- School of Pharmacy, Centre for Biomolecular Sciences, University of Nottingham, University Park, Nottingham, UK
The shortening of the poly(A) tail of cytoplasmic mRNA (deadenylation) is a pivotal step in the regulation of gene expression in eukaryotic cells. Deadenylation impacts on both regulated mRNA decay as well as the rate of mRNA translation. An important enzyme complex involved in poly(A) shortening is the Ccr4-Not deadenylase. In addition to at least six non-catalytic subunits, it contains two distinct subunits with ribonuclease activity: a Caf1 subunit, characterized by a DEDD (Asp-Glu-Asp-Asp) domain, and a Ccr4 component containing an endonuclease-exonuclease-phosphatase (EEP) domain. In vertebrate cells, the complexity of the complex is further increased by the presence of paralogs of the Caf1 subunit (encoded by either CNOT7 or CNOT8) and the occurrence of two Ccr4 paralogs (encoded by CNOT6 or CNOT6L). In plants, there are also multiple Caf1 and Ccr4 paralogs. Thus, the composition of the Ccr4-Not complex is heterogeneous. The potential differences in the intrinsic enzymatic activities of the paralogs will be discussed. In addition, the potential redundancy, cooperation, and/or the extent of unique roles for the deadenylase subunits of the Ccr4-Not complex will be reviewed. Finally, novel approaches to study the catalytic roles of the Caf1 and Ccr4 subunits will be discussed.
Introduction
Virtually all mature protein-coding mRNAs in eukaryotic cells contain a 5′ cap structure and a 3′ poly(A) tail, with the notable exception of mRNAs encoding histones. Both modifications play a critical role in translation and mRNA turnover. In mRNA turnover, shortening of the poly(A) tail (deadenylation) is the initial and often rate-limiting step (Parker and Song, 2004; Goldstrohm and Wickens, 2008; Wahle and Winkler, 2013). Following deadenylation, recruitment of the decapping enzyme complex is followed by exonucleolytic degradation from the 5′ end by Xrn1, or by the exosome complex with 3′-5′ polarity (Parker and Song, 2004; Garneau et al., 2007; Houseley and Tollervey, 2009).
Deadenylation may also influence protein synthesis. During translation, the efficiency of initiation is enhanced by interactions between the cap and the poly(A) tail, which are mediated by the poly(A)-binding protein (PABP), the cap-binding factor eIF4E, and the intermediary scaffold eIF4G (Munroe and Jacobson, 1990; Gallie, 1991; Wells et al., 1998). Consistent with the notion that the poly(A) tail contributes to initiation of translation is the observation that poly(A) tail length correlates with ribosome binding, a measure for translational efficiency, in Schizosaccharomyces pombe (Lackner et al., 2007).
Several enzyme complexes are implicated in deadenylation. These include the conserved, trimeric Pan2-Pan3 complex, composed of a single Pan2 catalytic subunit bound to a Pan3 dimer, the PARN deadenylase, which is absent in single cellular eukaryotes, and less-well characterized enzymes, such as the circadian deadenylase Nocturnin, and the Caf1z-Ccr4d complex (reviewed in Godwin et al., 2013; Wahle and Winkler, 2013). However, in all model systems examined (Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, and human cells), the multisubunit Ccr4-Not complex has been identified as the main deadenylase (Tucker et al., 2001; Temme et al., 2004; Yamashita et al., 2005; Nousch et al., 2013).
The activity of the deadenylase enzymes appears to be targeted to specific mRNAs by RNA-binding proteins. The following paradigms have been established for recruitment of the Ccr4-Not deadenylase: targeting mediated by direct interactions between Ccr4-Not and the RNA-binding protein recognizing a linear RNA sequence, as exemplified by pumilio proteins, or the ARE-binding protein tristetraprolin (TTP; Goldstrohm et al., 2006; Sandler et al., 2011); by RNA-binding partners recognizing structural elements, such as an RNA stem loop structure (Roquin; Leppek et al., 2013); via ternary complexes, as indicated by recruitment involving the RNA-binding protein CPEB3, which is mediated via Tob1, thus forming a ternary CPEB3/Tob1/Ccr4-Not complex (Hosoda et al., 2011). Finally, an important mechanism for the recruitment of both Ccr4-Not and the Pan2-Pan3 complex to specific mRNAs involves interactions with GW182/TNRC6 proteins, which are part of the miRNA repression complex (Chekulaeva et al., 2011; Fabian et al., 2011; Kuzuoglu-Ozturk et al., 2012; Huntzinger et al., 2013). In addition to these mechanisms of recruitment, the Pan2-Pan3 and Ccr4-Not deadenylases can also bind to the poly(A)-binding protein (PABP). In the case of the Pan2-Pan3 complex, this is mediated by the presence of a short PAM2 motif in the Pan3 protein, which can interact directly with the PABP C-terminal domain (Siddiqui et al., 2007). By contrast, none of the subunits of the Ccr4-Not complex contain a PAM2 motif. However, this motif is present in both the Tob1 and Tob2 protein, which directly interact with the Ccr4-Not complex (Ezzeddine et al., 2007). Thus, where the Pan2-Pan3 complex can interact directly with the PABP, this interaction is indirect in case of the Ccr4-Not complex.
Overview of the Ccr4-Not Complex
The Ccr4-Not complex contains two components that are associated with deadenylase activity: the Ccr4 and Caf1 subunits (Table 1). Carbon catabolite repression (ccr) 4 was originally identified as a regulator of alcohol dehydrogenase II in the yeast Saccharomyces cerevisiae (Denis, 1984). Likewise, Ccr4-associated factor (Caf) 1 was originally isolated in yeast as PGK promoter directed over production (pop) 2, a mutant overproducing mouse α-amylase under control of a PGK promoter (Sakai et al., 1992; Draper et al., 1995). In addition to its nuclease subunits, several non-catalytic components have been identified (reviewed in Collart and Panasenko, 2012; Wahle and Winkler, 2013). Most of the genes encoding these subunits, including NOT1, NOT2, NOT3 as well as NOT4, were first identified in yeast using a genetic screen for transcriptional regulators (Collart and Struhl, 1994).
In Saccharomyces cerevisiae, two variant complexes with a distinct molecular weight of 1.0 and 1.9 MDa are present (Liu et al., 1997; Liu et al., 1998; Chen et al., 2001). The overall structure of the yeast 1.0 MDa complex is L-shaped with the Caf1 and Ccr4 components located in the hinge connecting the two arms (Nasertorabi et al., 2011). In Homo sapiens, the Ccr4-Not complex has an estimated molecular weight of 1.2 MDa. Importantly, while the Not4 subunit appears to be an integral component of the yeast complex, CNOT4, its human ortholog, resides in a separate complex with an estimated molecular weight of 200 kDa (Lau et al., 2009).
