Halophilic Methylotrophic Methanogens May Contribute to the High Ammonium Concentrations Found in Shale Oil and Shale Gas Reservoirs
- 1Division 4.1 Biodeterioration and Reference Organisms, Federal Institute for Materials Research and Testing, Berlin, Germany
- 2Petroleum Microbiology Research Group, Department of Biological Sciences, University of Calgary, Calgary, AB, Canada
Flow-back and produced waters from shale gas and shale oil fields contain high ammonium, which can be formed by methanogenic degradation of methylamines into methane and ammonium. Methylamines are added to fracturing fluid to prevent clay swelling or can originate from metabolism of the osmolyte triglycinebetaine (GB). We analyzed field samples from a shale gas reservoir in the Duvernay formation and from a shale oil reservoir in the Bakken formation in Canada to determine the origin of high ammonium. Fresh waters used to make fracturing fluid, early flow-back waters, and late flow back waters from the shale gas reservoir had increasing salinity of 0.01, 0.58, and 2.66 Meq of NaCl, respectively. Microbial community analyses reflected this fresh water to saline transition with halophilic taxa including Halomonas, Halanaerobium, and Methanohalophilus being increasingly present. Early and late flow-back waters had high ammonium concentrations of 32 and 15 mM, respectively. Such high concentrations had also been found in the Bakken produced waters. Enrichment cultures of Bakken produced waters in medium containing mono, di-, or trimethylamine, or triglycinebetaine (GB) converted these substrates into ammonium (up to 20 mM) and methane. The methylotrophic methanogen Methanohalophilus, which uses methylamines for its energy metabolism and uses GB as an osmolyte, was a dominant community member in these enrichments. Halanaerobium was also a dominant community member that metabolizes GB into trimethylamine, which is then metabolized further by Methanohalophilus. However, the micromolar concentrations of GB measured in shale reservoirs make them an unlikely source for the 1,000-fold higher ammonium concentrations in flow-back waters. This ammonium either originates directly from the reservoir or is formed from methylamines, which originate from the reservoir, or are added during the hydraulic fracturing process. These methylamines are then converted into ammonium and methane by halophilic methylotrophic methanogens, such as Methanohalophilus, present in flow-back waters.
Introduction
Halophilic microorganisms (halophiles) have been studied for decades, especially with respect to their biotechnology potential for osmolyte synthesis (Stan-Lotter and Fendrihan, 2012). Halophiles adapt to high external NaCl concentrations by accumulation of KCl or by production and accumulation of compatible solutes (osmolytes), such as trimethylglycine (also called glycine betaine; GB), in the cytoplasm (Oren, 2001). The first strategy requires that intracellular proteins are highly acidic (Oren, 2013). Halophiles are often isolated from highly saline environments such as the Great Salt Lake and the Dead Sea (Oren, 2001, 2008; Roberts et al., 2005). Recent exploration of highly saline shale oil and shale gas reservoirs has also indicated the presence of halophilic microbial communities (Cluff et al., 2014; Tucker et al., 2015; An et al., 2017).
The shale oil and shale gas industries have seen a rapid expansion due to development of hydraulic fracturing technologies (Brittingham et al., 2014; Mouser et al., 2016; Shrestha et al., 2017). In addition to high salinity in excess of 1 Meq NaCl, which is defined as the salinity of a solution of salts with a conductivity as that of 1 M NaCl, deep shale reservoirs have high temperatures in excess of 60°C. This bears the question whether microorganisms can survive in such conditions. Injection of fresh water-based fracturing fluids into shale reservoirs will decrease the salinity and temperature of the down-hole environment, allowing increased microbial activity, including of sulfate-reducing bacteria (SRB). Decreases in temperature and salinity due to hydraulic fracturing give rise to distinct microbial communities in different recovery stages (Cluff et al., 2014). Bowers and Wiegel (2011) showed that 110 halophilic Archaea had optimal growth temperatures between 30 and 40°C and optimal salinities between 2.5 and 3.5 M NaCl. But the maximum growth temperature for halophilic Archaea is 55°C, which limits their growths in down-hole environments.
