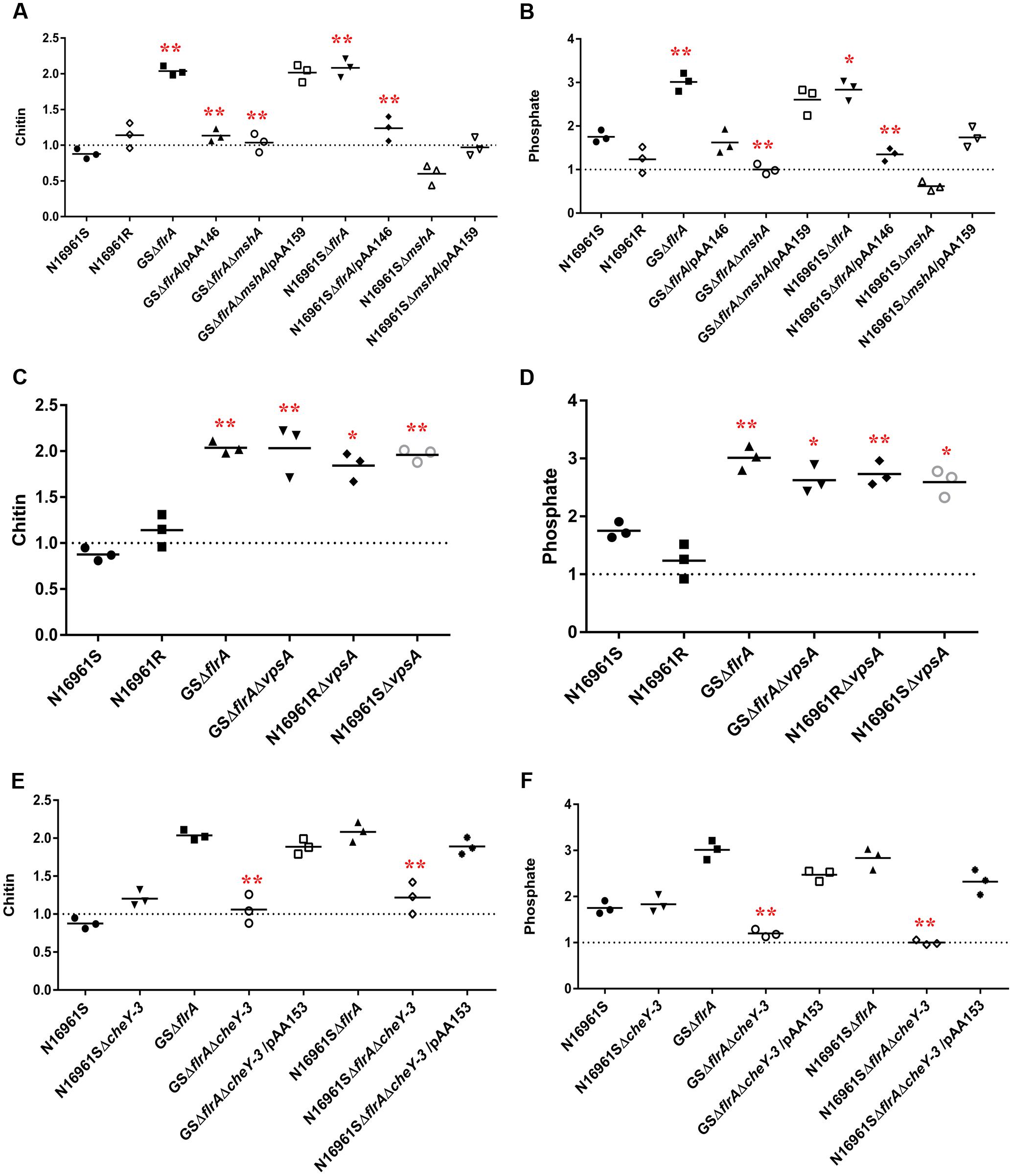
FIGURE 5. vps-independent biofilm promoted V. cholerae to move toward nutrient sources in FSLW as measured by chemotaxis. Chemotaxis assay was performed on V. cholerae strains as described previously (Butler et al., 2006) with modifications. Briefly, assay was performed in nutrient-deficient FSLW or FSLW supplemented with nutrients, including chitin or phosphate. The results represent three independent experiments and the data were expressed as a comparative index (CI) for each strain. CI was calculated by the number of bacterial counts (colony forming units per milliliter) present in FSLW in the capillary tube supplemented with nutrient divided by the number of bacterial counts (colony forming units per milliliter) present in FSLW in the capillary tube not supplemented with nutrient. The horizontal bar indicates the mean of the results from three capillary tube experiments. The observations resulting from an experiment pointing at or below the dashed line reflect no measurable response between control (-nutrient) and experiment (+nutrient) for each strain examined. A p-value of <0.05 was considered statistically significant. Impact of ΔflrA and ΔmshA mutants and reverted mutants on: (A) chitin and (B) phosphate; impact of ΔcheY-3 mutants and reverted mutants on: (C) chitin and (D) phosphate; impact of ΔvpsA mutants on: (E) chitin and (F) phosphate; ∗∗p < 0.01,∗p < 0.05.
