- 1 Department of Bioinformatics, Biocenter, University of Würzburg, Würzburg, Germany
- 2 Division of Microbiology, University of Osnabrück, Osnabrück, Germany
The infectious intracellular lifestyle of Salmonella enterica relies on the adaptation to nutritional conditions within the Salmonella-containing vacuole (SCV) in host cells. We summarize latest results on metabolic requirements for Salmonella during infection. This includes intracellular phenotypes of mutant strains based on metabolic modeling and experimental tests, isotopolog profiling using 13C-compounds in intracellular Salmonella, and complementation of metabolic defects for attenuated mutant strains towards a comprehensive understanding of the metabolic requirements of the intracellular lifestyle of Salmonella. Helpful for this are also genomic comparisons. We outline further recent studies and which analyses of intracellular phenotypes and improved metabolic simulations were done and comment on technical required steps as well as progress involved in the iterative refinement of metabolic flux models, analyses of mutant phenotypes, and isotopolog analyses. Salmonella lifestyle is well-adapted to the SCV and its specific metabolic requirements. Salmonella metabolism adapts rapidly to SCV conditions, the metabolic generalist Salmonella is quite successful in host infection.
Introduction
Salmonella enterica serovar Typhimurium (S. Typhimurium) is an important human gastrointestinal pathogen with an invasive and facultative intracellular lifestyle (Neidhardt, 1996; Eisenreich et al., 2010). Among the various habitats that can be colonized by Salmonella, the adaptation to life inside the host cell is of specific interest, since this ability is considered as crucial for systemic infections with fatal outcome. The World Health Organization estimated 1.4 million cases of non-typhoidal Salmonella infections. Furthermore, these cause 580 deaths annually even in the United States (World Health Organization, 2005). Infections are often associated with selected subgroups as elderly or patients suffering from HIV and connective tissue disorders (Cummings et al., 2010).
Throughout the intracellular life, Salmonella remains in a membrane-bound compartment, which is termed Salmonella-containing vacuole or SCV. The SCV is probably a unique compartment that is formed by the combined action of a large number of bacterial virulence factors (Figure 1). Virulent Salmonellae are able to modify this vacuole in order to escape killing in the endocytic pathway, and to proliferate within host cells (Haraga et al., 2008). The ability to survive and replicate within host cells is closely related to the systemic pathogenesis of Salmonella in a murine model of typhoid fever. Mutant strains defective in intracellular replication due to auxotrophies are also attenuated in virulence in an animal murine model of typhoid fever (Fields et al., 1986). Salmonella is able to rapidly multiply in various eukaryotic cell lines, but the proliferation appears to be far less rapid within cells in tissues of infected hosts, indicating a more restrictive situation in vivo (Mastroeni et al., 2009). The SCV is commonly considered as a nutritional deprived environment, and this notion is based on the phenotypes of auxotrophic strains, analyses of bacterial reporter strains, and microarray analyses. However, the fact that Salmonella replicates within the SCV indicates the successful adaptation to this intracellular environment.
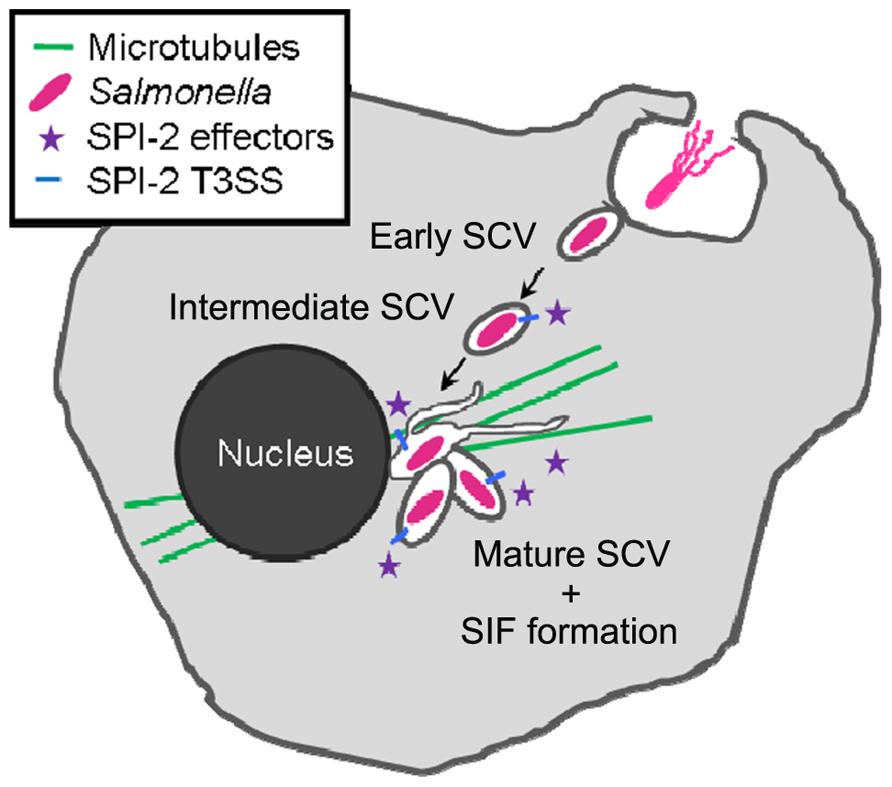
FIGURE 1. Intracellular lifestyle of Salmonella. Salmonella is taken up by host cells either by Salmonella-induced invasion (T3SS-1 triggered macropinocytosis) or via phagocytosis. By translocating effector proteins via the T3SS-2 into the host cell, the SCV undergoes an altered maturation process characterized by specific endosomal markers. The onset of Salmonella-induced filaments (SIFs) formation coincides with the start of Salmonella replication between 4 and 6 h after infection. SIFs develop on a microtubule scaffold.
Despite the remarkable increase in understanding of the cellular microbiology of Salmonella infections and the molecular functions of virulence factors required for intracellular life, the nutritional basis of life of Salmonella within the SCV is still not completely understood.
Understanding how Salmonella survives and thrives within this compartment and how nutrients are acquired is not only essential for the understanding of the intracellular lifestyle, but might as well open new avenues to therapeutic interference with Salmonella infections.
Methods and Approaches to get Insights into Intracellular Nutrition
To target metabolism of intracellular Salmonella, several approaches have been established covering in vitro approaches from analyzing simple growth behavior in full or minimal medium and changes in morphology (Paterson et al., 2009), intracellular replication ability (Bowden et al., 2009,2010) to more complex transcriptome analysis (Eriksson et al., 2003) or 13C-isotopolog profiling analysis (13C-IPA; Götz and Goebel, 2010) in a cell culture model. Experiments to test if a gene of interest contributes to virulence are mainly done in macrophage cell lines as strains unable to replicate within macrophages proved to be avirulent (Fields et al., 1986). In addition, epithelial cell infection models, such as HeLa or CaCo-2 cells (Götz and Goebel, 2010) which are targeted first in Salmonella infection are under investigation.
The importance of certain metabolic functions and their corresponding pathways can be investigated by intracellular replication assays performed in murine macrophage cell lines with Salmonella wild-type (WT) and mutant strains with metabolic defects (Bowden et al., 2009; Lim et al., 2010). Mutant strains can be easily generated by the λ-Red mediated mutagenesis approach (Datsenko and Wanner, 2000). The intracellular replication ability of these mutant strains compared to the WT strain indicates the importance of the respective metabolic pathway for intracellular nutrition and, in turn, intracellular survival and replication. Other useful ways to “scan” for the complete setup of essential genes and metabolic enzymes are transcriptome and proteome analyses of Salmonella strains isolated from infected murine macrophage cell lines (Eriksson et al., 2003; Shi et al., 2006) giving direct evidence of genes and proteins expressed under intracellular conditions. The application of 13C-IPA to follow metabolic fluxes in the host and the bacteria cells in vitro is on the rise (Figure 2). It has been widely used for analyzing metabolic fluxes in different bacterial species like Escherichia coli (Fischer and Sauer, 2003) or intracellular pathogens like Salmonella, enteroinvasive E. coli (EIEC; Götz and Goebel, 2010), Listeria (Eylert et al., 2008), and Legionella (Eylert et al., 2010).