Sulfate-reducing bacteria reduce sulfate to sulfide, which partitions into the gas phase as H2S. This decreases the value of produced gas. SRB, methanogens and other microbes can also contribute to microbially-influenced corrosion (MIC; Enning and Garrelfs, 2014). Polymer added to fracturing fluid to keep proppant (sand) in suspension may also be microbially degraded. Hence, there are multiple incentives for controlling microbial activities in shale gas reservoirs. Creating multiple environments by changing salinity and temperature through injection of cold fresh water and by storing produced water above ground complicates the problem of microbial control, e.g., when temperature and/or salinity are kept high nitrate-reducing bacteria (NRB) reduce nitrate only to nitrite, which is a powerful SRB inhibitor (Fida et al., 2016; An et al., 2017). Hence, SRB control with nitrate is easier if environments with low salinity and temperature are not allowed to emerge in shale gas or shale oil operations.
Halophilic SRB and methanogens, such as Desulfohalobium and Methanohalophilus, are found in shale reservoirs. The latter is a methylotrophic methanogen, which produces methane and ammonium from methylated amines (Boone et al., 1993; Katayama et al., 2014; Daly et al., 2016) and uses GB as an osmolyte (Lai and Lai, 2011). This can be metabolized by other microorganisms (King, 1984), such as Halanaerobium, for energy production (Cluff et al., 2014). Halanaerobium metabolizes GB into methylated amines that can again be metabolized by halophilic methylotrophic methanogens to methane and ammonium (Daly et al., 2016). GB is also used by some SRB as electron donor to reduce sulfate to sulfide (Oren, 1990). Organic amines, including tetramethylammonium chloride, are also often added in shale operations in concentrations of up to 0.3% v/v to inhibit clay swelling (Horton and Jones, 1998).
The multiple effects from additives like methylamines on the microbial communities in shale reservoirs are as yet poorly understood and require further analyses. The aim of the present study was therefore to provide insight into the origin of high ammonium concentrations in shale reservoirs using water samples from Canadian shale oil and shale gas reservoirs.
Materials and Methods
Sample Collection
Samples were collected from a shale gas reservoir from the Duvernay formation in Alberta, Canada. This included samples of source water (SW), flow back water (FBW), and C-ring storage tank water (STW). SW1, SW2, and SW3 were obtained from fresh water lakes, whereas SW4 was an industrial waste water. These source waters were combined with other components to make fracturing fluid, which was used for hydraulic fracturing by injection in horizontal wells. Following fracturing FBWs were produced together with produced gas at multiple sites (FBW1 to FBW8). These were a mixture of fracturing fluid and waters or other components (e.g., salts) from the shale formation with the fraction of the latter increasing with time. Following production FBWs were stored in multiple C-ring containment systems, which were open to the air. Samples from these systems are referred to as storage tank water (STW) and were received from STW1 to STW4. Samples were received in either September 2015, November 2016, or January 2017 (Table 1). Sample bottles were filled to the brim to exclude air and were shipped on ice. Samples were received within 2 days and were stored in the anaerobic hood with 90% v/v N2 and 10% v/v CO2 (N2-CO2) upon arrival. Samples from the Bakken shale oil field were as described previously (An et al., 2017).