The Ccr4 Subunit, Orthologs, and Paralogs
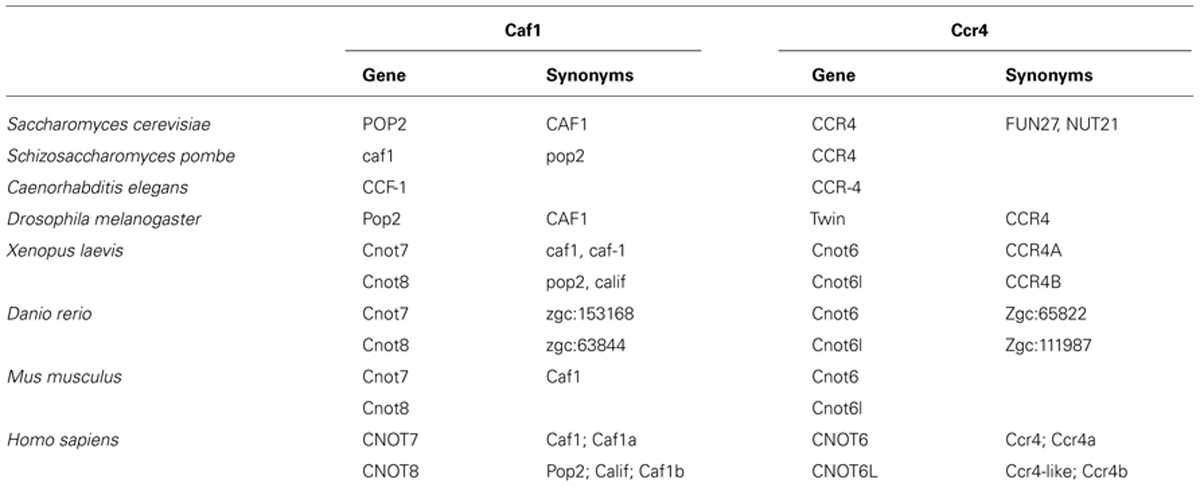
The Ccr4 subunit is characterized by the presence of two domains: an amino-terminal leucine-rich repeat (LRR) domain, and a carboxy-terminal endonuclease-exonuclease-phosphatase (EEP) domain. The latter is associated with its ribonuclease activity. Analysis of the enzymatic activity of human and yeast Ccr4 indicates that the enzyme has a strong preference for poly(A) residues in vitro (Chen et al., 2002; Wang et al., 2010). A crystal structure of the nuclease domain indicates that the C-terminus forms an α/β-sandwich fold, which is very similar to that of other hydrolyses such as apurinic/apyrimidinic endonuclease (APE) 1 (Figure 1A; Wang et al., 2010). Two Mg(II) ions are required for hydrolysis of the phosphoester backbone. The metal ions are coordinated by an asparagine, glutamate, two aspartate, and a histidine residue, which are part of short sequence motifs that are highly conserved in the ExoIII/APE1 family of DNA and RNA nucleases. Substitutions of amino acids involved in the coordination of the metal ions abolish the enzymatic activity (Chen et al., 2002; Wang et al., 2010). The Mg(II) ions are located at the bottom of a narrow cleft in which the nucleic acid substrate is inserted (Figure 1B). The crystal structure of the nuclease domain in complex with an oligo(A) DNA molecule highlights some features of the selective interaction with poly(A), which involve a specific interaction between a backbone carbonyl and the 6′ amino group of the adenosine base (Wang et al., 2010).
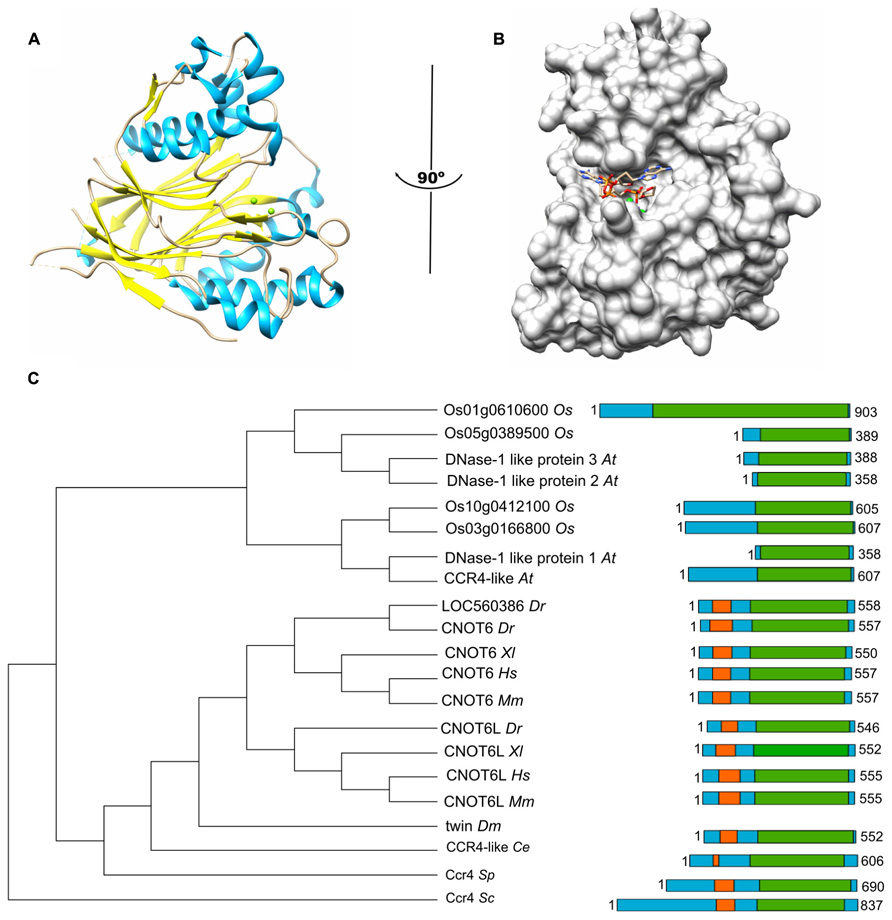
FIGURE 1. The Ccr4 nuclease subunit. (A) Structure of the human Ccr4/CNOT6L catalytic domain. The nuclease domain forms an α/β sandwich typical for the endonuclease-exonuclease-phosphatase (EEP) domain. The Mg(II) ions located in the active site are indicated in green; α-helical regions, blue; β-strands, yellow. (B) Binding of poly(A) by human Ccr4/CNOT6L. The Mg(II) ions located in the active site are indicated in green. Structures (using PDB accession number 3NGO) were visualized using the UCSF Chimera package (http://www.cgl.ucsf.edu/chimera; Pettersen et al., 2004). (C) Evolutionary conservation of Ccr4 across the eukaryotic kingdom. Sequence analysis was carried out using Basic local alignment search tool (BLAST) in combination with the Reference protein database. Only homologs with >75% sequence coverage as compared to Saccharomyces cerevisiae (Sc) Ccr4 were selected from the following species: Schizosaccharomyces pombe (Sp), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Danio rerio (Dr), Xenopus laevis (Xl), Mus musculus (Mm), Homo sapiens (Hs), Arabidopsis thaliana (At), and Oryza sativa (Os). The EEP domain is indicated in green; the leucine-rich repeat domain is highlighted in orange.
In addition to the nuclease domain, Ccr4 contains an amino-terminal LRR domain (Malvar et al., 1992; Dupressoir et al., 2001). The LRR repeats are composed of alternating α-helices and β-sheets, which form a moderately curved solenoid structure with the β-sheets located on the concave side, and the α-helices exposed on the convex curvature (Basquin et al., 2012). The LRR domain provides an interaction surface with the Caf1 subunit (Dupressoir et al., 2001; Clark et al., 2004).
In the single cellular yeasts Saccharomyces cerevisiae, Schizosaccharomyces pombe, as well as the metazoans Drosophila melanogaster and Caenorhabditis elegans, a single Ccr4 subunit is present. By contrast, two paralogs are present in vertebrates, including Danio rerio, Xenopus laevis, Mus musculus, and Homo sapiens (Figure 1C; Dupressoir et al., 2001; Morita et al., 2007; Cooke et al., 2010). Both Ccr4 paralogs (encoded by CNOT6 or CNOT6L) can associate with the Ccr4-Not complex, but the two paralogs cannot co-exist in the same complex (see below for further details). In addition, inspection of the non-redundant protein databank using the Basic local alignment search tool (BLAST) identified several homologs of yeast Ccr4 in Oryza sativa and Arabidopsis thaliana >75% sequence coverage as compared to Saccharomyces cerevisiae Ccr4. However, none of these plant homologs contain the characteristic amino-terminal LRR domain. Indeed, these proteins appear to be more related to eukaryotic Ccr4 homologs that are not associated with the Ccr4-Not complex. For instance, Saccharomyces cerevisiae contains three non-essential homologs of Ccr4 (Ngl1, Ngl2, and Ngl3). While Ngl2 is involved in the processing of the 5.8S ribosomal precursor RNA (Faber et al., 2002), Ngl3 is proposed to play a role in cellular deadenylation independent of Ccr4-Not (Feddersen et al., 2012). Several human Ccr4 homologs lacking an LRR domain (Nocturnin/NOC/CCRN4L, Angel, Angel2, and PDE12) are also implicated in deadenylation (Godwin et al., 2013). Nocturnin is a circadian deadenylase that may associate with subunits of the Ccr4-Not complex in the absence of an LRR (Baggs and Green, 2003). Moreover, Angel homolog 2 (Ccr4d) binds to a distant homolog of Caf1, Caf1z/TOE1 (Wagner et al., 2007). This complex has deadenylase activity, but its biological function is yet unclear. Thus, the fact that the plant homologs do not contain the LRR domain, which is a characteristic for the yeast and vertebrate Ccr4 subunits, may suggest that plant Ccr4-Not may have a fundamentally different architecture.