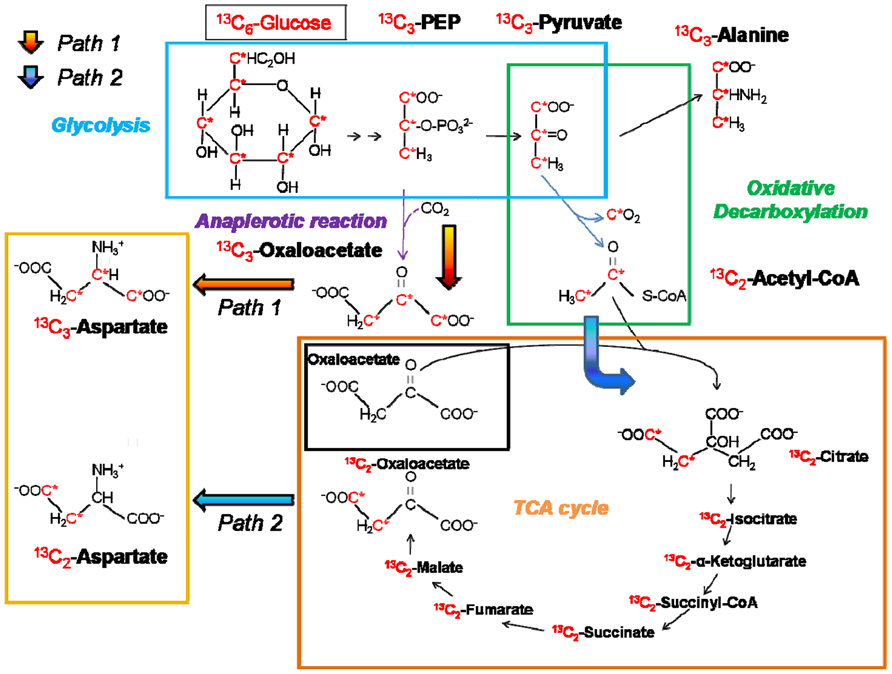
FIGURE 2. Principles of isotopolog profiling with labeled [U-13C6] glucose. The fate of labeled glucose via different metabolic routes can be followed by isotopolog profiling in analyzing resulting metabolites or products (in this case amino acids). An example is given for the biosynthetic routes of the two aspartate isotopologs 13C2-aspartate and 13C3-aspartate (represented by orange and blue arrows, respectively) and the origin of a 13C3-alanine. Isotopolog studies are described in Eisenreich et al. (2006), Eylert et al. (2008), and Eylert et al. (2010). 13C-atoms are marked in red and indicated by an asterisk.
In vitro approaches provide essential clues about the nutritional status of intracellular Salmonella. However, the gained in vitro data are often supported by, or compared to, data received from animal models, e.g., mouse (Bowden et al., 2009). A large number of different mutant strains with metabolic defects has been tested for virulence in mouse models (Tchawa Yimga et al., 2006; Bowden et al., 2009; Paterson et al., 2009) and a proteome analysis with Salmonella isolated from infected mice (recovered from cecum and spleen) has been performed (Becker et al., 2006) providing information about the essential metabolic enzymes and pathways. Data on metabolite levels would provide important complementary information, but are often difficult to obtain for intracellular bacteria and require complex experimental setups. Another complementary approach is proteome analysis, a modern technique to directly determine enzyme type and amount as well as modifications (e.g., regulatory phosphorylation of metabolic enzymes). The fact that there may be differences between results obtained by in vitro and in vivo approaches does not render data from in vitro experiments questionable. The data may very well be reliable (i.e., reproducible) and of importance for understanding of a limiting number of specific factors. The differences in results from in vivo analyses can best be explained by the presence of a large number of additional factors (immune responses, different concentration gradients of nutrients in different tissues, cytokines, etc.) that are often interrelated and affect pathogen survival and replication in host tissue. For example, comparing experimental evidence including proteomics, metabolomics, and survival data on mutants with in vivo and in vitro conditions, conflicting results can be observed for mutant strains of Salmonella with defects in enzymes of the tricarboxylic acid (TCA) cycle. The mutant strains showed even increased replication in a murine RAW macrophage cell line, but were highly reduced in virulence in an animal model (Bowden et al., 2010).
As they are easier to study and analyze, cell culture models for Salmonella infections will remain the essential basis for the understanding of the cellular and molecular changes and mechanisms of the intracellular bacterial nutrition. Cell culture experiments are less complex and laborious than animal experiments and offer the possibility to study such important aspects of intracellular nutrition as access of Salmonella to host cell nutrients or to nutrients in extracellular medium. Attenuation of certain mutant strains can be directly linked to the lack of a certain metabolite in the cell culture media. Supplementation experiments with labeled carbon sources (C-sources) allow to analyze if and how this nutrients reach Salmonella inside its SCV. Data gained from in vitro experiments help to establish and improve mathematical models of Salmonella during infection. By contrast, the large number of additional and partially unknown factors affecting the pathogen in in vivo infection studies would lead to models of extreme complexity.
Salmonella Intracellular Lifestyle in Comparison to Lifestyles of Other Intracellular Pathogens
Salmonella is well-adapted to the SCV compartment as seen if compared to cytoplasmic lifestyle (e.g., Listeria) or Legionella as a further pathogen in a membrane-bound compartment. In Salmonella, glucose represents a major C-source. However, gluconeogenesis rather than glycolysis is observed for cytoplasmic listerial intracellular metabolism. This is required to synthesize glucose from available substrates. Glucose is then predominantly degraded in the pentose phosphate pathway (PPP). A complete TCA cycle is observed for Salmonella. However, in cytoplasmic Listeria the enzyme 2-oxoglutarate dehydrogenase is missing (Glaser et al., 2001; Eisenreich et al., 2006), oxaloacetate is formed by carboxylation of C3 compounds like pyruvate and the anaplerotic enzyme pyruvate carboxylase becomes crucial (Schar et al., 2010), major C-source is glycerol. De novo synthesized listerial amino acids are Ala, Asp, Glu, Ser, Thr, Val, and Gly (Eisenreich et al., 2010), indicating that other amino acids are taken up in the host cell cytoplasm (Schauer et al., 2010).
For comparison, Legionella pneumophila, a gram-negative intracellular pathogen and causative agent for Legionnaire’s disease was long time supposed to feed solely on amino acids while residing in the host alveolar macrophages (Tesh and Miller, 1981). Indeed, the amino acids Cys, Gln, Ser, and Arg are efficiently used as carbon and energy sources in vivo (Wieland et al., 2005). However, recent studies highlight that also glucose is metabolized by Legionella during infection of eukaryotic cells. In contrast to Salmonella, Legionella predominantly degrades glucose by the 2-keto-3-deoxy-phosphogluconate pathway (KDPGP) and only in small quantities by glycolysis. The non-oxidative branch of PPP also accounts for small amounts of glucose catabolism (Harada et al., 2010). 13C-IPA further revealed that there is no evidence for a functional glyoxylate bypass (Eylert et al., 2010), confirming earlier models build on genome sequence analysis (Cazalet et al., 2004). Furthermore, 13C-IPA results indicate that a complete and active TCA cycle occurs in Legionella and that the inability to synthesize amino acids de novo is only valid for Ile, Leu, Val, Phe, Met, Arg, and Tyr.