We previously reported that vpsA mutants inhibiting vps-dependent biofilm promoted vps-independent biofilm in FSLW (Jubair et al., 2014). We questioned if ΔvpsA-driven biofilm can also move toward nutrients in FSLW. To test our idea we compared the ability of vpsA mutants to move toward nutrients relative to their corresponding wild-type strains. Data presented in Figures 5C,D indicated that ΔvpsA-driven biofilm akin to ΔflrA-driven biofilm moved to chitin and phosphate compared to their corresponding wild-type strains. Our data clearly indicate that in contrast to vps-dependent biofilm, vps-independent biofilm elicited by both ΔflrA- and ΔvpsA-driven biofilm can promote V. cholerae to move toward nutrients, including chitin and phosphate in FSLW.
Previous reports suggested that bacterial chemotaxis genes, including cheY-3 are involved in chemotaxis (Lee et al., 2001). To determine if cheY-3 plays any role to promote V. cholerae to move toward nutrients in FSLW, we created a null mutation in cheY-3 gene in the background of N16961S, GSΔflrA, and N16961SΔflrA. As depicted in Figures 5E,F, compared to wild-type strain N16961S, GSΔflrAΔcheY-3 and N16961SΔflrAΔcheY-3 significantly (p < 0.01) repressed their movement toward chitin and phosphate; complementation of mutation with the wild-type cheY-3 gene restored movement to chitin and phosphate (Figures 5E,F). Interestingly, we observed that there was no significant difference in biofilm production between N16961SΔflrA and N16961SΔflrAΔcheY-3 (Supplementary Figure 2). Our data indicate that CheY-3 of V. cholerae significantly (p < 0.01) contributed to movement toward nutrients, including chitin and phosphate in FSLW.
Discussion
Vibrio cholerae produces two types of biofilm, including vps-dependent and vps-independent biofilms (Kierek and Watnick, 2003a,b; Moorthy and Watnick, 2004). vps-dependent biofilm, encoded by vps gene clusters, promotes “rugose” colony morphology associated with three-dimensional biofilm matrix consisting of pillars interspersed with water channels (Yildiz and Schoolnik, 1999). The formation of vps-dependent biofilm has been investigated in poorly defined or undefined commercially available media, including L-broth containing monosaccharides, aminoacids, and vitamins among other ingredients (Ali et al., 2002; Beyhan and Yildiz, 2007). In addition to undefined media, vps-dependent biofilm has also been studied in minimal media supplemented with monosaccharides (Wai et al., 1998; Moorthy and Watnick, 2004). In contrast to undefined media or defined media contributing to vps-dependent biofilm formation, previous studies (Kierek and Watnick, 2003a,b) have demonstrated that only vps-dependent biofilm, not vps-independent biofilm, can be induced in fresh surface water supplemented with monosaccharides. In contrast, V. cholerae produced both vps-dependent and vps-independent biofilm in commercially available artificial seawater (ASW) or real seawater supplemented either with monosaccharide sources (yeast extract) or vitamin assay casamino acids (CAA), also referred as defined media (Kierek and Watnick, 2003b,a; Moorthy and Watnick, 2004). Furthermore, early investigations have reported that Ca+2 ions of seawater contributed to vps-independent biofilm, and that V. cholerae flagella and MSHA pilus are required in concert for vps-independent biofilm as seen with vps-dependent biofilm (Kierek and Watnick, 2003b).