The Caf1 Subunit, Orthologs, and Paralogs
The Caf1 subunit is characterized by the presence of an RNAse D domain, which belongs to the DEDD (Asp-Glu-Asp-Asp) superfamily of proteins associated with ribonuclease (RNAse) and deoxyribonuclease (DNAse) activity. Crystal structures from Saccharomyces cerevisiae, Schizosaccharomyces pombe, and H. sapiens Caf1 proteins indicate a central core composed of β-sheets surrounded by α-helices (Figure 2A; Thore et al., 2003; Jonstrup et al., 2007; Horiuchi et al., 2009; Petit et al., 2012). As is the case for the catalytic EEP domain of the Ccr4 subunit, the Caf1 protein contains two Mg(II) ions, which are required for its enzymatic activity. The metal ions are coordinated by a single glutamate and three aspartate residues, and substitution of any of these amino acids abrogates the enzymatic activity (Thore et al., 2003; Jonstrup et al., 2007; Horiuchi et al., 2009). Other bivalent cations may also bind in the active site and modulate enzyme activity (Andersen et al., 2009). The Saccharomyces cerevisiae Caf1 protein is unusual as compared to its homologs in other species, because it contains a long amino-terminal extension. In addition, the metal-binding region contains a non-canonical sequence. Although the purified yeast protein displays ribonuclease activity, it has a broad specificity and no preference for poly(A) (Thore et al., 2003). Moreover, mutations of active site residues do not cause phenotypes in yeast (Viswanathan et al., 2004). Thus, the importance of the enzymatic activity of the Saccharomyces cerevisiae Caf1 protein is not unambiguous. By contrast, the Schizosaccharomyces pombe and H. sapiens Caf1 orthologs contain highly conserved active site residues and the ribonuclease activity has a preference for poly(A) residues (Bianchin et al., 2005; Jonstrup et al., 2007; Horiuchi et al., 2009). Moreover, mutation of a residue involved in Mg(II) coordination results in sensitivity to hydroxyurea of Schizosaccharomyces pombe (Takahashi et al., 2007). The activity of Caf1 is distributive and AMP is released as the reaction product (Bianchin et al., 2005).
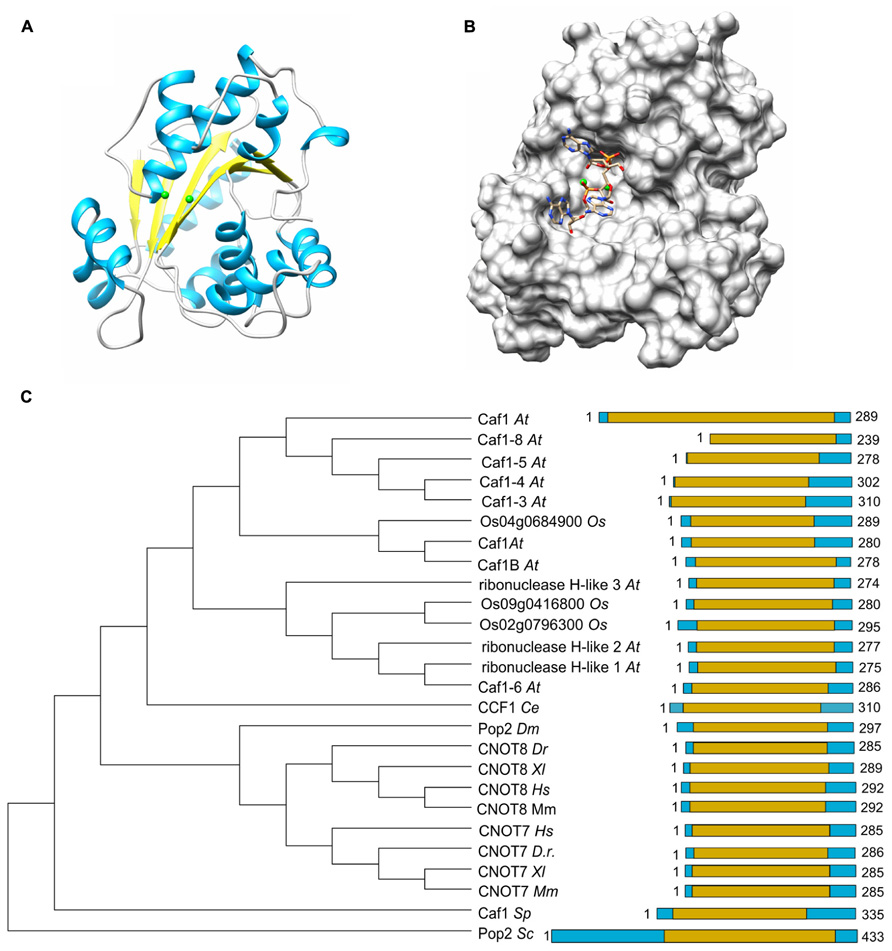
FIGURE 2. The Caf1 nuclease component. (A) Structure of the human Caf1/CNOT7 catalytic domain (PDB accession number 4GMJ). The Mg(II) ions located in the active site are indicated in green; α-helical regions, blue; β-strands, yellow. (B) Model of poly(A) binding by human Caf1/CNOT7. The model was derived from superposition the structure of the PARN deadenyase in complex with RNA (PDB accession number 2A1R). The Mg(II) ions located in the active site are indicated in green. Structures were visualized using the UCSF Chimera package (http://www.cgl.ucsf.edu/chimera; Pettersen et al., 2004). (C) Evolutionary conservation of Caf1 across the eukaryotic kingdom. Sequence analysis was carried out using Basic local alignment search tool (BLAST) in combination with the Reference protein database. Only homologs with >75% sequence coverage as compared to Saccharomyces cerevisiae (Sc) Caf1 were selected from the following species: Schizosaccharomyces pombe (Sp), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Danio rerio (Dr), Xenopus laevis (Xl), Mus musculus (Mm), Homo sapiens (Hs), Arabidopsis thaliana (At), and Oryza sativa (Os). The DEDD domain is highlighted in yellow.
The poly(A) binding site of Caf1 has a different shape and is significantly wider as compared to the substrate binding pocket of Ccr4 (Figure 2B). No structures are available of Caf1-RNA complexes, but a model for RNA recognition was proposed based on the superposition of the DEDD domains of Caf1 and PARN in complex with poly(A) (Wu et al., 2005; Jonstrup et al., 2007; Andersen et al., 2009). This led to the identification of conserved serine and leucine residues for selectivity and processivity, respectively (Andersen et al., 2009).