Computational and System Biological Analyses of the Metabolism of Intracellular Salmonella
A number of recent studies highlight the importance of system biological modeling of Salmonella during infection (Bumann, 2009b; McDermott et al., 2011). Besides the genome information, the analysis of the pathogen proteomes during infection provides an important basis for infection research, as well as for devising novel control strategies including antibiotics and vaccines (Bumann, 2009a). On this, independent models for Salmonella metabolism are built (Becker et al., 2006; AbuOun et al., 2009; Raghunathan et al., 2009; Eisenreich et al., 2010). In such models the flow of metabolites is modeled in terms of pathways. Enzyme chains are calculated such that metabolites are balanced, i.e., consumed and produced in equilibrium, this is called flux balance analysis (FBA). If such a chain of metabolic enzymes can not be dissected any further, this is named an elementary mode. Helpful software tools include the COBRA Toolbox (Schellenberger et al., 2011) and YANAsquare (Schwarz et al., 2007) which can compile available biochemical and genome data systematically and calculate flux distributions by FBA or elementary mode analysis (EMA). Specific tools such as the KEGGbrowser (Schwarz et al., 2007) simplify the direct import of available biochemical data into metabolic network reconstructions even on genome-scale basis. Furthermore, metabolic gaps and dubious annotations for enzymes of central metabolism often occur and therefore different genome annotation software and comparisons are strongly recommended to improve the network reconstruction (Gaudermann et al., 2006).
Recently, a community effort towards a knowledge-base and mathematical model of S. Typhimurium strain LT2 has been initiated, resulting in the BiGG knowledge-base of Salmonella metabolism (Thiele et al., 2011). A consensus metabolic reconstruction was obtained from two independently developed (Ruppin et al., 2010) metabolic reconstructions for S. Typhimurium. The joined reconstruction effort included, furthermore, the development and implementation of a community-based workflow for annotation and corrections including incorporation of thermodynamic information (to decide on reversible and irreversible reactions). By this, metabolite transporters and reactions are more accurately identified and considered. Higher reliable consensus models improve, furthermore, the potential of multi-target drug therapy approaches for specific strains though of course the host response is another important factor to consider.
Our metabolic modeling approach calculating elementary flux modes on central carbon and amino acid metabolism in S. Typhimurium indicates that the anaplerotic reactions around phosphoenolpyruvate (PEP) to oxaloacetate are pivotal and occur in many flux modes. For modeling growth on glucose as the sole C-source, PEP carboxylase (ppc) plays a central role in directing the flux to the TCA cycle. However, disruption of PEP carboxylase can be partly compensated by alternative routes in the network regarding carbohydrate metabolism. A ppc-deficient mutant showed no reduced virulence in vivo (Tchawa Yimga et al., 2006), indicating the availability of further C-sources such as amino acids which, through transaminase reactions, also can feed into the TCA cycle. This would give support to a model that contains both glucose and amino acids as C-sources. Here, the flux through PEP carboxylase decreased and the flux through PEP carboxykinase (pckA) increased, compared to a medium with only glucose. PEP carboxykinase catalyzes a reaction in the opposite direction of the normal flow cycle, i.e., increasing the flow from oxaloacetate to PEP. The direct conversion of oxaloacetate to aspartate cannot be easily compensated by alternative flux modes. Transaminase activity can be compensated, for instance, loss of two transaminases (aspC and tyrB) is required before aspartate auxotrophy appears. Moreover, aspartate can also be acquired from the host. However, the production and availability of oxaloacetate is critical, requiring increased PEP carboxylase activity or a reversal of the flow from oxaloacetate to aspartate. This and similar other modeling results suggest that amino acid metabolism is easier impaired and more critical in the SCV than in intracytoplasmic survival (Schauer et al., 2010).
Flux balance analysis and EMA calculate metabolic pathways but the integration of experimental “omics” data allows to better determine metabolic flux strengths (Covert et al., 2004). Tools have thus recently been developed that help to integrate experimental data into metabolic models such as YANAvergence (Liang et al., 2011) fit experimental flux measurements, enzyme activities, gene expression data (Cecil et al., 2011), as well as extracellular metabolite ratios to computational predictions.
Due to the intracellular lifestyle, the host provides further nutrients by transporters (Figure 3). Overall, there is an intensive exchange of metabolites during growth in the host cell. Game theoretical approaches treat such interdependencies in a novel mathematical way showing advantages as well as limitations for any specific survival strategy (bacterial pathogen) or opposing strategy from the host (Ruppin et al., 2010; Schuster et al., 2010b,2011). This becomes even more important as the robust Salmonella metabolism including its redundant, overlapping pathway organization limits possibilities for new antimicrobials interfering with its metabolic processes (Becker et al., 2006). Moreover, the potential of central carbon metabolism as a target for microbial defense depends on the environmental factors of the specific niche and the genetic and phenotypic traits of infecting bacteria. For instance, in dormant sub-populations, or “persisters,” the uptake of glucose, mannitol, or fructose implies a direct influence in preparatory steps of glycolysis. This potentiates the killing by aminoglycosides (Allison et al., 2011). Thus, for an iterative refinement of computational modeling the integration of transporter reactions (Raghunathan et al., 2009) and a strain-specific analysis (Liang et al., 2011) is crucial. In this regard, a global gene expression analysis of S. Typhimurium (Harvey et al., 2011) during colonization of the chicken’s cecal lumen and cecal mucosa demonstrates very specific Salmonella metabolic adaptations to its environment. For comparison, differences in expression of transporters and in the usage of C-sources regarding three specific niches (cecal lumen, mucosal wall, and the SCV in macrophages and epithelial cells) are illustrated in Figure 3.
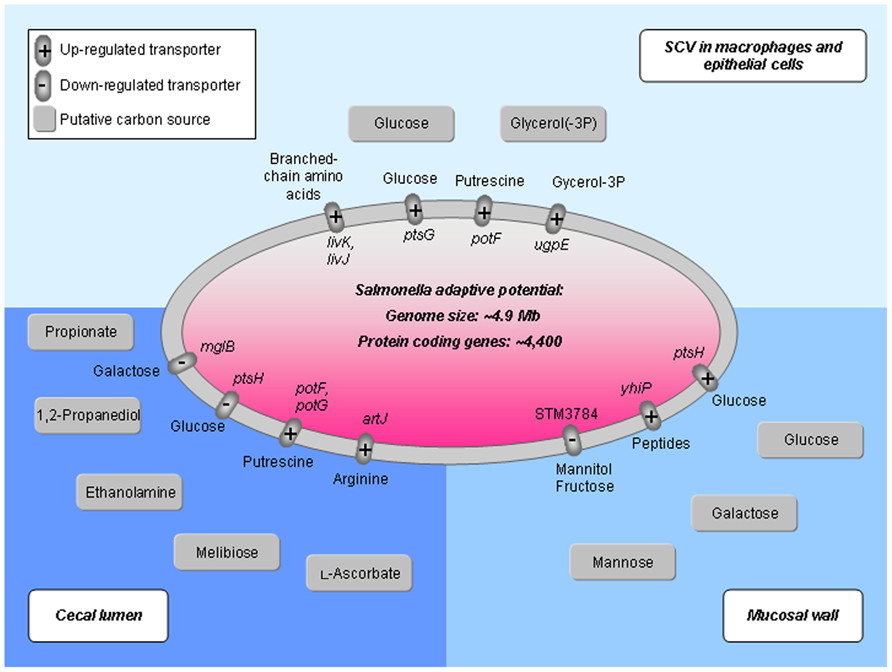
FIGURE 3. Activity of nutrient transport systems in various host habitats colonized by Salmonella. Transporter gene expression and carbon source usage is exemplified according to Hautefort et al. (2008), Eisenreich et al. (2010), and Harvey et al. (2011). During the colonization of the intestine, Salmonella first resides in the cecal lumen (bottom left), then attaches to the mucosal layer (bottom right) until it invades macrophages and epithelial cells forming the SCV (upper part). Transporter proteins differentially expressed in the specific niches are represented as small ovals with transported metabolites at the outside of the Salmonella cell (central oval) and gene names on the inside. Carbon sources suggested according to literature are illustrated as gray boxes. Salmonella genome information is according to McClelland et al. (2001).
The Role of Central Carbon Metabolism Pathways During Intracellular Survival
Salmonella is in fact a pathogen with a very broad and versatile metabolism and as already seen by its comparatively large genome, is a generalist among gram-negative bacteria (Figure 4). For instance, Salmonella can easily metabolize glucose combining various pathways to supply both energy and amino acids and the same applies for most nutrient sources.