Contrary to previous investigations described above, we observed that, compared to wild-type V. cholerae N16961S strain, mutation in the flrA gene in V. cholerae promoted vps-independent biofilm formation in FSLW, not supplemented with any extraneous nutrients (Figures 3B, 4B). In addition to the ΔflrA, inactivation of vpsA (Figures 3C,D; Jubair et al., 2014), inhibiting vps-dependent biofilm formation, promoted vps-independent biofilm in FSLW. In summary, our current observation and a previous report (Jubair et al., 2014) suggest that vps-independent biofilm is produced in FSLW either by inactivation of flrA or vpsA. Our observation is in contrast to a previous study (Kierek and Watnick, 2003a) indicating that a vps mutant was unable to form vps-independent biofilm in fresh water even with supplementation of nutrients. The observed differences between our study and that previous report (Kierek and Watnick, 2003a) could be attributed to V. cholerae strain differences: while we used V. cholerae N16961S strain (a serogroup O1 biotype El Tor strain), previous studies used a V. cholerae MO10 strain (a serogroup O139 strain possessing a capsule unlike serogroup O1 strain). Indeed, V. cholerae strains with distinct genetic and phenotypic traits exhibited distinct vps-dependent biofilm formation as reported previously (Watnick et al., 2001; Ali et al., 2002). We primarily used GSΔflrA and N16961SΔflrA for this study. In contrast to N16961SΔflrA, we were only able to partially restore motility and biofilm phenotypes in GSΔflrA using a complementing vector implying that GSΔflrA may have sustained additional mutation(s) affecting these phenotypes. In future studies, we will perform whole-genome sequencing and bioinformatic analysis to determine if GSΔflrA indeed sustained additional mutation(s). Consistent with a previous study (Kierek and Watnick, 2003b), we found that MSHA is required for ΔflrA-driven vps-independent biofilm formation in FSLW (Figures 3F, 4E).
In vps-dependent mature biofilm, V. cholerae inhibits flagella synthesis while promoting VPS biosynthesis; thus, it is tempting to speculate that vps-independent biofilm could be a component of the vps-dependent mature biofilm. Indeed, previous studies have demonstrated that both vps-dependent and vps-independent biofilm are produced in real seawater and ASW supplemented with nutrients and vitamins (Kierek and Watnick, 2003a,b). Given the very high energy cost associated with the production of exopolysaccharide and biofilm, why would V. cholerae prefer to synthesize two types of biofilm at a given time? Does V. cholerae alternate between vps-dependent and -independent biofilm pathways after sensing environmental cues requiring a specific biofilm formation pathway? These are important questions that are yet to be fully elucidated. However, available data suggest that V. cholerae employs vps-dependent biofilm to confer resistance to chlorine, oxidative and osmotic stresses (Morris et al., 1996; Wai et al., 1998; Yildiz and Schoolnik, 1999); furthermore, this biofilm protects the bacterium from predators (Matz et al., 2005). Based on these studies, it has been inferred that vps-dependent biofilm promotes V. cholerae’s environmental persistence by evading environmental stressors. So, what is the role of vps-independent biofilm? In our recent study (Jubair et al., 2014), we reported that vps-independent biofilm resists only oxidative stress, not chlorine or osmotic stresses. Furthermore, we demonstrated that vps-independent biofilm is scattered and dispersed in nature (Jubair et al., 2014). Based on these observations, we hypothesized that vps-independent biofilm attracts or scavenges scarce nutrients present in aquatic reservoirs. Our hypothesis is based in part of our earlier finding (Jubair et al., 2012) that very starved and aggregated V. cholerae (identified as a novel “persister” phenotype) acquired chitin and phosphate from FSLW microcosms supplemented with these nutrients. Consistent with our hypothesis we have demonstrated that strains having vps-independent, but not vps-dependent, biofilm moved toward nutrients, including chitin and phosphate, as determined by a standard chemotaxis assay (Figures 5A,B). Intriguingly, strains with vps-independent biofilm resulting from either inactivation of flrA or vpsA were able to move toward nutrients in FSLW suggesting that V. cholerae has retained flexibility in attracting nutrients (Figures 5A–D). For example, the microorganism can retain functional vps-dependent biofilm while promoting vps-independent biofilm by repressing flrA. Alternatively, the bacterium may repress vps genes facilitating down-regulation of vps-dependent biofilm while promoting vps-independent biofilm without affecting flrA (retain motility). Generally, bacterial motility and chemotaxis are interdependent or work in concert (Butler and Camilli, 2005); however, we report here that non-motile ΔflrA-driven vps-independent biofilm promotes movement of V. cholerae toward nutrients. Currently, we do not have evidence to show how non-motile bacterium moves to nutrients; however, we speculate that non-motile cells move as biofilm (aggregate) toward nutrient. As vps-independent biofilm is mshA dependent and vpsA independent, it is possible that biofilm matrix deposition (adhesin) on the microorganism’s cell surface and/or bacterial cell–cell interaction via MSHA pilus contribute to the movement of biofilm bacteria toward nutrients.