As is the case for Ccr4, Caf1 is conserved with a single subunit present in Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster, and C. elegans. By contrast, two closely related paralogs (encoded by CNOT7 and CNOT8) are present in the vertebrates Danio rerio, Xenopus laevis, and Mus musculus as well as Homo sapiens (Figure 2C). While both Caf1 paralogs are subunits of Ccr4-Not, their presence in the complex is mutually exclusive (see below for further details). In plants, there are multiple homologs of Caf1, which are distantly related to their counterparts in the fruit fly and vertebrates. Interestingly, the human Caf1z/TOE1, which is part of a complex with the distant Angel2/Ccr4d homolog that lacks an LRR domain, is more closely related to plant Caf1 homologs (Wagner et al., 2007). This may suggest that the plant Caf1 homologs form complexes with their respective Ccr4 partners via interactions that do not involve an LRR domain. It may be speculated that some of these complexes may contain the plant orthologs of the non-catalytic Ccr4-Not subunits.
The Non-Catalytic Subunits
The large Not1 (CNOT1) subunit is the central platform of the complex, on to which several modules are attached (Bai et al., 1999). The nuclease sub-complex is anchored to the central MIF4G domain, which is composed of several α-helices and which binds to the Caf1 subunit (Figures 3A,C; Bai et al., 1999; Basquin et al., 2012; Petit et al., 2012). Interactions between Caf1 and the LRR domain of Ccr4 are essential for stable interaction of the Ccr4 subunit to the complex (Draper et al., 1995; Dupressoir et al., 2001; Mittal et al., 2011; Basquin et al., 2012). The nuclease module appears to contain a single Caf1 and Ccr4 subunit bound to a single Not1 (CNOT1) MIF4G domain. In vertebrate organisms, the presence of two paralogs of Caf1 as well as the occurrence of two highly related Ccr4 subunits thus suggests that the Ccr4-Not complex is heterogeneous. Both human Caf1 paralogs can bind to the human CNOT1 subunit. In addition, both Caf1 proteins can interact with either one of the Ccr4 paralogs and no clear binding preference has been identified (Figure 3B).
FIGURE 3. Modular architecture of the Ccr4-Not complex. (A) Schematic overview of the modular architecture of the mammalian Ccr4-Not complex. (B) Heterogeneity of the Ccr4-Not complex in vertebrate cells. The duplication of the catalytic subunits Caf1 (light blue, CNOT7; dark blue, CNOT8) and Ccr4 (yellow, CNOT6; orange, CNOT6L) may result in the formation of four highly related complexes. The non-catalytic components are represented in a L-shaped form (gray). (C) Structural overview of the nuclease sub-complex. Indicated are the MIF4G domain of human CNOT1 (yellow), the BTG domain of TOB1 (green), Caf1/CNOT7 (blue), yeast Ccr4 (khaki), and the catalytic EEP domain of human Ccr4/CNOT6L (cyan). The location and orientation of the active sites of Caf1 and Ccr4 are indicated and Mg(II) ions are shown (dark gray). The model was generated by superimposing the Tob-Caf1 structure (PDB accession number 2D5R), the MIF4G domain of human CNOT1 in complex with Caf1/CNOT7 (PDB accession number 4GMJ), and the EEP nuclease domain of human Ccr4/CNOT6L (PDB accession number 3NGO) on the structure of the yeast Not1-Caf1-Ccr4 complex (PDB accession number 4B8C). Molecular graphics and analyses were performed with the UCSF Chimera package (http://www.cgl.ucsf.edu/chimera; Pettersen et al., 2004).
Other modules of the Ccr4-Not complex include the CNOT11/CNOT10 module, which is attached to the amino-terminus of CNOT1 (Not1) that largely contains α-helical HEAT repeats (Basquin et al., 2012; Bawankar et al., 2013; Mauxion et al., 2013). A central region encompassing a DUF3819 domain interacts with the RQCD1/RCD1/Caf40/CNOT9 subunit, which is composed of Armadillo repeats forming a bundle of α-helices (Garces et al., 2007; Bawankar et al., 2013). This subunit has the ability to bind single and double stranded nucleic acids (Garces et al., 2007). Interestingly, the affinity for sequences containing G/C/T is much greater than oligo(A). The carboxy-terminus of CNOT1 is bound by CNOT2 (Not2), which interacts with CNOT3 (Not3/Not5) via their carboxy-terminal Not-Box region (Bai et al., 1999; Bawankar et al., 2013; Bhaskar et al., 2013; Boland et al., 2013). In yeast, this region also interacts with the Not4 ubiquitin protein ligase subunit (Bai et al., 1999). However, in human cells, the homologs CNOT4 subunit does not bind stably to the CNOT1 protein (Lau et al., 2009).
In addition to the nuclease subunits, several non-catalytic subunits are implicated in mRNA deadenylation (Tucker et al., 2002; Temme et al., 2004; Ito et al., 2011a,b; Takahashi et al., 2012). However, genetic analysis in Saccharomyces cerevisiae has shown that the Not-module has additional roles as compared to the Caf1 and Ccr4 subunits (Bai et al., 1999). The additional role of the Ccr4-Not complex is less well defined as compared to its function in deadenylation, and may include transcriptional regulation, and/or an involvement in the co-translational control of protein folding (Collart and Panasenko, 2012).
Specialized or Redundant Roles for the Caf1 and Ccr4 Paralogs?
The duplication of the genes encoding the Caf1 and Ccr4 subunits in vertebrate cells led to the suggestion that the paralogs might have slightly specialized roles in mRNA deadenylation. So far, this question has been addressed in most detail by studying the human paralogs. The human Caf1 proteins (76% identity, 89% similarity at the amino acid level) are encoded by the CNOT7 and CNOT8 genes (Figure 4A). The surface residues in the active site are completely conserved (Figure 4B). Despite the high sequence conservation, however, the enzymatic activity of the purified Caf1/CNOT7 and Caf1/CNOT8 proteins differs substantially. Whereas both proteins display selectivity toward poly(A), the Caf1/CNOT8 protein appeared to have a significantly higher turnover rate (Bianchin et al., 2005). By contrast, following a systematic approach to identify interacting proteins of Ccr4-Not subunits, a more limited role was proposed for the Caf1/CNOT8 subunit as compared to Caf1/CNOT7. In this study, proteins involved in splicing were not found to associate with Caf1/CNOT8, and a less stable association with Ccr4/CNOT6 was proposed (Lau et al., 2009). However, the role of Caf1/CNOT7 and Caf1/CNOT8 in the regulation of mRNA levels is essentially identical when studied in human breast cancer cells using knockdown approaches in combination with genome-wide expression profiling (Aslam et al., 2009). Upon knockdown of either CNOT7 or CNOT8 mRNA, modest changes were observed on a limited set of mRNAs. By contrast, quantitatively significant effects were observed upon combined knockdown of CNOT7 and CNOT8. Thus, from this study it appears that the Caf1/CNOT7 and Caf1/CNOT8 paralogs do not have intrinsically different properties in the regulation of mRNA levels in cells.