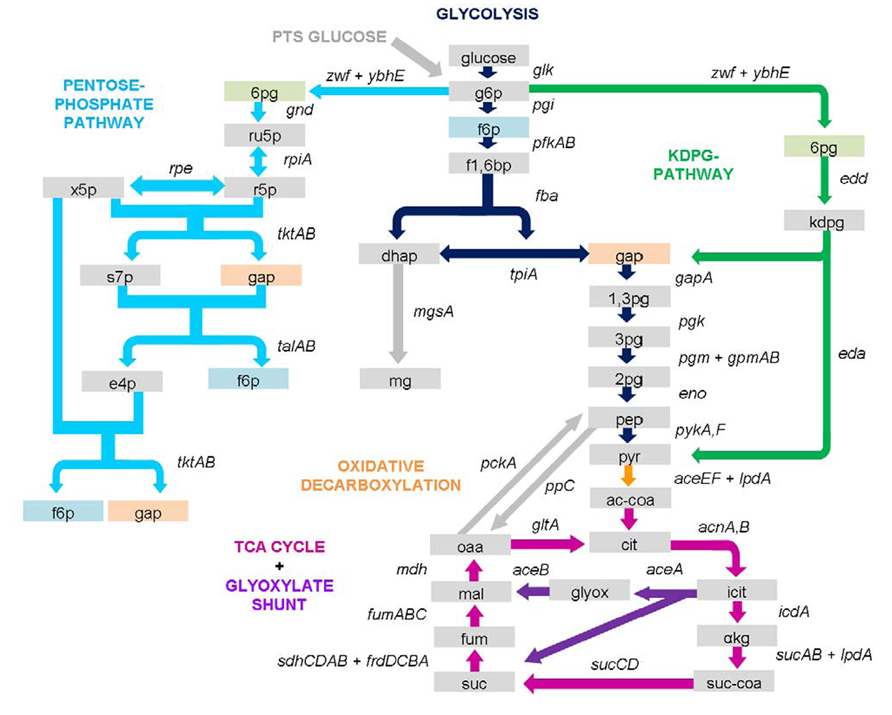
FIGURE 4. Salmonella central carbon metabolism. Shown are genes and enzymes of the central carbon metabolism covering glycolysis, PPP, KDPG/Entner–Doudoroff pathway, oxidative carboxylation, TCA cycle, and anaplerotic reactions. Boxes represent metabolites, arrows indicate enzyme reactions. Metabolites and enzymes are colored according to the color of their pathway name. Arrows are directed according flux direction under glucose metabolism but may vary under different conditions. Detailed information on importance of the illustrated pathways in intracellular infection is outlined in the text including behavior of an array of different enzyme mutations. 1,3pg, 1,3-bisphospho-D-glycerate; 2pg, 2-phospho-D-glycerate; 3pg, 3-phospho-D-glycerate; 6pg, 6-phospho-D-gluconate; αkg, α-ketoglutarate; ac-coa, acetyl-CoA; cit, citrate; dhap, dihydroxyacetone phosphate; e4p, D-erythrose-4-phosphate; f1,6bp, fructose-1,6-bisphosphate; f6p, D-fructose-6-phosphate; fum, fumarate; gap, D-glyceraldehyde-3-phosphate; glyox, glyoxylate; icit, isocitrate; kdpg, 2-dehydro-3-deoxy-D-gluconate-6-phosphate; mal, (S)-malate; mg, methylglyoxal; oaa, oxaloacetate; pep, phosphoenolpyruvate; pyr, pyruvate; r5p, D-ribose-5-phosphate; ru5p, D-ribulose-5-phosphate; s7p, D-sedoheptulose-7-phosphate; suc, succinate; suc-coa, succinyl-CoA; x5p, D-xylulose-5-phosphate.
Furthermore, the combination of different pathways guarantees a fine-tuned balance of internal metabolites. Due to this redundancy it is not easy to block growth of Salmonella by antibiotics targeted against key metabolic enzymes of primary metabolism (Becker et al., 2006). In the following we discuss specific Salmonella pathways.
The Role of Catabolism of Glucose
There are three routes for the catabolism of glucose: (i) glycolysis, (ii) the PPP, and (iii) the Entner–Doudoroff pathway also known as the KDPGP. The two latter pathways for glucose utilization seem to be of lesser importance for Salmonella. A Salmonella mutant strain deficient in zwf (encoding glucose-6-phosphate dehydrogenase), catalyzing the first step of both PPP and KDPGP, and a double mutant strain in gnd (PPP) and edd (KDPGP), or a double mutant in gnd (PPP) and edd (KDPGP) are not attenuated in proliferation in the murine macrophage cell line RAW 264.7 (own unpublished results). Nevertheless, it was reported that a zwf mutant shows reduced virulence in a mouse model of systemic infection (Lundberg et al., 1999). This group refers to the importance of NADPH production in the PPP which is used as electron donor for reductases required for oxidative stress response. However, we think that in our cell culture models the superoxide levels should be less high than in the mouse infection model and this may explain the decreased need for such reductases and for NADPH. Other important PPP products like ribose used for nucleoside synthesis can still be produced by the non-oxidative part of PPP. We suggest that PPP and KDPGP play no important role if glucose is the major substrate. Isotopolog profiling experiments in CaCo-2 cells showed that the internalized glucose is mainly converted by glycolysis and/or KDPGP pathway and excluded PPP as a major route for glucose catabolism (Götz and Goebel, 2010). Salmonella studied in epithelial cell lines are significantly less challenged with reactive oxygen intermediates (ROI) and reactive nitrogen intermediates (RNI) responses, hence, the generation of NADPH would be less important than in an animal model. In particular, the activity of the PPP may become important if Salmonella is challenged with higher degrees of oxidative stress.
The significance of glucose as one of the major C-sources, and glycolysis as the main route for utilization has recently been shown (Bowden et al., 2009). Extending the set of glycolysis mutant strains analyzed by Bowden et al. (2009), work in our groups showed that eno, fba, pgk, gapA, or tpiA deficient strains are strongly attenuated in intracellular replication and survival in RAW 264.7 cells (unpublished results). In case of a tpiA mutant it was demonstrated that reduced growth in rich medium (lysogeny broth, LB) and decreased fitness in mice seems to depend on accumulation of the toxic electrophile methylglyoxal from accumulated dihydroxyacetone phosphate by reaction of the methylglyoxal synthetase (Paterson et al., 2009). Overproduction of methylglyoxal as a result of tpiA mutation for E. coli has been shown before (Cooper and Anderson, 1970; Cooper, 1984) and although the concentration is lower in the tpiA mutant of Salmonella, it still leads to reduced growth in medium and lower fitness in mice (Paterson et al., 2009). Mutant strains deficient in eno, fba, gapA, or pgk show similar growth characteristics in medium containing just traces of glucose or other C-sources ending up in glycolytic intermediates, e.g., in LB or minimal medium containing ribose. None of these mutant strains was able to grow in these media, but growth was observed on minimal medium with glycerol or PEP as C-source. The growth defect in the presence of sugars is due to accumulation of phosphorylated sugars, i.e., fructose-6-phosphate (f6p), glucose-6-phosphate (g6p), and fructose-1,6-bisphosphate (f1,6bp), that appear to influence RNA stability as observed in E. coli mutant strains (Bock and Neidhardt, 1966; Singer et al., 1991; Morita et al., 2003). Glucose is the main C-source in common cell culture media for eukaryotic host cells along with amino acids. The inability of these mutants to survive intracellularly suggests that the metabolism of glucose through glycolysis and the accumulation of phosphorylated sugars is harmful, caused by the knockout of the aforementioned glycolytic genes. Nevertheless, further analytical approaches such as quantification of metabolites by GC–MS are required to determine the presence and accumulation of these glycolytic intermediates.