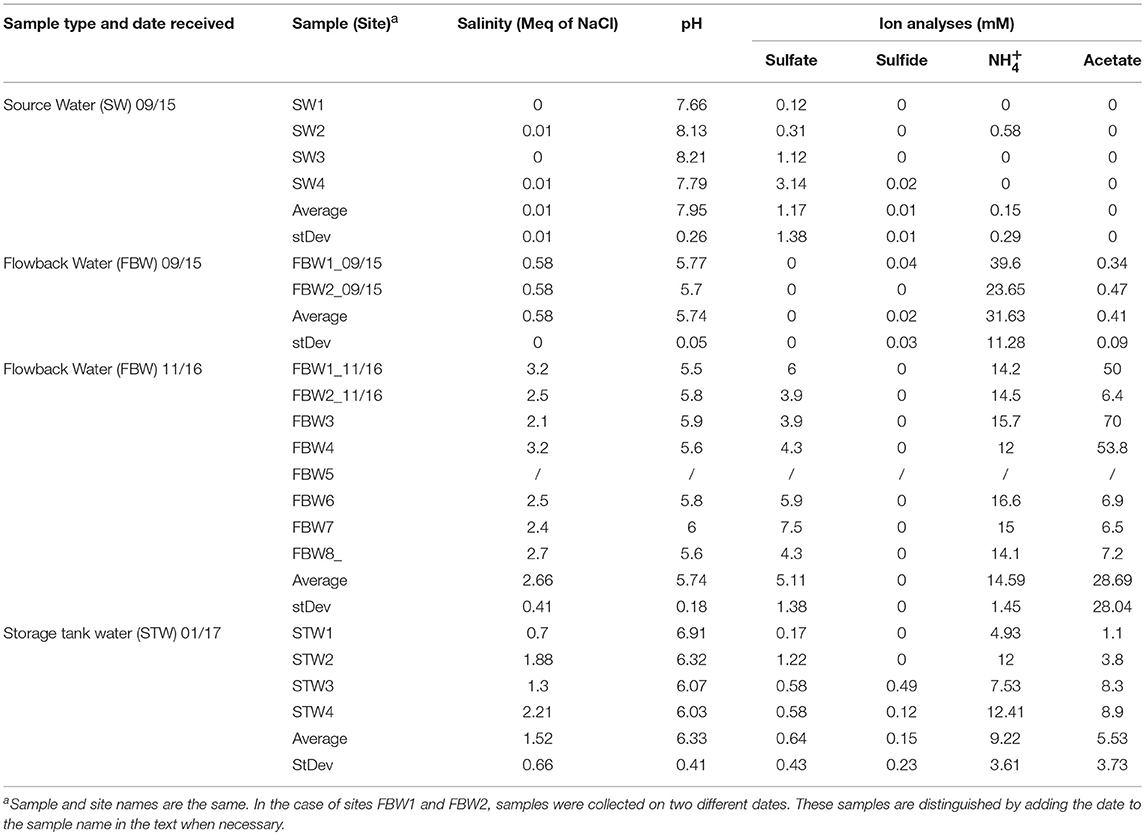
Table 1. Water chemistry of source, flow-back, and C-ring storage tank water samples from the Duvernay formation received in 09/15, 11/16, and 01/17 as indicated.
Water Analyses
Water chemistry analyses were carried out using 50 mL of field sample for measurement of pH, salinity, sulfate, sulfide, organic acids, and ammonium concentrations. The pH was measured using an Orion pH meter (Model 370; VWR International, Mississauga, ON). Salinity in molar equivalent (Meq) of NaCl was analyzed with an Orion conductivity cell (model 013005MD; Thermo Scientific, Beverly, MA). Conductivity and pH of the field water samples were averages of triplicate measurements. The concentration of dissolved sulfide was measured using the diamine method (Trüper and Schlegel, 1964). Samples were diluted to 1 Meq of NaCl using MilliQ-filtered water (Millipore, Etobicoke, ON), before analyzing sulfate, nitrate and nitrite with the Waters 600E high performance liquid chromatography (HPLC) instrument. Sulfate was measured using a conductivity detector (Waters 423) and IC-PAK anion column (4 × 150 mm, Waters). Nitrate and nitrite were eluted from the same column with the same buffer but were measured with an UV detector (UV/VIS-2487, Waters) at 220 nm. Concentrations of sulfate, nitrate and nitrite were calculated from appropriate standard lines taking dilution factors into account. Concentrations of lactate and of volatile fatty acids (VFA) acetate, propionate and butyrate, were measured using a HPLC system (Waters, model 515) with the UV detector at 220 nm and an organic acids column (Alltech, 250 × 4.6 mm). Ammonium concentrations were measured using spectrophotometry with the indophenol method (Aminot et al., 1997).