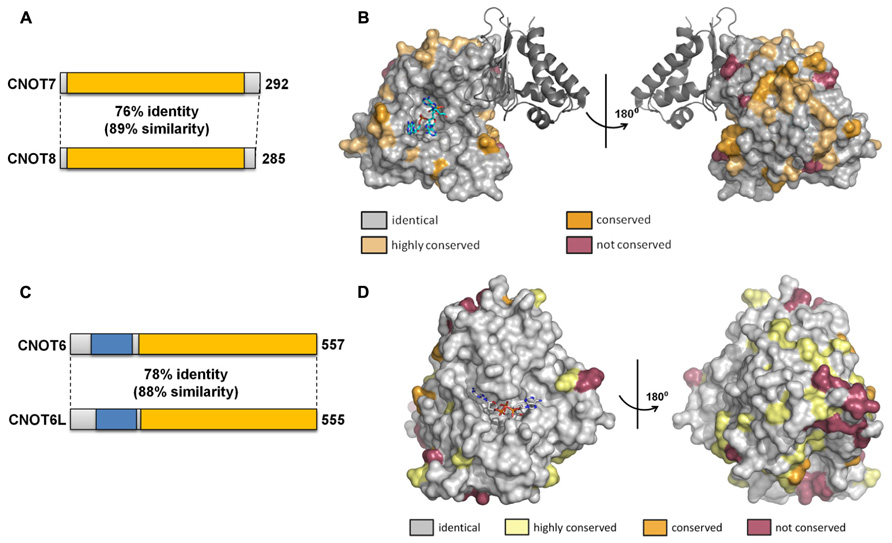
FIGURE 4. Similarity of the Caf1 and Ccr4 paralogs. (A) Schematic overview of similarity between the Caf1 paralogs encoded by CNOT7 and CNOT8. The conserved DEDD nuclease domain is indicated in yellow. (B) Conservation of surface residues between the Caf1/CNOT7 and Caf1/CNOT8 subunits. The surface of the human Caf1/CNOT7 protein is shown with residues identical in Caf1/CNOT7 and Caf1/CNOT8 shown in gray, conserved residues in tan or dark yellow, and non-conserved residues in purple. The BTG domain of TOB1 is shown in dark gray. The image was generated using PDB file 2D5R. (C) Schematic overview of similarity between the Ccr4 paralogs encoded by CNOT6 and CNOT6L. The conserved EEP nuclease domain is indicated in yellow, the LRR domain in blue. (D) Conservation of surface residues between the Ccr4/CNOT6 and Ccr4/CNOT6L subunits. The surface of the human Ccr4/CNOT6L protein is shown with residues identical in Ccr4/CNOT6 and Ccr4/CNOT6L shown in gray, conserved residues in tan or dark yellow, and non-conserved residues in purple. The image was generated using PDB file 3NGO.
The human Ccr4 proteins (78% identity, 88% similarity at the amino acid level) are encoded by the CNOT6 and CNOT6L genes (Figure 4C). As is the case for the human Caf1 paralogs, the surface residues in the active site are completely conserved in Ccr4/CNOT6 and Ccr4/CNOT6L (Figure 4D). In cells, these paralogs appear to have essentially identical properties as determined using a knockdown approach in combination with microarray-based expression profiling (Mittal et al., 2011). Whereas CNOT6 knockdown has little effect on mRNA levels as compared to CNOT6L knockdown, the combined knockdown of CNOT6 and CNOT6L quantitatively changes the differential expression pattern observed upon CNOT6L knockdown.
The importance of duplications of the genes encoding Caf1 and Ccr4 remains unclear. It may be speculated that, whilst the intrinsic properties of the Caf1 paralogs on the one hand, and of the Ccr4 paralogs on the other, appear to be essentially identical, duplication of the genes encoding the Caf1 and Ccr4 subunits may have allowed fine-tuning of tissue-specific expression of these proteins in vertebrata. Therefore, whilst the CNOT7 and CNOT8 genes, as well as CNOT6 and CNOT6L, respectively, may be largely redundant, they may have cell-type specific functions. For instance, Ccr4/CNOT6L, but not Ccr4/CNOT6, is required for proliferation of mouse 3T3 fibroblast cells (Morita et al., 2007). By contrast, both Ccr4/CNOT6 and Ccr4/CNOT6L are required for proliferation of human MCF-7 breast carcinoma cells (Mittal et al., 2011). Moreover, while both Ccr4 paralogs are involved in the maintenance of MCF-7 cell viability, no such role has been documented for Ccr4/CNOT6L in mouse 3T3 cells (Morita et al., 2007; Mittal et al., 2011).
The BTG/TOB Proteins
The BTG/TOB proteins are the best characterized interaction partners of the Caf1 proteins in vertebrates (Mauxion et al., 2009; Winkler, 2010). These proteins interact with the Caf1 protein via their conserved amino-terminal BTG domain, which interacts at a site away from the Caf1 active site residues (Figure 3C; Horiuchi et al., 2009). When over-expressed, these proteins inhibit cell cycle progression, which is dependent on interactions with the Caf1 proteins (Horiuchi et al., 2009; Doidge et al., 2012; Ezzeddine et al., 2012). Interestingly, the number of these proteins appears to have expanded during evolution. Single cellular eukaryotes do not contain a gene encoding a BTG/TOB protein, but BTG/TOB proteins are present in Caenorhabditis elegans (1), Drosophila melanogaster (2), mouse (6), and human (6) cells. In their carboxy-termini, the TOB1 and TOB2 proteins are highly similar and contain a conserved PAM2 motif that is able to interact with the PABP C-terminal domain (Ezzeddine et al., 2007). Similarly, the BTG1 and BTG2 proteins, which contain a short carboxy-terminal extension, are highly related. Thus, an intriguing number of possible interactions may occur between TOB1/TOB2 or BTG1/BTG2 and the Caf1 paralogs in vertebrate cells. Although the use of genetically engineered mice lacking the Tob1, Tob2, Btg1, or Cnot7 gene have started to uncover the significance of specific roles of the paralog genes, the fundamental importance of this network remains presently unclear (Yoshida et al., 2000; Nakamura et al., 2004; Park et al., 2004; Ajima et al., 2008).
In vitro, BTG2 can inhibit the activity of purified Caf1, although the conserved BTG domain of TOB1 does not affect the activity of Caf1 (Yang et al., 2008; Horiuchi et al., 2009). In addition, it was reported that TOB1 can inhibit the deadenylase activity of immunopurified GFP-CNOT6L (Miyasaka et al., 2008). However, functional studies based on reporter genes indicate that BTG2, TOB1, and TOB2 are factors promoting deadenylation and mRNA degradation in cellular transfection assays (Ezzeddine et al., 2007; Mauxion et al., 2008; Doidge et al., 2012; Ezzeddine et al., 2012).
Collaboration or Specialization of Caf1 and Ccr4 Subunits?
At first glance, several findings suggest a collaborative mode of action for Caf1 and Ccr4: firstly, both proteins have a preference for poly(A), and, secondly, the subunits directly interact. However, as explained above, it is ambiguous whether the Saccharomyces cerevisiae Caf1 is an active deadenylase in vivo, indicating that the yeast Ccr4 subunit may be the only active catalytic subunit (Tucker et al., 2002; Thore et al., 2003; Viswanathan et al., 2004). Moreover, the (partial) crystal structure of the yeast Not1-Caf1-Ccr4 sub-complex shows that the catalytic centers are distant and pointing in different directions (Figure 3C; Basquin et al., 2012). Functional analysis of Caf1 and Ccr4 in human cells also indicate that the proteins do not have identical roles. Comparison of the expression profile of Caf1 knockdown cells with that of Ccr4 knockdown cells shows qualitative differences in the differentially regulated mRNA sets (Mittal et al., 2011). Moreover, the phenotypes of Caf1 knockdown cells are different to that of Ccr4 knockdown cells. Whereas cell proliferation is reduced upon knockdown of either Caf1 or Ccr4, Ccr4 is required for cell viability and the prevention of cellular senescence. Thus, these findings suggest that the Caf1 and Ccr4 subunits may have specialized roles.
It is unclear what the mechanistic basis is for the differential role in mRNA deadenylation of the Caf1 and Ccr4 subunits. It may be speculated that the Ccr4-Not complex may be distinctly arranged depending on the mechanism of recruitment by protein–protein interactions. Thus, depending on the particular proteins involved, either the Caf1 or the Ccr4 subunit may be placed in a suitable orientation for deadenylation.