Oxidative Decarboxylation of Pyruvate – The Connection to the TCA Cycle
Pyruvate as final product of glycolysis or KDPGP can be further metabolized to acetyl-CoA by reaction of pyruvate dehydrogenase. This production of acetyl-CoA and the connection to the TCA cycle is of great importance as indicated by the reduced growth of a pyruvate dehydrogenase subunit I deficient strain (ΔaceE) in rich media (LB) and strong attenuation of intracellular replication. An aceE mutant of S. enterica serovar Enteritidis tested in a chicken infection model also showed reduced growth compared to a WT strain, a lower invasion rate in HeLa cells and was less resistant to ROI. Chicken macrophages (HD-11) were able to eliminate this strain completely within 24 h after infection (Pang et al., 2011). In non-activated RAW 264.7 cells that produce low amounts of ROI, reduced replication was observed (unpublished observations). The loss of aceE may also have influence on the expression of virulence genes important for the defense against ROI. The attenuation of the ΔaceE strain may be due to the metabolic burden of virulence gene expression and the lower energy production due to this knockout. Energy is probably mainly produced by glycolysis resulting in less ATP than via respiratory chain of the reduction agents produced in the TCA cycle.
The Role of the TCA Cycle and Anaplerotic Reactions
The TCA cycle plays an important role as a source for precursors for anabolic pathways, e.g., amino acids, and reducing agents used as electron donors in the respiratory chain or for biosynthesis. Due to this central role, it is obvious that in the last years research is focused on this part of the central carbon metabolism. Some groups already showed loss of virulence in a murine model for mutants with defects in the TCA cycle (Tchawa Yimga et al., 2006; Bowden et al., 2010), concluding that the ability to run the full TCA cycle is critical for virulence. The latter group could show that for some of these TCA cycle mutant strains showing reduced virulence in the murine model (mdh, sdhCDAB, and sucCD) an increased replication in resting and activated macrophages (RAW 264.7) could be observed that is probably due to differences in the in vitro and in vivo environment (available nutrients, etc.). An sdhA mutant strain of E. coli had an expanded life span due to a lower production of superoxide in contrast to the WT (Gonidakis et al., 2011). The phenomenon may explain the observed increased intracellular replication for the mdh, sdhCDAB, and sucCD strains of Salmonella in vitro. A TCA cycle mutant strain with an icdA deletion was not tested in this experimental setup but we could observe a strong attenuation in inactivated murine RAW macrophages. The strong attenuation of the icdA mutant could be due to the accumulation of an inhibitory product like citrate or isocitrate as was shown for an E. coli icd mutant (Lakshmi and Helling, 1976).
In view of the significance of the anaplerotic reaction for survival in macrophages (Figure 4), it seems quite convincing that the glyoxylate shunt, which is used during growth on acetate and fatty acids is important for chronic infections but not for acute infections (Fang et al., 2005; Tchawa Yimga et al., 2006). This supports the suggestion that acetate and fatty acids are of no importance as C-sources in acute infection either due to their absence or the presence of more favorable C-sources like glucose. This is confirmed by transcriptome data observing no up-regulation in expression of the isocitrate lyase gene aceA (Eriksson et al., 2003) and by observations that Salmonella strains with a defect in β-oxidation of fatty acids (fadD) also showed the same virulence as the WT strain in a murine model (Tchawa Yimga et al., 2006).
The direct conversion of PEP to oxaloacetate, an important precursor for the amino acids aspartate and asparagine, is another anaplerotic reaction preventing the TCA cycle from idling and is catalyzed by the enzyme PEP carboxylase (Sabe et al., 1984). Loss of this enzyme does not lead to reduced virulence in a murine model (Tchawa Yimga et al., 2006) indicating the presence of C-sources other than glucose, e.g., amino acids, in amounts required for intracellular replication of Salmonella. This would make redundant the replenishment of the TCA cycle by this route.
Towards a Comprehensive Understanding of the Metabolic Requirements of the Intracellular Lifestyle of Salmonella
The environmental and nutritional conditions encountered by Salmonella within the SCV are still a matter of debate. In the process of SCV formation an acidification of the intravacuolar environment takes place (Rathman et al., 1996). Salmonella metabolism during infection adapts to this intracellular life style in the host cell. It is regulated by different transcription factors (Eisenreich et al., 2010; Götz and Goebel, 2010). However, to what extent can the SCV be reached from outside? Are there macrophage-specific strategies of adaptation? A number of recent efforts were stimulated by these questions (Eriksson et al., 2003; Becker et al., 2006; Bowden et al., 2009; Götz and Goebel, 2010) and gene knockout strategies as well as metabolite measurements and modeling were applied to better understand Salmonella life inside the cell.
Limitation of magnesium, phosphate, and iron occur as genes for their uptake are up-regulated in Salmonella during infection. By gene expression analysis it could also be shown that glycolysis, KDPGP, and TCA cycle are highly expressed in SCV of macrophages and epithelial cells (Hautefort et al., 2008). In contrast, gluconeogenesis is not required for a full virulent phenotype in mice (Tchawa Yimga et al., 2006).
In a number of reports, the nutrient-poor, underfed conditions of the SCV were stressed (Eriksson et al., 2003; Ibarra et al., 2009). This applies even more for the early phases of the infection. Furthermore, detailed studies suggest that intracellular Salmonella deploys specific mechanisms to redirect vacuolar transport to make use of host-derived nutrients (Kuhle et al., 2006; Drecktrah et al., 2007; Rajashekar et al., 2008).
Direct comparison of intracellular lifestyles of bacterial pathogens such as Salmonella and Legionella that reside within a membrane-bound compartment, to those with survival and replication within the cytoplasm of host cells is of interest. For the latter, survival depends on the availability of cytoplasmic host cell metabolites and tight regulation is necessary such as by the pathogenicity factor PrfA in Listeria (Eylert et al., 2008) to survive surprisingly poor nutritional conditions (Eriksson et al., 2003; Ibarra et al., 2009). For the Salmonella lifestyle in the SCV, also strong metabolic adaptations are necessary, in particular regarding low abundant nutrients and ions, however, carbohydrates are not that limiting. The survival under these different conditions of intracellular life in the host cell indicates successful adaptation to different metabolic limitations.
The expression of virulence factors by Salmonella does not come without a price. For example, in a recent study retarded growth of S. Typhimurium cells expressing the type III secretion system 1 (T3SS-1) compared to the minus phenotype was observed and the effects on growth kinetics were modeled (Sturm et al., 2011). Growth retardation was at least partially attributable to the expression of the T3SS-1 effector and/or translocon proteins. In spite of this growth penalty, the T3SS-1(+) sub-population increased from <10% to approximately 60% during the late logarithmic growth phase of an LB batch culture. As shown by experimental data and mathematical modeling, this was attributable to an increasing initiation rate of expression of T3SS-1 genes, in response to environmental cues accumulating during this growth phase. The key is here to mathematically describe the whole system, correctly quantify responses and obtain results from the model which agree with observation. Such models and methods can, furthermore, also be transferred to better understand other genetic variations pertaining, for instance, to effector proteins and estimate the cost of virulence regarding growth and metabolism. Such systems biology approaches to Salmonella lifestyle and pathogenicity are expected to grow (Helaine et al., 2010).
Furthermore, a number of technical developments contributed to the understanding of bacterial metabolism during infection. The modeling of genome-scale metabolic networks is now feasible, originally pioneered by the Palsson group (Price et al., 2004a). However, even with strong computational power at hand, detailed modeling requires additional methods to handle the combinatorial explosion of different pathways concerned, for instance, by dividing the metabolic network (Schuster et al., 2002) or sampling averages over high numbers of different modes (Price et al., 2004b). Furthermore, a number of recent developments tackle new software solutions for metabolic modeling including modifications of extreme pathway analysis (Kaleta et al., 2009; Schuster et al., 2010a). Moreover, the analysis of isotopolog data profits from advances in software development (Nanchen et al., 2007). Achieving gene knockout combinations experimentally has been advanced technically (Datsenko and Wanner, 2000; Gerlach et al., 2009) and this has been complemented by in silico prediction studies on gene knockouts agreeing well with these experiments. This can also be applied to study survival during infection (Raghunathan et al., 2009).