DNA Extraction and Microbial Community Analyses
DNA was extracted from field samples and from sample enrichments. For field samples 250 mL was centrifuged at 14,000 × g for 20 min at 4°C. For enrichments, 5 mL was centrifuged at 14,000 × g for 10 min at 4°C. DNA was extracted from the pellets using the FastDNA extraction kit for soil (MP Biomedicals). DNA was quantified with a Qubit fluorimeter (Invitrogen) using the Quant-iT double-stranded DNA (dsDNA) HS assay kit (Invitrogen).
DNAs were amplified using a two-step PCR process with Illumina Miseq non-barcoded primers 926Fi5 and 1392RiF (An et al., 2017) for the first PCR. PCR was performed for 3 min at 95°C, followed by 25 cycles of 30 s at 95°C, 45 s at 55°C, and 2 min at 72°C, and then 10 min at 72°C. For the second PCR (10 cycles), forward primer P5-S50X-OHAF and reverse primer P7-N7XX-OHAF were used (Fida et al., 2016; An et al., 2017). The final PCR product was purified and quantified using the same procedures as above and sent for Illumina Miseq sequencing at the University of Calgary.
Illumina Miseq sequences were analyzed with the MetaAmp software, (http://ebg.ucalgary.ca/metaamp/) (Dong et al., 2017). Sequences were merged using PEAR 0.9.8 and merged reads were uploaded to MetaAmp, which used a cut-off quality control (QC) score for each sequence of 50 and a minimum length of each sequence of 420 base pairs. The QC sequences were clustered into operational taxonomic units (OTUs) using average neighbor clustering at a distance of 3%. Each OTU was assigned to a taxon by comparison with the latest version of the non-redundant 16S rRNA small subunit SILVA database. Amplicon sequence libraries were clustered into a dendrogram using the unweighted pair group method algorithm (UPGMA) and the distance between these was calculated using the Bray-Curtis coefficient in the Mothur software. The dendrogram was visualized using the MEGA5.2.2. Program (Tamura et al., 2011).
Enrichment of Halophilic Methylotrophic Methanogens
Coleville Synthetic Brine K (CSBK) medium (Fida et al., 2016) with 2.0 M of NaCl and 20 mM of methylamine (MA), dimethylamine (DMA), or trimethylamine (TMA) or 10 mM of GB was used. Bakken field samples (10% v/v) were inoculated into 60 mL of fresh anaerobic saline CSBK medium in 120 ml serum bottles with a headspace of N2-CO2, all enrichments were grown in duplicate. Enrichments were incubated at 30°C in the dark. Headspace methane was measured by anaerobically removing 200 μL of headspace from the serum bottles and injecting into a Hewlett-Packard 5890 (USA) Gas Chromatograph (GC) equipped with a flame ionization detector (FID).
Corrosion Experiments and Analyses
Ball bearings (BBs) made from a36 carbon steel with a diameter Ø = 0.238 ± 0.003 cm were obtained from Thomson Precision Ball. These were prepared using the NACE protocol RP0775-2005 (Voordouw et al., 2016) in which BBs were polished using grit size 400 sand paper, placed in dibutylthiourea HCl solution for 2 min, neutralized with 1.2 M NaHCO3 for 2 min, rinsed with deionized water, rinsed with acetone, and dried with N2. The BBs were stored anaerobically prior to use. BBs were weighed thrice and the average weight was recorded as the starting weight. Post corrosion experiments, BBs were cleaned and weighed using the same procedure as above. The general corrosion rate CR (mm/year) was calculated from the weight loss (ΔW in g) as: CR = 87,600 × ΔW/ (D × A × T), where D is the density of the carbon steel (7.85 g/cm3), A is the surface area of the BBs (cm2) and T is the incubation time in h. The factor 87,600 converts the measured corrosion rate from cm/h to mm/year.