Cooperation and Redundancy with other Deadenylase Complexes
Whilst the Ccr4-Not complex is described as the main deadenylase in a variety of organisms, including Saccharomyces cerevisiae and human cells (Tucker et al., 2001; Yamashita et al., 2005), it is clear that other deadenylase complexes, notably the Pan2-Pan3 complex, also play an important role. Using transcriptional pulse experiments of reporter mRNAs in human cells, it has been established that Pan2-Pan3 is important for the early stage of deadenylation in human cells, with the Ccr4-Not components involved in a distinct, second stage (Yamashita et al., 2005; Zheng et al., 2008). Interestingly, interactions between Pan2-Pan3 and Ccr4-Not can be detected suggesting that both deadenylase complexes can reside in a higher order complex that may include other components (Zheng et al., 2008). Thus, these experiments suggest that Pan2-Pan3 and Ccr4-Not may have unique, but cooperative roles, in mRNA decay. On the other hand, genetic analyses in the budding yeast Saccharomyces cerevisiae indicate that yeast pan2Δ or ccr4Δ cells lacking a single catalytic deadenylase subunit are viable and do not display significant growth phenotypes (Boeck et al., 1996; Brown et al., 1996; Tucker et al., 2001). By contrast, yeast pan2Δ ccr4Δ cells lacking both catalytic subunits display a strong synthetic phenotype indicating that the Pan2-Pan3 and Ccr4-Not complexes are largely redundant and can compensate for each other’s loss of function (Boeck et al., 1996; Brown et al., 1996; Tucker et al., 2001).
Perspective and Concluding Remarks
Recently, significant progress has been made to understand the importance of deadenylation by the Ccr4-Not complex. Despite this, fundamental questions remain regarding the significance of the duplications of genes encoding the Ccr4-Not nuclease subunits, and the enzymatic mechanism of deadenylation by Ccr4-Not. The functional significance of paralogs encoding the nuclease subunits and of the (potential) redundancy with other deadenylases may be addressed by using mouse knockout models and mice engineered to have point mutations resulting in catalytically inactive deadenylase subunits. Such experiments may be facilitated by advances in genome engineering tools, such as the use of zinc finger nucleases or RNA-guided genome editing using the CRISPR/Cas system. Additionally, the use of novel approaches, such as the use of chemical probes of deadenylase enzymes that specifically inhibit their enzymatic activity without affecting structural roles, should be explored to further the understanding of deadenylation by the Ccr4-Not complex (Maryati et al., 2013).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ajima, R., Akiyama, T., Usui, M., Yoneda, M., Yoshida, Y., Nakamura, T., et al. (2008). Osteoporotic bone formation in mice lacking tob2; involvement of Tob2 in RANK ligand expression and osteoclasts differentiation. FEBS Lett. 582, 1313–1318. doi: 10.1016/j.febslet.2008.03.012
Andersen, K. R., Jonstrup, A. T., Van, L. B., and Brodersen, D. E. (2009). The activity and selectivity of fission yeast Pop2p are affected by a high affinity for Zn2+ and Mn2+ in the active site. RNA 15, 850–861. doi: 10.1261/rna.1489409
Aslam, A., Mittal, S., Koch, F., Andrau, J. C., and Winkler, G. S. (2009). The Ccr4-not deadenylase subunits CNOT7 and CNOT8 have overlapping roles and modulate cell proliferation. Mol. Biol. Cell 20, 3840–3850. doi: 10.1091/mbc.E09-02-0146
Baggs, J. E., and Green, C. B. (2003). Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr. Biol. 13, 189–198. doi: 10.1016/S0960-9822(03)00014-9
Bai, Y., Salvadore, C., Chiang, Y. C., Collart, M. A., Liu, H. Y., and Denis, C. L. (1999). The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol. Cell. Biol. 19, 6642–6651.
Basquin, J., Roudko, V. V., Rode, M., Basquin, C., Seraphin, B., and Conti, E. (2012). Architecture of the nuclease module of the yeast Ccr4-not complex: the Not1-Caf1-Ccr4 interaction. Mol. Cell 48, 207–218. doi: 10.1016/j.molcel.2012.08.014
Bawankar, P., Loh, B., Wohlbold, L., Schmidt, S., and Izaurralde, E. (2013). NOT10 and C2orf29/NOT11 form a conserved module of the CCR4-NOT complex that docks onto the NOT1 N-terminal domain. RNA Biol. 10, 228–244. doi: 10.4161/rna.23018
Bhaskar, V., Roudko, V., Basquin, J., Sharma, K., Urlaub, H., Seraphin, B., et al. (2013). Structure and RNA-binding properties of the Not1-Not2-Not5 module of the yeast Ccr4-Not complex. Nat. Struct. Mol. Biol. 20, 1281–1288. doi: 10.1038/nsmb.2686
Bianchin, C., Mauxion, F., Sentis, S., Seraphin, B., and Corbo, L. (2005). Conservation of the deadenylase activity of proteins of the Caf1 family in human. RNA 11, 487–494. doi: 10.1261/rna.7135305
Boeck, R., Tarun, S. Jr., Rieger, M., Deardorff, J. A., Muller-Auer, S., and Sachs, A. B. (1996). The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J. Biol. Chem. 271, 432–438. doi: 10.1074/jbc.271.1.432
Boland, A., Chen, Y., Raisch, T., Jonas, S., Kuzuoglu-Ozturk, D., Wohlbold, L., et al. (2013). Structure and assembly of the NOT module of the human CCR4-NOT complex. Nat. Struct. Mol. Biol. 20, 1289–1297. doi: 10.1038/nsmb.2681
Brown, C. E., Tarun, S. Z. Jr., Boeck, R., and Sachs, A. B. (1996). PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 5744–5753.
Chekulaeva, M., Mathys, H., Zipprich, J. T., Attig, J., Colic, M., Parker, R., et al. (2011). miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat. Struct. Mol. Biol. 18, 1218–1226. doi: 10.1038/nsmb.2166
Chen, J., Chiang, Y. C., and Denis, C. L. (2002). CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 21, 1414–1426. doi: 10.1093/emboj/21.6.1414
Chen, J., Rappsilber, J., Chiang, Y. C., Russell, P., Mann, M., and Denis, C. L. (2001). Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J. Mol. Biol. 314, 683–694. doi: 10.1006/jmbi.2001.5162
Clark, L. B., Viswanathan, P., Quigley, G., Chiang, Y. C., McMahon, J. S., Yao, G., et al. (2004). Systematic mutagenesis of the leucine-rich repeat (LRR) domain of CCR4 reveals specific sites for binding to CAF1 and a separate critical role for the LRR in CCR4 deadenylase activity. J. Biol. Chem. 279, 13616–13623. doi: 10.1074/jbc.M313202200
Collart, M. A., and Panasenko, O. O. (2012). The Ccr4 – not complex. Gene 492, 42–53. doi: 10.1016/j.gene.2011.09.033
Collart, M. A., and Struhl, K. (1994). NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 8, 525–537. doi: 10.1101/gad.8.5.525
Cooke, A., Prigge, A., and Wickens, M. (2010). Translational repression by deadenylases. J. Biol. Chem. 285, 28506–28513. doi:10.1074/jbc.M110.150763
Denis, C. L. (1984). Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics 108, 833–844.
Doidge, R., Mittal, S., Aslam, A., and Winkler, G. S. (2012). The anti-proliferative activity of BTG/TOB proteins is mediated via the Caf1a (CNOT7) and Caf1b (CNOT8) deadenylase subunits of the Ccr4-not complex. PLoS ONE 7:e51331. doi: 10.1371/journal.pone.0051331
Draper, M. P., Salvadore, C., and Denis, C. L. (1995). Identification of a mouse protein whose homolog in Saccharomyces cerevisiae is a component of the CCR4 transcriptional regulatory complex. Mol. Cell. Biol. 15, 3487–3495.