Implications and Conclusions
Pathogenicity can arise from a bacterial specialization where intrinsic cellular functions are especially adapted to the host’s environment such as in exclusive human pathogens like Mycobacterium tuberculosis (Moller and Hoal, 2010), Neisseria gonorrhoeae (Criss and Seifert, 2012), N. meningitides (Criss and Seifert, 2012), and Mycoplasma pneumoniae (Dumke et al., 2011). In contrast, Salmonella is a surprisingly successful intracellular pathogen that is less specialized but rather a generalist with an extraordinary metabolic versatility. Furthermore, E. coli, Pseudomonas spp., Klebsiella spp., and further bacteria are common pathogens with a similar metabolic background which are not highly specialized for a specific host environment.
To underline this, a broad spectrum of central carbon metabolism routes constitutes the repertoire of the Salmonella lifestyle (Figure 4). Glucose, the predominant C-source during SCV colonization can either be degraded in the glycolytic pathway or the KDPG route, the former one preferentially used. The TCA cycle as the major biosynthetic origin of precursor and provider of reductive agents is complete and is supported by important anaplerotic reactions that lead the metabolic flux to the TCA cycle, mainly by the PEP carboxylase. However, this enzyme can be compensated by other flux modes as shown by EMA. In systemic infections, the PPP generates NADPH required for reductases in oxidative stress response. Salmonella is well-equipped to rapidly adapt to various environments during the passage through the host’s body.
The flexible metabolic abilities of Salmonella make it challenging to elucidate targets to inhibit metabolic functions during infection. The pathogen efficiently modifies its unique vacuolar compartment for its benefits. Additionally, the transport of nutrients to the SCV in response of Salmonella effector proteins is currently under investigation. Host–pathogen metabolism is intertwined.
Is there an Achilles’ heel of Salmonella metabolism in infection? To answer this we suggest a combination of a thorough analysis of central metabolic enzymes such as mutant phenotypes of glycolytic enzymes with systemic analysis such of transporter expression profiles, strain-specific analysis, and projecting those on the very specific environmental conditions under study. Thus, the loss of pyruvate dehydrogenase may have an influence on the expression of virulence genes important for ROI defense. Further, mutants with a defect in the TCA cycle showed reduced virulence in a murine infection model. In Salmonella, the T3SS-2 is responsible for translocation of over 20 virulence proteins into the host cell cytoplasm (Haraga et al., 2008). SifA is probably the most prominent among these effectors in maintaining the integrity of the SCV and induction of extensive networks of tubular membrane compartments including Salmonella-induced filaments and recently identified further tubular compartment (Schroeder et al., 2011).
Future work will take up the challenge of this surprisingly well-adapted pathogen by looking more closely at the host–pathogen interaction, not only regarding metabolic interactions for further pathways including secondary metabolism, iso-enzymes, and intertwined links between pathways (including supplementation experiments), but also regarding the regulatory and immune response from the host, for instance, potential immune modifiers as novel approaches to boost host response in severe infections by Salmonella. Infectious disease burden in developing countries will doubtless profit from improved hygiene and clean water supply but also protective nutrient additives (e.g., vitamins; golden rice, and similar recent developments) and regarding trace elements such as selenocysteine. Challenges in industrialized countries include persistent infection which can again be coped with metabolic approaches relying, for instance, on elicitors of growth which in turn make the previous persister Salmonella vulnerable to standard antibiotics. In general, the exploration of the link between metabolism and infection has to be explored further to improve medical options against Salmonella.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was supported by the German Research Foundation (DFG), grants Da 208/13-2 and He 1964/14-1.
References
AbuOun, M., Suthers, P. F., Jones, G. I., Carter, B. R., Saunders, M. P., Maranas, C. D., Woodward, M. J., and Anjum, M. F. (2009). Genome scale reconstruction of a Salmonella metabolic model: comparison of similarity and differences with a commensal Escherichia coli strain. J. Biol. Chem. 284, 29480–29488.
Allison, K. R., Brynildsen, M. P., and Collins, J. J. (2011). Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473, 216–220.
Becker, D., Selbach, M., Rollenhagen, C., Ballmaier, M., Meyer, T. F., Mann, M., and Bumann, D. (2006). Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440, 303–307.
Bock, A., and Neidhardt, F. C. (1966). Properties of a mutant of Escherichia coli with a temperature-sensitive fructose-1,6-diphosphate aldolase. J. Bacteriol. 92, 470–476.
Bowden, S. D., Ramachandran, V. K., Knudsen, G. M., Hinton, J. C., and Thompson, A. (2010). An incomplete TCA cycle increases survival of Salmonella Typhimurium during infection of resting and activated murine macrophages. PLoS ONE 5, e13871. doi: 10.1371/journal.pone.0013871
Bowden, S. D., Rowley, G., Hinton, J. C., and Thompson, A. (2009). Glucose and glycolysis are required for the successful infection of macrophages and mice by Salmonella enterica serovar Typhimurium. Infect. Immun. 77, 3117–3126.
Bumann, D. (2009a). Pathogen proteomes during infection: a basis for infection research and novel control strategies. J. Proteomics 73, 2267–2276.
Bumann, D. (2009b). System-level analysis of Salmonella metabolism during infection. Curr. Opin. Microbiol. 12, 559–567.
Cazalet, C., Rusniok, C., Bruggemann, H., Zidane, N., Magnier, A., Ma, L., Tichit, M., Jarraud, S., Bouchier, C., Vandenesch, F., Kunst, F., Etienne, J., Glaser, P., and Buchrieser, C. (2004). Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36, 1165–1173.
Cecil, A., Rikanovic, C., Ohlsen, K., Liang, C., Bernhardt, J., Oelschlaeger, T. A., Gulder, T., Bringmann, G., Holzgrabe, U., Unger, M., and Dandekar, T. (2011). Modeling antibiotic and cytotoxic effects of the dimeric isoquinoline IQ-143 on metabolism and its regulation in Staphylococcus aureus, Staphylococcus epidermidis and human cells. Genome Biol. 12, R24.
Cooper, R. A. (1984). Metabolism of methylglyoxal in microorganisms. Annu. Rev. Microbiol. 38, 49–68.
Cooper, R. A., and Anderson, A. (1970). The formation and catabolism of methylglyoxal during glycolysis in Escherichia coli. FEBS Lett. 11, 273–276.
Covert, M. W., Knight, E. M., Reed, J. L., Herrgard, M. J., and Palsson, B. O. (2004). Integrating high-throughput and computational data elucidates bacterial networks. Nature 429, 92–96.
Criss, A. K., and Seifert, H. S. (2012). A bacterial siren song: intimate interactions between Neisseria and neutrophils. Nat. Rev. Microbiol. 10, 178–190.
Cummings, P. L., Sorvillo, F., and Kuo, T. (2010). Salmonellosis-related mortality in the United States, 1990–2006. Foodborne Pathog. Dis. 7, 1393–1399.
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645.
Drecktrah, D., Knodler, L. A., Howe, D., and Steele-Mortimer, O. (2007). Salmonella trafficking is defined by continuous dynamic interactions with the endolysosomal system. Traffic 8, 212–225.
Dumke, R., Hausner, M., and Jacobs, E. (2011). Role of Mycoplasma pneumoniae glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in mediating interactions with the human extracellular matrix. Microbiology 157, 2328–2338.
Eisenreich, W., Dandekar, T., Heesemann, J., and Goebel, W. (2010). Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat. Rev. Microbiol. 8, 401–412.
Eisenreich, W., Slaghuis, J., Laupitz, R., Bussemer, J., Stritzker, J., Schwarz, C., Schwarz, R., Dandekar, T., Goebel, W., and Bacher, A. (2006). 13C isotopologue perturbation studies of Listeria monocytogenes carbon metabolism and its modulation by the virulence regulator PrfA. Proc. Natl. Acad. Sci. U.S.A. 103, 2040–2045.
Eriksson, S., Lucchini, S., Thompson, A., Rhen, M., and Hinton, J. C. (2003). Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47, 103–118.
Eylert, E., Herrmann, V., Jules, M., Gillmaier, N., Lautner, M., Buchrieser, C., Eisenreich, W., and Heuner, K. (2010). Isotopologue profiling of Legionella pneumophila: role of serine and glucose as carbon substrates. J. Biol. Chem. 285, 22232–22243.