Vibrio cholerae is ubiquitous to aquatic environments where it can persist for decades. Microorganism can use chitin, the most abundant source of carbon in aquatic reservoirs (Dang and Lovell, 2016), as a sole carbon source using genetic elements present in the bacterium’s chromosome (Meibom et al., 2004). However, to maintain optimal growth, bacteria require balanced nutrients; unfortunately, inorganic phosphate, a key growth-promoting factor for V. cholerae, is very limited or absent in both fresh and ocean water limiting the optimal growth of bacteria, including culturable forms of toxigenic V. cholerae (Smith and Schindler, 2009; Schindler, 2012). Because of nutrient limitation, the isolation of culturable V. cholerae from aquatic reservoirs has been very low even during ongoing outbreaks in a country where cholera is epidemic (Huq et al., 2005; Alam et al., 2015). However, available data suggest that agricultural runoff, particularly during rainfall, enriches surface and estuary waters with increased inorganic phosphate (Smith and Schindler, 2009). Based on the data presented in this paper we propose that strains expressing the vps-independent biofilm of toxigenic V. cholerae are able to move to acquire phosphate promoting the bacterium’s growth at least transiently in aquatic reservoirs. Consumption of water with increased toxigenic V. cholerae counts following rainfall may serve as a trigger for cholera epidemics, in keeping with the observation that epidemics often follow rainfall events in cholera endemic countries (Alam et al., 2006, 2015).
In summary, we provide evidence in this paper that vps-independent biofilm plays a major role in attraction to nutrients in FSLW. Our observations could have major implications for our understanding of the persistence of V. cholerae in aquatic reservoirs, the potential impact of nutrient (phosphate and chitin) recycling (Dang and Lovell, 2016), and factors responsible for initiation of cholera epidemics in cholera-endemic countries.
Author Contributions
AA designed and supervised the research experiments; SS-R performed the experiments and processed the data; AA and SS-R interpreted data and performed statistical analysis; and AA and SS-R wrote, critically revised, and approved the manuscript.
Funding
This work was supported by funding from National Institute of Health (NIH) grants R01 AI097405 awarded to J. Glenn Morris at University of Florida and R01 AI039129 awarded to R. Bradley Sack at Johns Hopkins University. AA is the University of Florida subcontract PI of the R01 AI039129 grant. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Mohammad Jubair and Meer T. Alam for helping us with the biofilm and chemotaxis assay. We thank Dr. Glenn Morris for critically reviewing the manuscript. We acknowledge the assistance of Dr. Hedwin Kitdorlang Dkhar (College of Veterinary Medicine), Rodolfo Alvarado, and Karen Kelly of Electron Microscopy Core, ICBR, University of Florida at Gainesville for helping us with confocal microscopy and Dr. Mohammed H. Rashid of Emerging Pathogens Institute for his technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01770/full#supplementary-material
References
Alam, M., Sultana, M., Nair, G. B., Sack, R. B., Sack, D. A., Siddique, A. K., et al. (2006). Toxigenic Vibrio cholerae in the aquatic environment of Mathbaria, Bangladesh. Appl. Environ. Microbiol. 72, 2849–2855. doi: 10.1128/aem.72.4.2849-2855.2006
Alam, M. T., Weppelmann, T. A., Longini, I., De Rochars, V. M., Morris, J. G. Jr., and Ali, A. (2015). Increased isolation frequency of toxigenic Vibrio cholerae O1 from environmental monitoring sites in Haiti. PLOS ONE 10:e0124098. doi: 10.1371/journal.pone.0124098
Ali, A., Johnson, J. A., Franco, A. A., Metzger, D. J., Connell, T. D., Morris, J. G. J., et al. (2000a). Mutations in the extracellular protein secretion pathway genes (eps) interfere with rugose polysaccharide production in and motility of Vibrio cholerae. Infect. Immun. 68, 1967–1974.