Dupressoir, A., Morel, A.-P., Barbot, W., Loireau, M.-P., Corbo, L., and Heidmann, T. (2001). Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics 2:9. doi: 10.1186/1471-2164-2-9
Ezzeddine, N., Chang, T. C., Zhu, W., Yamashita, A., Chen, C. Y., Zhong, Z., et al. (2007). Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol. Cell. Biol. 27, 7791–7801. doi: 10.1128/MCB.01254-07
Ezzeddine, N., Chen, C. Y., and Shyu, A. B. (2012). Evidence providing new insights into TOB-promoted deadenylation and supporting a link between TOB’s deadenylation-enhancing and antiproliferative activities. Mol. Cell. Biol. 32, 1089–1098. doi: 10.1128/MCB.06370–6311
Faber, A. W., Van Dijk, M., Raue, H. A., and Vos, J. C. (2002). Ngl2p is a Ccr4p-like RNA nuclease essential for the final step in 3′-end processing of 5.8S rRNA in Saccharomyces cerevisiae. RNA 8, 1095–1101. doi: 10.1017/S1355838202021027
Fabian, M. R., Cieplak, M. K., Frank, F., Morita, M., Green, J., Srikumar, T., et al. (2011). miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat. Struct. Mol. Biol. 18, 1211–1217. doi: 10.1038/nsmb.2149
Feddersen, A., Dedic, E., Poulsen, E. G., Schmid, M., Van, L. B., Jensen, T. H., et al. (2012). Saccharomyces cerevisiae Ngl3p is an active 3′-5′ exonuclease with a specificity towards poly-A RNA reminiscent of cellular deadenylases. Nucleic Acids Res. 40, 837–846. doi: 10.1093/nar/gkr782
Gallie, D. R. (1991). The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 5, 2108–2116. doi: 10.1101/gad.5.11.2108
Garces, R. G., Gillon, W., and Pai, E. F. (2007). Atomic model of human Rcd-1 reveals an armadillo-like-repeat protein with in vitro nucleic acid binding properties. Protein Sci. 16, 176–188. doi: 10.1110/ps.062600507
Garneau, N. L., Wilusz, J., and Wilusz, C. J. (2007). The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8, 113–126. doi: 10.1038/nrm2104
Godwin, A. R., Kojima, S., Green, C. B., and Wilusz, J. (2013). Kiss your tail goodbye: the role of PARN, nocturnin, and angel deadenylases in mRNA biology. Biochim. Biophys. Acta 1829, 571–579. doi: 10.1016/j.bbagrm.2012.12.004
Goldstrohm, A. C., Hook, B. A., Seay, D. J., and Wickens, M. (2006). PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 13, 533–539. doi: 10.1038/nsmb1100
Goldstrohm, A. C., and Wickens, M. (2008). Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 9, 337–344. doi: 10.1038/nrm2370
Horiuchi, M., Takeuchi, K., Noda, N., Muroya, N., Suzuki, T., Nakamura, T., et al. (2009). Structural basis for the antiproliferative activity of the Tob-hCaf1 complex. J. Biol. Chem. 284, 13244–13255. doi: 10.1074/jbc.M809250200
Hosoda, N., Funakoshi, Y., Hirasawa, M., Yamagishi, R., Asano, Y., Miyagawa, R., et al. (2011). Anti-proliferative protein Tob negatively regulates CPEB3 target by recruiting Caf1 deadenylase. EMBO J. 30, 1311–1323. doi: 10.1038/emboj.2011.37
Houseley, J., and Tollervey, D. (2009). The many pathways of RNA degradation. Cell 136, 763–776. doi: 10.1016/j.cell.2009.01.019
Huntzinger, E., Kuzuoglu-Ozturk, D., Braun, J. E., Eulalio, A., Wohlbold, L., and Izaurralde, E. (2013). The interactions of GW182 proteins with PABP and deadenylases are required for both translational repression and degradation of miRNA targets. Nucleic Acids Res. 41, 978–994. doi: 10.1093/nar/gks1078
Ito, K., Inoue, T., Yokoyama, K., Morita, M., Suzuki, T., and Yamamoto, T. (2011a). CNOT2 depletion disrupts and inhibits the CCR4-NOT deadenylase complex and induces apoptotic cell death. Genes Cells 16, 368–379. doi: 10.1111/j.1365-2443.2011.01492.x
Ito, K., Takahashi, A., Morita, M., Suzuki, T., and Yamamoto, T. (2011b). The role of the CNOT1 subunit of the CCR4-NOT complex in mRNA deadenylation and cell viability. Protein Cell 2, 755–763. doi: 10.1007/s13238-011-1092–1094
Jonstrup, A. T., Andersen, K. R., Van, L. B., and Brodersen, D. E. (2007). The 1.4-A crystal structure of the S. pombe Pop2p deadenylase subunit unveils the configuration of an active enzyme. Nucleic Acids Res. 35, 3153–3164. doi: 10.1093/nar/gkm178
Kuzuoglu-Ozturk, D., Huntzinger, E., Schmidt, S., and Izaurralde, E. (2012). The Caenorhabditis elegans GW182 protein AIN-1 interacts with PAB-1 and subunits of the PAN2-PAN3 and CCR4-NOT deadenylase complexes. Nucleic Acids Res. 40, 5651–5665. doi: 10.1093/nar/gks218
Lackner, D. H., Beilharz, T. H., Marguerat, S., Mata, J., Watt, S., Schubert, F., et al. (2007). A network of multiple regulatory layers shapes gene expression in fission yeast. Mol. Cell 26, 145–155. doi: 10.1016/j.molcel.2007.03.002
Lau, N. C., Kolkman, A., Van Schaik, F. M., Mulder, K. W., Pijnappel, W. W., Heck, A. J., et al. (2009). Human Ccr4-Not complexes contain variable deadenylase subunits. Biochem. J. 422, 443–453. doi: 10.1042/BJ20090500
Leppek, K., Schott, J., Reitter, S., Poetz, F., Hammond, M. C., and Stoecklin, G. (2013). Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell 153, 869–881. doi: 10.1016/j.cell.2013.04.016
Liu, H. Y., Badarinarayana, V., Audino, D. C., Rappsilber, J., Mann, M., and Denis, C. L. (1998). The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 17, 1096–1106. doi: 10.1093/emboj/17.4.1096
Liu, H. Y., Toyn, J. H., Chiang, Y. C., Draper, M. P., Johnston, L. H., and Denis, C. L. (1997). DBF2, a cell cycle-regulated protein kinase, is physically and functionally associated with the CCR4 transcriptional regulatory complex. EMBO J. 16, 5289–5298. doi: 10.1093/emboj/16.17.5289
Malvar, T., Biron, R. W., Kaback, D. B., and Denis, C. L. (1992). The CCR4 protein from Saccharomyces cerevisiae contains a leucine-rich repeat region which is required for its control of ADH2 gene expression. Genetics 132, 951–962.
Maryati, M., Kaur, I., Gopal, J., Olotu-Umoren, L., Oveh, B., Hashmi, L., et al. (2013). A fluorescence-based assay suitable for quantitative analysis of deadenylase enzyme activity. Nucleic Acids Res. doi: 10.1093/nar/gkt972 [Epub ahead of print].
Mauxion, F., Chen, C. Y., Seraphin, B., and Shyu, A. B. (2009). BTG/TOB factors impact deadenylases. Trends Biochem. Sci. 34, 640–647. doi: 10.1016/j.tibs.2009.07.008
Mauxion, F., Faux, C., and Seraphin, B. (2008). The BTG2 protein is a general activator of mRNA deadenylation. EMBO J. 27, 1039–1048. doi: 10.1038/emboj.2008.43
Mauxion, F., Preve, B., and Seraphin, B. (2013). C2ORF29/CNOT11 and CNOT10 form a new module of the CCR4-NOT complex. RNA Biol. 10, 267–276. doi: 10.4161/rna.23065
Mittal, S., Aslam, A., Doidge, R., Medica, R., and Winkler, G. S. (2011). The Ccr4a (CNOT6) and Ccr4b (CNOT6L) deadenylase subunits of the human Ccr4-Not complex contribute to the prevention of cell death and senescence. Mol. Biol. Cell 22, 748–758. doi: 10.1091/mbc.E10-11-0898
Miyasaka, T., Morita, M., Ito, K., Suzuki, T., Fukuda, H., Takeda, S., et al. (2008). Interaction of antiproliferative protein Tob with the CCR4-NOT deadenylase complex. Cancer Sci. 99, 755–761. doi: 10.1111/j.1349-7006.2008.00746.x
Morita, M., Suzuki, T., Nakamura, T., Yokoyama, K., Miyasaka, T., and Yamamoto, T. (2007). Depletion of mammalian CCR4b deadenylase triggers elevation of the p27Kip1 mRNA level and impairs cell growth. Mol. Cell. Biol. 27, 4980–4990. doi: 10.1128/MCB.02304-06
Munroe, D., and Jacobson, A. (1990). mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol. Cell. Biol. 10, 3441–3455.