Eylert, E., Schar, J., Mertins, S., Stoll, R., Bacher, A., Goebel, W., and Eisenreich, W. (2008). Carbon metabolism of Listeria monocytogenes growing inside macrophages. Mol. Microbiol. 69, 1008–1017.
Fang, F. C., Libby, S. J., Castor, M. E., and Fung, A. M. (2005). Isocitrate lyase (AceA) is required for Salmonella persistence but not for acute lethal infection in mice. Infect. Immun. 73, 2547–2549.
Fields, P. I., Swanson, R. V., Haidaris, C. G., and Heffron, F. (1986). Mutants of Salmonella Typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. U.S.A. 83, 5189–5193.
Fischer, E., and Sauer, U. (2003). Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC–MS. Eur. J. Biochem. 270, 880–891.
Gaudermann, P., Vogl, I., Zientz, E., Silva, F. J., Moya, A., Gross, R., and Dandekar, T. (2006). Analysis of and function predictions for previously conserved hypothetical or putative proteins in Blochmannia floridanus. BMC Microbiol. 6, 1. doi: 10.1186/1471-2180-6-1
Gerlach, R. G., Jäckel, D., Holzer, S. U., and Hensel, M. (2009). Rapid oligonucleotide-based recombineering of the chromosome of Salmonella enterica. Appl. Environ. Microbiol. 75, 1575–1580.
Glaser, P., Frangeul, L., Buchrieser, C., Rusniok, C., Amend, A., Baquero, F., Berche, P., Bloecker, H., Brandt, P., Chakraborty, T., Charbit, A., Chetouani, F., Couve, E., De Daruvar, A., Dehoux, P., Domann, E., Dominguez-Bernal, G., Duchaud, E., Durant, L., Dussurget, O., Entian, K. D., Fsihi, H., Garcia-Del Portillo, F., Garrido, P., Gautier, L., Goebel, W., Gomez-Lopez, N., Hain, T., Hauf, J., Jackson, D., Jones, L. M., Kaerst, U., Kreft, J., Kuhn, M., Kunst, F., Kurapkat, G., Madueno, E., Maitournam, A., Vicente, J. M., Ng, E., Nedjari, H., Nordsiek, G., Novella, S., De Pablos, B., Perez-Diaz, J. C., Purcell, R., Remmel, B., Rose, M., Schlueter, T., Simoes, N., Tierrez, A., Vazquez-Boland, J. A., Voss, H., Wehland, J., and Cossart, P. (2001). Comparative genomics of Listeria species. Science 294, 849–852.
Gonidakis, S., Finkel, S. E., and Longo, V. D. (2011). Lifespan extension and paraquat resistance in a ubiqui-deficient Escherichia coli mutant depend on transcription factors ArcA and TdcA. Aging (Albany NY) 3, 291–303.
Götz, A., and Goebel, W. (2010). Glucose and glucose 6-phosphate as carbon sources in extra- and intracellular growth of enteroinvasive Escherichia coli and Salmonella enterica. Microbiology 156, 1176–1187.
Harada, E., Iida, K., Shiota, S., Nakayama, H., and Yoshida, S. (2010). Glucose metabolism in Legionella pneumophila: dependence on the Entner–Doudoroff pathway and connection with intracellular bacterial growth. J. Bacteriol. 192, 2892–2899.
Haraga, A., Ohlson, M. B., and Miller, S. I. (2008). Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6, 53–66.
Harvey, P. C., Watson, M., Hulme, S., Jones, M. A., Lovell, M., Berchieri, A. Jr., Young, J., Bumstead, N., and Barrow, P. (2011). Salmonella enterica serovar Typhimurium colonizing the lumen of the chicken intestine grows slowly and upregulates a unique set of virulence and metabolism genes. Infect. Immun. 79, 4105–4121.
Hautefort, I., Thompson, A., Eriksson-Ygberg, S., Parker, M. L., Lucchini, S., Danino, V., Bongaerts, R. J., Ahmad, N., Rhen, M., and Hinton, J. C. (2008). During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell. Microbiol. 10, 958–984.
Helaine, S., Thompson, J. A., Watson, K. G., Liu, M., Boyle, C., and Holden, D. W. (2010). Dynamics of intracellular bacterial replication at the single cell level. Proc. Natl. Acad. Sci. U.S.A. 107, 3746–3751.
Ibarra, J. A., Knodler, L. A., Sturdevant, D. E., Virtaneva, K., Carmody, A. B., Fischer, E. R., Porcella, S. F., and Steele-Mortimer, O. (2009). Induction of Salmonella pathogenicity island 1 under different growth conditions can affect Salmonella-host cell interactions in vitro. Microbiology 156, 1120–1133.
Kaleta, C., De Figueiredo, L. F., and Schuster, S. (2009). Can the whole be less than the sum of its parts? Pathway analysis in genome-scale metabolic networks using elementary flux patterns. Genome Res. 19, 1872–1883.
Kuhle, V., Abrahams, G. L., and Hensel, M. (2006). Intracellular Salmonella enterica redirect exocytic transport processes in a Salmonella pathogenicity island 2-dependent manner. Traffic 7, 716–730.
Lakshmi, T. M., and Helling, R. B. (1976). Selection for citrate synthase deficiency in icd mutants of Escherichia coli. J. Bacteriol. 127, 76–83.
Liang, C., Liebeke, M., Schwarz, R., Zuhlke, D., Fuchs, S., Menschner, L., Engelmann, S., Wolz, C., Jaglitz, S., Bernhardt, J., Hecker, M., Lalk, M., and Dandekar, T. (2011). Staphylococcus aureus physiological growth limitations: insights from flux calculations built on proteomics and external metabolite data. Proteomics 11, 1915–1935.
Lim, S., Kim, M., Choi, J., and Ryu, S. (2010). A mutation in tdcA attenuates the virulence of Salmonella enterica serovar Typhimurium. Mol. Cells 29, 509–517.
Lundberg, B. E., Wolf, R. E. Jr., Dinauer, M. C., Xu, Y., and Fang, F. C. (1999). Glucose 6-phosphate dehydrogenase is required for Salmonella Typhimurium virulence and resistance to reactive oxygen and nitrogen intermediates. Infect. Immun. 67, 436–438.
Mastroeni, P., Grant, A., Restif, O., and Maskell, D. (2009). A dynamic view of the spread and intracellular distribution of Salmonella enterica. Nat. Rev. Microbiol. 7, 73–80.
McClelland, M., Sanderson, K. E., Spieth, J., Clifton, S. W., Latreille, P., Courtney, L., Porwollik, S., Ali, J., Dante, M., Du, F., Hou, S., Layman, D., Leonard, S., Nguyen, C., Scott, K., Holmes, A., Grewal, N., Mulvaney, E., Ryan, E., Sun, H., Florea, L., Miller, W., Stoneking, T., Nhan, M., Waterston, R., and Wilson, R. K. (2001). Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413, 852–856.
McDermott, J. E., Yoon, H., Nakayasu, E. S., Metz, T. O., Hyduke, D. R., Kidwai, A. S., Palsson, B. O., Adkins, J. N., and Heffron, F. (2011). Technologies and approaches to elucidate and model the virulence program of Salmonella. Front. Microbiol. 2, 121. doi: 10.3389/fmicb.2011.00121
Moller, M., and Hoal, E. G. (2010). Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis. Tuberculosis (Edinb) 90, 71–83.
Morita, T., El-Kazzaz, W., Tanaka, Y., Inada, T., and Aiba, H. (2003). Accumulation of glucose 6-phosphate or fructose 6-phosphate is responsible for destabilization of glucose transporter mRNA in Escherichia coli. J. Biol. Chem. 278, 15608–15614.
Nanchen, A., Fuhrer, T., and Sauer, U. (2007). Determination of metabolic flux ratios from 13C-experiments and gas chromatography-mass spectrometry data: protocol and principles. Methods Mol. Biol. 358, 177–197.
Neidhardt, F. C. (1996). Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: ASM Press.
Pang, E., Tien-Lin, C., Selvaraj, M., Chang, J., and Kwang, J. (2011). Deletion of the aceE gene (encoding a component of pyruvate dehydrogenase) attenuates Salmonella enterica serovar Enteritidis. FEMS Immunol. Med. Microbiol. 63, 108–118.