Ali, A., Mahmud, Z. H., Morris, J. G. JR., Sozhamannan, S., and Johnson, J. A. (2000b). Sequence analysis of TnphoA insertion sites in Vibrio cholerae mutants defective in rugose polysaccharide production. Infect. Immun. 68, 6857–6864.
Ali, A., Rashid, M. H., and Karaolis, D. K. R. (2002). High-frequency rugose exopolysaccharide production by Vibrio cholerae. Appl. Environ. Microbiol. 68, 5773–5778. doi: 10.1128/AEM.68.11.5773-5778.2002
Beyhan, S., and Yildiz, F. H. (2007). Smooth to rugose phase variation in Vibrio cholerae can be mediated by a single nucleotide change that targets c-di-GMP signalling pathway. Mol. Microbiol. 63, 995–1007. doi: 10.1111/j.1365-2958.2006.05568.x
Bourret, R. B., and Stock, A. M. (2002). Molecular information processing: lessons from bacterial chemotaxis. J. Biol. Chem. 277, 9625–9628. doi: 10.1074/jbc.R100066200
Butler, S. M., and Camilli, A. (2005). Going against the grain: chemotaxis and infection in Vibrio cholerae. Nat. Rev. Microbiol. 3, 611–620. doi: 10.1038/nrmicro1207
Butler, S. M., Nelson, E. J., Chowdhury, N., Faruque, S. M., Calderwood, S. B., and Camilli, A. (2006). Cholera stool bacteria repress chemotaxis to increase infectivity. Mol. Microbiol. 60, 417–426. doi: 10.1111/j.1365-2958.2006.05096.x
Colwell, R. R., and Huq, A. (1994). “Vibrios in the environment: viable but nonculturable Vibrio cholerae,” in Vibrio cholerae and cholera: molecular to global perspectives, eds I. K. Wachsmuth, P. A. Blake, and Ø. Olsvik (Washington, DC: American Society for Microbiology).
Dang, H., and Lovell, C. R. (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 80, 91–138. doi: 10.1128/MMBR.00037-15
Donnenberg, M. S., and Kaper, J. B. (1991). Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59, 4310–4317.
Faruque, S. M., Albert, M. J., and Mekalanos, J. J. (1998). Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62, 1301–1314.
Finkel, S. E. (2006). Long-term survival during stationary phase: evolution and the GASP phenotype. Nat. Rev. Microbiol. 4, 113–120. doi: 10.1038/nrmicro1340
Finkel, S. E., and Kolter, R. (1999). Evolution of microbial diversity during prolonged starvation. Proc. Natl. Acad. Sci. U.S.A. 96, 4023–4027. doi: 10.1073/pnas.96.7.4023
Gaddy, J. A., and Actis, L. A. (2009). Regulation of Acinetobacter baumannii biofilm formation. Future Microbiol. 4, 273–278. doi: 10.2217/fmb.09.5
Gardel, C. L., and Mekalanos, J. J. (1996). Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 64, 2246–2255.
Gosink, K. K., Kobayashi, R., Kawagishi, I., and Hase, C. C. (2002). Analyses of the roles of the three cheA homologs in chemotaxis of Vibrio cholerae. J. Bacteriol. 184, 1767–1771. doi: 10.1128/JB.184.6.1767-1771.2002
Heidelberg, J. F., Eisen, J. A., Nelson, W. C., Clayton, R. A., Gwinn, M. L., Dodson, R. J., et al. (2000). DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406, 477–483. doi: 10.1038/35020000
Herrington, D. A., Hall, R. H., Losonsky, G. A., Mekalanos, J. J., Taylor, R. K., and Levine, M. M. (1988). Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 168, 1487–1492. doi: 10.1084/jem.168.4.1487
Hsiao, A., Toscano, K., and Zhu, J. (2008). Post-transcriptional cross-talk between pro- and anti-colonization pili biosynthesis systems in Vibrio cholerae. Mol. Microbiol. 67, 849–860. doi: 10.1111/j.1365-2958.2007.06091.x
Hsiao, A., Xu, X., Kan, B., Kulkarni, R. V., and Zhu, J. (2009). Direct regulation by the Vibrio cholerae regulator ToxT to modulate colonization and anticolonization pilus expression. Infect. Immun. 77, 1383–1388. doi: 10.1128/iai.01156-08
Huq, A., Colwell, R. R., Rahman, R., Ali, A., Chowdhury, M. A. R., Parveen, S., et al. (1990). Detection of Vibrio cholerae O1 in the aquatic environment by fluorescent-monoclonal antibody and culture methods. Appl. Environ. Microbiol. 56, 2370–2373.