Scanning electron microscope (SEM) images of incubated carbon steel BBs were obtained at the Instrumentation Facility for Analytical Electron Microscopy (IFAEM) at the University of Calgary using an FEI Quanta 250 FEG variable pressure/environmental field emission SEM with an Everhart Thornley Detector (ETD) for high-vacuum secondary electron imaging and a two-segment semiconductor Backscatter Electron Detector (BSE). Images were obtained using an acceleration voltage of 15 kV and a working distance of 10 mm.
Results
Water Chemistry Analyses of Field Samples
The water chemistry analyses of Duvernay shale gas samples are summarized in Table 1. The average salinity of the source water samples was 0.01 ± 0.01 Meq of NaCl (Table 1). These fresh water samples also had low concentrations of sulfate (1.2 ± 1.4 mM), sulfide (0.01 ± 0.01 mM), and (0.2 ± 0.3 mM) (Table 1). Flow-back water samples FBW1_09/15 and FBW2_09/15 had a higher salinity (0.58 Meq of NaCl) and a very high concentration (31.6 ± 11.3 mM). But sulfate, sulfide, and acetate concentrations were low, as for the source water samples (Table 1). The salinity of flow-back waters increased with time with values of 3.2 and 2.5 Meq of NaCl measured for FBW1_11/16 and FBW2_11/16, respectively (Table 1). The average for eight flow-back waters collected in November of 2016 was 2.66 ± 0.41 Meq of NaCl. These samples also had a higher average concentration of sulfate of 5.1 mM, whereas that of decreased to 14.6 mM (Table 1). FWB3, FWB4, and FBW1_11/16 had high concentrations of acetate (Table 1). FBW5 was a water-in-oil emulsion and sterile MilliQ water was added to obtain an aqueous layer for DNA extraction, which was not used for water chemistry analyses (Table 1). The C-ring storage tanks, used to contain flow-back waters, are mildly heated (45°C). The salinity of STW1 and STW3 was lower than that for STW2 and STW4 (Table 1; 1 and 2 Meq of NaCl, respectively). All C-ring water samples had lower sulfate concentrations (0.17–1.22 mM) than found in the flow-back water samples. STW3 and STW4 had detectable concentrations of aqueous hydrogen sulfide (Table 1: 0.49 and 0.12 mM). The lower sulfate and higher sulfide concentrations in the C-ring storage tank waters, as compared to the flow-back waters, suggested active sulfate reduction. C-ring water samples had lower ammonium concentrations of 9.2 ± 13.6 mM (Table 1) than found in flow-back waters. Produced waters from a Bakken shale oil field, which was injected with water with 0.6 Meq of NaCl, 30 mM sulfate, and 2 mM of ammonium, had high salinity (average 1.6 Meq of NaCl), sulfate (average 24 mM) and ammonium (average 17 mM). Values for individual produced waters depended on the degree of injection water breakthrough (An et al., 2017). A high ammonium concentration thus appears to be shared by shale gas field flow-back waters (Table 1) and shale oil field produced waters (An et al., 2017).
Microbial Community Analyses of Field Samples
Results for Illumina sequencing of 16S rRNA gene amplicons obtained for the 09/15 samples, including numbers of QC reads and of derived OTUs, taxa and the Shannon diversity index are shown in Table S1. The microbial communities in SW1, SW2, and SW3 lake waters were dominated by freshwater microorganisms, such as Limnohabitans, Flavobacterium, Polynucleobacter, and Actinobacteria/hgcI_clade, which are well-known taxa in freshwater lakes. The microbial community in SW4, which was an industrial waste water, also contained these fresh water microorganisms but in addition contained taxa which were shared with flow-back water FBW1_09/15, including Hydrogenophaga, GKS98_freshwater_group, Acidibacter, and Mycobacterium. Flow-back water FBW2_09/15 had high fractions of Thermus, Anoxybacillus, Rubrivivax, Shewanella, and Thauera, whereas flow-back water FBW1_09/15 had high fractions of Thermus, Flavobacterium, and Mesorhizobium with smaller fractions of the fresh water taxa Limnohabitans, Polynucleobacter, Hydrogenophaga, GKS98_freshwater_group, Acidibacter, and Mycobacterium (Table S1).