Paterson, G. K., Cone, D. B., Northen, H., Peters, S. E., and Maskell, D. J. (2009). Deletion of the gene encoding the glycolytic enzyme triosephosphate isomerase (tpi) alters morphology of Salmonella enterica serovar Typhimurium and decreases fitness in mice. FEMS Microbiol. Lett. 294, 45–51.
Price, N. D., Reed, J. L., and Palsson, B. O. (2004a). Genome-scale models of microbial cells: evaluating the consequences of constraints. Nat. Rev. Microbiol. 2, 886–897.
Price, N. D., Schellenberger, J., and Palsson, B. O. (2004b). Uniform sampling of steady-state flux spaces: means to design experiments and to interpret enzymopathies. Biophys. J. 87, 2172–2186.
Raghunathan, A., Reed, J., Shin, S., Palsson, B., and Daefler, S. (2009). Constraint-based analysis of metabolic capacity of Salmonella typhimurium during host-pathogen interaction. BMC Syst. Biol. 3, 38. doi: 10.1186/1752-0509-3-38
Rajashekar, R., Liebl, D., Seitz, A., and Hensel, M. (2008). Dynamic remodeling of the endosomal system during formation of Salmonella-induced filaments by intracellular Salmonella enterica. Traffic 9, 2100–2116.
Rathman, M., Sjaastad, M. D., and Falkow, S. (1996). Acidification of phagosomes containing Salmonella typhimurium in murine macrophages. Infect. Immun. 64, 2765–2773.
Ruppin, E., Papin, J. A., De Figueiredo, L. F., and Schuster, S. (2010). Metabolic reconstruction, constraint-based analysis and game theory to probe genome-scale metabolic networks. Curr. Opin. Biotechnol. 21, 502–510.
Sabe, H., Miwa, T., Kodaki, T., Izui, K., Hiraga, S., and Katsuki, H. (1984). Molecular cloning of the phosphoenolpyruvate carboxylase gene, ppc, of Escherichia coli. Gene 31, 279–283.
Schar, J., Stoll, R., Schauer, K., Loeffler, D. I., Eylert, E., Joseph, B., Eisenreich, W., Fuchs, T. M., and Goebel, W. (2010). Pyruvate carboxylase plays a crucial role in carbon metabolism of extra- and intracellularly replicating Listeria monocytogenes. J. Bacteriol. 192, 1774–1784.
Schauer, K., Geginat, G., Liang, C., Goebel, W., Dandekar, T., and Fuchs, T. M. (2010). Deciphering the intracellular metabolism of Listeria monocytogenes by mutant screening and modelling. BMC Genomics 11, 573. doi: 10.1186/1471-2164-11-573
Schellenberger, J., Que, R., Fleming, R. M., Thiele, I., Orth, J. D., Feist, A. M., Zielinski, D. C., Bordbar, A., Lewis, N. E., Rahmanian, S., Kang, J., Hyduke, D. R., and Palsson, B. O. (2011). Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat. Protoc. 6, 1290–1307.
Schroeder, N., Mota, L. J., and Meresse, S. (2011). Salmonella-induced tubular networks. Trends Microbiol. 19, 268–277.
Schuster, S., De Figueiredo, L. F., and Kaleta, C. (2010a). Predicting novel pathways in genome-scale metabolic networks. Biochem. Soc. Trans. 38, 1202–1205.
Schuster, S., Kreft, J. U., Brenner, N., Wessely, F., Theissen, G., Ruppin, E., and Schroeter, A. (2010b). Cooperation and cheating in microbial exoenzyme production – theoretical analysis for biotechnological applications. Biotechnol. J. 5, 751–758.
Schuster, S., De Figueiredo, L. F., Schroeter, A., and Kaleta, C. (2011). Combining metabolic pathway analysis with Evolutionary Game Theory: explaining the occurrence of low-yield pathways by an analytic optimization approach. Biosystems 105, 147–153.
Schuster, S., Pfeiffer, T., Moldenhauer, F., Koch, I., and Dandekar, T. (2002). Exploring the pathway structure of metabolism: decomposition into subnetworks and application to Mycoplasma pneumoniae. Bioinformatics 18, 351–361.
Schwarz, R., Liang, C., Kaleta, C., Kuhnel, M., Hoffmann, E., Kuznetsov, S., Hecker, M., Griffiths, G., Schuster, S., and Dandekar, T. (2007). Integrated network reconstruction, visualization and analysis using YANAsquare. BMC Bioinformatics 8, 313. doi: 10.1186/1471-2105-8-313
Shi, L., Adkins, J. N., Coleman, J. R., Schepmoes, A. A., Dohnkova, A., Mottaz, H. M., Norbeck, A. D., Purvine, S. O., Manes, N. P., Smallwood, H. S., Wang, H., Forbes, J., Gros, P., Uzzau, S., Rodland, K. D., Heffron, F., Smith, R. D., and Squier, T. C. (2006). Proteomic analysis of Salmonella enterica serovar Typhimurium isolated from RAW 264.7 macrophages: identification of a novel protein that contributes to the replication of serovar Typhimurium inside macrophages. J. Biol. Chem. 281, 29131–29140.
Singer, M., Walter, W. A., Cali, B. M., Rouviere, P., Liebke, H. H., Gourse, R. L., and Gross, C. A. (1991). Physiological effects of the fructose-1,6-diphosphate aldolase ts8 mutation on stable RNA synthesis in Escherichia coli. J. Bacteriol. 173, 6249–6257.
Sturm, A., Heinemann, M., Arnoldini, M., Benecke, A., Ackermann, M., Benz, M., Dormann, J., and Hardt, W. D. (2011). The cost of virulence: retarded growth of Salmonella Typhimurium cells expressing type III secretion system 1. PLoS Pathog. 7, e1002143. doi: 10.1371/journal.ppat.1002143
Tchawa Yimga, M., Leatham, M. P., Allen, J. H., Laux, D. C., Conway, T., and Cohen, P. S. (2006). Role of gluconeogenesis and the tricarboxylic acid cycle in the virulence of Salmonella enterica serovar Typhimurium in BALB/c mice. Infect. Immun. 74, 1130–1140.
Tesh, M. J., and Miller, R. D. (1981). Amino acid requirements for Legionella pneumophila growth. J. Clin. Microbiol. 13, 865–869.
Thiele, I., Hyduke, D. R., Steeb, B., Fankam, G., Allen, D. K., Bazzani, S., Charusanti, P., Chen, F. C., Fleming, R. M., Hsiung, C. A., De Keersmaecker, S. C., Liao, Y. C., Marchal, K., Mo, M. L., Ozdemir, E., Raghunathan, A., Reed, J. L., Shin, S. I., Sigurbjornsdottir, S., Steinmann, J., Sudarsan, S., Swainston, N., Thijs, I. M., Zengler, K., Palsson, B. O., Adkins, J. N., and Bumann, D. (2011). A community effort towards a knowledge-base and mathematical model of the human pathogen Salmonella Typhimurium LT2. BMC Syst. Biol. 5, 8. doi: 10.1186/1752-0509-5-8
Keywords: metabolism, Salmonella-containing vacuole, regulation, virulence
Citation: Dandekar T, Fieselmann A, Popp J and Hensel M (2012) Salmonella enterica: a surprisingly well-adapted intracellular lifestyle. Front. Microbio. 3:164. doi: 10.3389/fmicb.2012.00164
Received: 15 December 2011; Accepted: 12 April 2012;
Published online: 03 May 2012.
Edited by:
Reinhard Guthke, Leibniz Institute for Natural Product Research and Infection Biology - Hans-Knoell-Institute, GermanyReviewed by:
Michael Lalk, University of Greifswald, GermanyKarsten Tedin, Freie Universität Berlin, Germany
Copyright: © 2012 Dandekar, Fieselmann, Popp and Hensel. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Thomas Dandekar, Department of Bioinformatics, Biocenter, University of Würzburg, Am Hubland, 97074 Würzburg, Germany. e-mail: dandekar@biozentrum.uni-wuerzburg.de