Huq, A., Sack, R. B., Nizam, A., Longini, I. M., Nair, G. B., Ali, A., et al. (2005). Critical factors influencing the occurrence of Vibrio cholerae in the environment of Bangladesh. Appl. Environ. Microbiol. 71, 4645–4654. doi: 10.1128/aem.71.8.4645-4654.2005
Huq, A., Small, E. B., West, P. A., Huq, M. I., Rahman, R., and Colwell, R. R. (1983). Ecology of Vibrio cholerae O1 with special reference to planktonic crustacean copepods. Appl. Environ. Microbiol. 45, 275–283.
Islam, M. S., Drasar, B. S., and Bradley, D. J. (1990). Long-term persistence of toxigenic Vibrio cholerae 01 in the mucilaginous sheath of a blue-green alga, Anabaena variabilis. J. Trop. Med. Hyg. 93, 133–139.
Jubair, M., Atanasova, K. R., Rahman, M., Klose, K. E., Yasmin, M., Yilmaz, O., et al. (2014). Vibrio cholerae persisted in microcosm for 700 days inhibits motility but promotes biofilm formation in nutrient-poor lake water microcosms. PLOS ONE 9:e92883. doi: 10.1371/journal.pone.0092883
Jubair, M., Morris, J. G. Jr., and Ali, A. (2012). Survival of Vibrio cholerae in nutrient-poor environments is associated with a novel “persister” phenotype. PLOS ONE 7:e45187. doi: 10.1371/journal.pone.0045187
Kierek, K., and Watnick, P. I. (2003a). Environmental determinants of Vibrio cholerae biofilm development. Appl. Environ. Microbiol. 69, 5079–5088.
Kierek, K., and Watnick, P. I. (2003b). The Vibrio cholerae O139 O-antigen polysaccharide is essential for Ca2+-dependent biofilm development in sea water. Proc. Natl. Acad. Sci. U.S.A. 100, 14357–14362. doi: 10.1073/pnas.2334614100
Klose, K. E., and Mekalanos, J. J. (1998). Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28, 501–520. doi: 10.1046/j.1365-2958.1998.00809.x
Lee, S. H., Butler, S. M., and Camilli, A. (2001). Selection for in vivo regulators of bacterial virulence. Proc. Natl. Acad. Sci. U.S.A. 98, 6889–6894. doi: 10.1073/pnas.111581598
Matz, C., McDougald, D., Moreno, A. M., Yung, P. Y., Yildiz, F. H., and Kjelleberg, S. (2005). Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 102, 16819–16824. doi: 10.1073/pnas.0505350102
Meibom, K. L., Li, X. B., Nielsen, A. T., Wu, C. Y., Roseman, S., and Schoolnik, G. K. (2004). The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. U.S.A. 101, 2524–2529. doi: 10.1073/pnas.0308707101
Miller, V. L., and Mekalanos, J. J. (1988). A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170, 2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988
Moorthy, S., and Watnick, P. I. (2004). Genetic evidence that the Vibrio cholerae monolayer is a distinct stage in biofilm development. Mol. Microbiol. 52, 573–587. doi: 10.1111/j.1365-2958.2004.04000.x
Morris, J. G. Jr. (2011). Cholera - modern pandemic disease of ancient lineage. Emerg. Infect. Dis. 17, 2099–2104. doi: 10.3201/eid1711.111109
Morris, J.G. Jr., Sztein, M. B., Rice, E. W., Nataro, J. P., Losonsky, G. A., Panigrahi, P., et al. (1996). Vibrio cholerae O1 can assume a chlorine-resistant rugose survival form that is virulent for humans. J. Infect. Dis. 174, 1364–1368. doi: 10.1093/infdis/174.6.1364
Nelson, E. J., Chowdhury, A., Flynn, J., Schild, S., Bourassa, L., Shao, Y., et al. (2008). Transmission of Vibrio cholerae is antagonized by lytic phage and entry into aquatic environment. PLOS Pathog. 4:e1000187. doi: 10.1371/journal.ppat.1000187
Nelson, E. J., Harris, J. B., Morris, J. G. Jr., Calderwood, S. B., and Camilli, A. (2009). Cholera transmission: the host, pathogen and bacteriophage dynamic. Nat. Rev. Microbiol. 7, 693–702. doi: 10.1038/nrmicro2204