Nakamura, T., Yao, R., Ogawa, T., Suzuki, T., Ito, C., Tsunekawa, N., et al. (2004). Oligo-astheno-teratozoospermia in mice lacking Cnot7, a regulator of retinoid X receptor beta. Nat. Genet. 36, 528–533. doi: 10.1038/ng1344
Nasertorabi, F., Batisse, C., Diepholz, M., Suck, D., and Bottcher, B. (2011). Insights into the structure of the CCR4-NOT complex by electron microscopy. FEBS Lett. 585, 2182–2186. doi: 10.1016/j.febslet.2011.05.071
Nousch, M., Techritz, N., Hampel, D., Millonigg, S., and Eckmann, C. R. (2013). The Ccr4-Not deadenylase complex constitutes the main poly(A) removal activity in C. elegans. J. Cell Sci. 126, 4274–4285. doi: 10.1242/jcs.132936
Park, S., Lee, Y. J., Lee, H. J., Seki, T., Hong, K. H., Park, J., et al. (2004). B-cell translocation gene 2 (Btg2) regulates vertebral patterning by modulating bone morphogenetic protein/smad signaling. Mol. Cell. Biol. 24, 10256–10262. doi: 10.1128/MCB.24.23.10256-10262.2004
Parker, R., and Song, H. (2004). The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11, 121–127. doi: 10.1038/nsmb724
Petit, A. P., Wohlbold, L., Bawankar, P., Huntzinger, E., Schmidt, S., Izaurralde, E., et al. (2012). The structural basis for the interaction between the CAF1 nuclease and the NOT1 scaffold of the human CCR4-NOT deadenylase complex. Nucleic Acids Res. 40, 11058–11072. doi: 10.1093/nar/gks883
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera – a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi: 10.1002/jcc.20084
Sakai, A., Chibazakura, T., Shimizu, Y., and Hishinuma, F. (1992). Molecular analysis of POP2 gene, a gene required for glucose-derepression of gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 20, 6227–6233. doi: 10.1093/nar/20.23.6227
Sandler, H., Kreth, J., Timmers, H. T., and Stoecklin, G. (2011). Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res. 39, 4373–4386. doi: 10.1093/nar/gkr011
Siddiqui, N., Mangus, D. A., Chang, T. C., Palermino, J. M., Shyu, A. B., and Gehring, K. (2007). Poly(A) nuclease interacts with the C-terminal domain of polyadenylate-binding protein domain from poly(A)-binding protein. J. Biol. Chem. 282, 25067–25075. doi: 10.1074/jbc.M701256200
Takahashi, A., Kikuguchi, C., Morita, M., Shimodaira, T., Tokai-Nishizumi, N., Yokoyama, K., et al. (2012). Involvement of CNOT3 in mitotic progression through inhibition of MAD1 expression. Biochem. Biophys. Res. Commun. 419, 268–273. doi: 10.1016/j.bbrc.2012.02.007
Takahashi, S., Kontani, K., Araki, Y., and Katada, T. (2007). Caf1 regulates translocation of ribonucleotide reductase by releasing nucleoplasmic Spd1-Suc22 assembly. Nucleic Acids Res. 35, 1187–1197. doi: 10.1093/nar/gkm015
Temme, C., Zaessinger, S., Meyer, S., Simonelig, M., and Wahle, E. (2004). A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 23, 2862–2871. doi: 10.1038/sj.emboj.7600273
Thore, S., Mauxion, F., Seraphin, B., and Suck, D. (2003). X-ray structure and activity of the yeast Pop2 protein: a nuclease subunit of the mRNA deadenylase complex. EMBO Rep. 4, 1150–1155. doi: 10.1038/sj.embor.7400020
Tucker, M., Staples, R. R., Valencia-Sanchez, M. A., Muhlrad, D., and Parker, R. (2002). Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 21, 1427–1436. doi: 10.1093/emboj/21.6.1427
Tucker, M., Valencia-Sanchez, M. A., Staples, R. R., Chen, J., Denis, C. L., and Parker, R. (2001). The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104, 377–386. doi: 10.1016/S0092-8674(01)00225-2
Viswanathan, P., Ohn, T., Chiang, Y. C., Chen, J., and Denis, C. L. (2004). Mouse CAF1 can function as a processive deadenylase/3′-5′-exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J. Biol. Chem. 279, 23988–23995. doi: 10.1074/jbc.M402803200
Wagner, E., Clement, S. L., and Lykke-Andersen, J. (2007). An unconventional human Ccr4-Caf1 deadenylase complex in nuclear cajal bodies. Mol. Cell. Biol. 27, 1686–1695. doi: 10.1128/MCB.01483-06
Wahle, E., and Winkler, G. S. (2013). RNA decay machines: deadenylation by the Ccr4-Not and Pan2-Pan3 complexes. Biochim. Biophys. Acta 1829, 561–570. doi: 10.1016/j.bbagrm.2013.01.003
Wang, H., Morita, M., Yang, X., Suzuki, T., Yang, W., Wang, J., et al. (2010). Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity. EMBO J. 29, 2566–2576. doi: 10.1038/emboj.2010.152
Wells, S. E., Hillner, P. E., Vale, R. D., and Sachs, A. B. (1998). Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2, 135–140. doi: 10.1016/S1097-2765(00)80122-7
Winkler, G. S. (2010). The mammalian anti-proliferative BTG/Tob protein family. J. Cell. Physiol. 222, 66–72. doi: 10.1002/jcp.21919
Wu, M., Reuter, M., Lilie, H., Liu, Y., Wahle, E., and Song, H. (2005). Structural insight into poly(A) binding and catalytic mechanism of human PARN. EMBO J. 24, 4082–4093. doi: 10.1038/sj.emboj.7600869
Yamashita, A., Chang, T. C., Yamashita, Y., Zhu, W., Zhong, Z., Chen, C. Y., et al. (2005). Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 12, 1054–1063. doi: 10.1038/nsmb1016
Yang, X., Morita, M., Wang, H., Suzuki, T., Yang, W., Luo, Y., et al. (2008). Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity. Nucleic Acids Res. 36, 6872–6881. doi: 10.1093/nar/gkn825
Yoshida, Y., Tanaka, S., Umemori, H., Minowa, O., Usui, M., Ikematsu, N., et al. (2000). Negative regulation of BMP/Smad signaling by Tob in osteoblasts. Cell 103, 1085–1097. doi: 10.1016/S0092-8674(00)00211-7
Keywords: deadenylase, mRNA turnover, poly(A), ribonuclease, Pop2, Calif
Citation: Winkler GS and Balacco DL (2013) Heterogeneity and complexity within the nuclease module of the Ccr4-not complex. Front. Genet. 4:296. doi: 10.3389/fgene.2013.00296
Received: 07 October 2013; Paper pending published: 04 November 2013;
Accepted: 04 December 2013; Published online: 23 December 2013.
Edited by:
Martine Anne Collart, University of Geneva, SwitzerlandReviewed by:
Yuichiro Watanabe, University of Tokyo, JapanMark Gerrard Bartlam, Nankai University, China
Copyright © 2013 Winkler and Balacco. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: G. Sebastiaan Winkler, School of Pharmacy, Centre for Biomolecular Sciences, University of Nottingham, University Park, Nottingham NG7 2RD, UK e-mail: sebastiaan.winkler@nottingham.ac.uk
 G. Sebastiaan Winkler
G. Sebastiaan Winkler Dario L. Balacco
Dario L. Balacco