Prouty, M. G., Correa, N. E., and Klose, K. E. (2001). The novel sigma54- and sigma28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39, 1595–1609. doi: 10.1046/j.1365-2958.2001.02348.x
Rhine, J. A., and Taylor, R. K. (1994). TcpA pilin sequences and colonization requirements for O1 and O139 Vibrio cholerae. Mol. Microbiol. 13, 1013–1020. doi: 10.1111/j.1365-2958.1994.tb00492.x
Schindler, D. W. (2012). The dilemma of controlling cultural eutrophication of lakes. Proc. Biol. Sci. 279, 4322–4333. doi: 10.1098/rspb.2012.1032
Shukla, B. N., Singh, D. V., and Sanyal, S. C. (1995). Attachment of non-culturable toxigenic Vibrio cholerae O1 and non-O1 and Aeromonas spp. to the aquatic arthropod Gerris spinolae and plants in the River Ganga, Varanasi. FEMS Immunol. Med. Microbiol. 12, 113–120. doi: 10.1111/j.1574-695X.1995.tb00182.x
Smith, V. H., and Schindler, D. W. (2009). Eutrophication science: where do we go from here? Trends Ecol. Evol. 24, 201–207. doi: 10.1016/j.tree.2008.11.009
Tamplin, M. L., Gauzens, A. L., Huq, A., Sack, D. A., and Colwell, R. R. (1990). Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl. Environ. Microbiol. 56, 1977–1980.
Thelin, K. H., and Taylor, R. K. (1996). Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect. Immun. 64, 2853–2856.
Tomaras, A. P., Flagler, M. J., Dorsey, C. W., Gaddy, J. A., and Actis, L. A. (2008). Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 154(Pt 11), 3398–3409. doi: 10.1099/mic.0.2008/0194710
Wadhams, G. H., and Armitage, J. P. (2004). Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5, 1024–1037. doi: 10.1038/nrm1524
Wai, S. N., Mizunoe, Y., Takade, A., Kawabata, S. I., and Yoshida, S. I. (1998). Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 64, 3648–3655.
Wang, R. F., and Kushner, S. R. (1999). Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100, 195–199. doi: 10.1016/0378-1119(91)90366-J
Watnick, P. I., Fullner, K. J., and Kolter, R. (1999). A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J. Bacteriol. 181, 3606–3609.
Watnick, P. I., and Kolter, R. (1999). Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34, 586–595. doi: 10.1046/j.1365-2958.1999.01624.x
Watnick, P. I., Lauriano, C. M., Klose, K. E., Croal, L., and Kolter, R. (2001). The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol. Microbiol. 39, 223–235. doi: 10.1046/j.1365-2958.2001.02195.x
Xu, H.-S., Roberts, N., Singleton, F. L., Attwell, R. W., Grimes, D. J., and Colwell, R. R. (1982). Survival and viability of non-culturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microb. Ecol. 8, 313–323. doi: 10.1007/BF02010671
Yildiz, F. H., and Schoolnik, G. K. (1999). Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 96, 4028–4033. doi: 10.1073/pnas.96.7.4028
Keywords: Vibrio cholerae, biofilm, motility, nutrient-poor, microcosm, chemotaxis
Citation: Sinha-Ray S and Ali A (2017) Mutation in flrA and mshA Genes of Vibrio cholerae Inversely Involved in vps-Independent Biofilm Driving Bacterium Toward Nutrients in Lake Water. Front. Microbiol. 8:1770. doi: 10.3389/fmicb.2017.01770
Received: 31 May 2017; Accepted: 31 August 2017;
Published: 13 September 2017.
Edited by:
Hongyue Dang, Xiamen University, ChinaReviewed by:
Kai Papenfort, Ludwig Maximilian University of Munich, GermanyAnne-Marie Krachler, University of Texas Health Science Center at Houston, United States
Juan M. Tomas, University of Barcelona, Spain
Copyright © 2017 Sinha-Ray and Ali. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Afsar Ali, aali@epi.ufl.edu
 Shrestha Sinha-Ray
Shrestha Sinha-Ray Afsar Ali
Afsar Ali