Microbial community compositions of the 11/16 flow-back waters showed three distinct clades (Figure 1). Clade I, consisting of communities in samples FBW7, FBW5, FBW6, and FWB3, was dominated by Halomonas. Anaerobic taxa including Desulfovibrio, Acetobacterium, and Methanocorposculum were also present (Table 2). Clade II, consisting of communities in samples FBW8, FBW1_11/16, and FWB4, was dominated by Desulfovibrio with Halomonas, Halanaerobium, Acetobacterium, and Arcobacter also being present (Table 2). Clade III consisted of a single community of FBW2_11/16, which was composed mostly of Halanaerobium (Table 2). The diversity as indicated by the Shannon index for the 2016 communities (0.5–3) was lower than that of the 2015 communities (2.6–4.8). Flow-back water samples from sites FBW1 and FBW2, received both in 11/16 and 09/15, showed very different microbial community compositions. The aerobic and non-halophilic taxa found in FBW1_09/15 and FBW2_09/15 (Table S1) were replaced with Halomonas and Halanaerobium in FBW1_11/16 and FBW2_11/16 (Table 2), which are halophilic or halotolerant (Vreeland et al., 1980); (Liang et al., 2016).
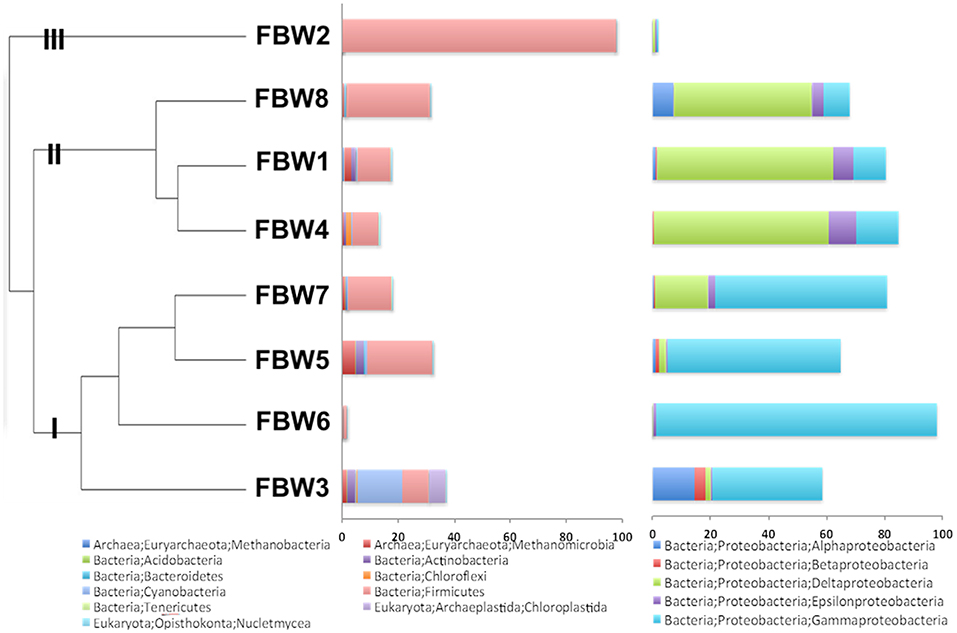
Figure 1. Microbial community compositions for 11/16 flow-back water (FBW) samples from the Duvernay formation; shown are a relational tree indicating clades I to III of the samples (left), the distribution of phyla other than Proteobacteria (middle) and the distribution of classes from the phylum Proteobacteria (right).
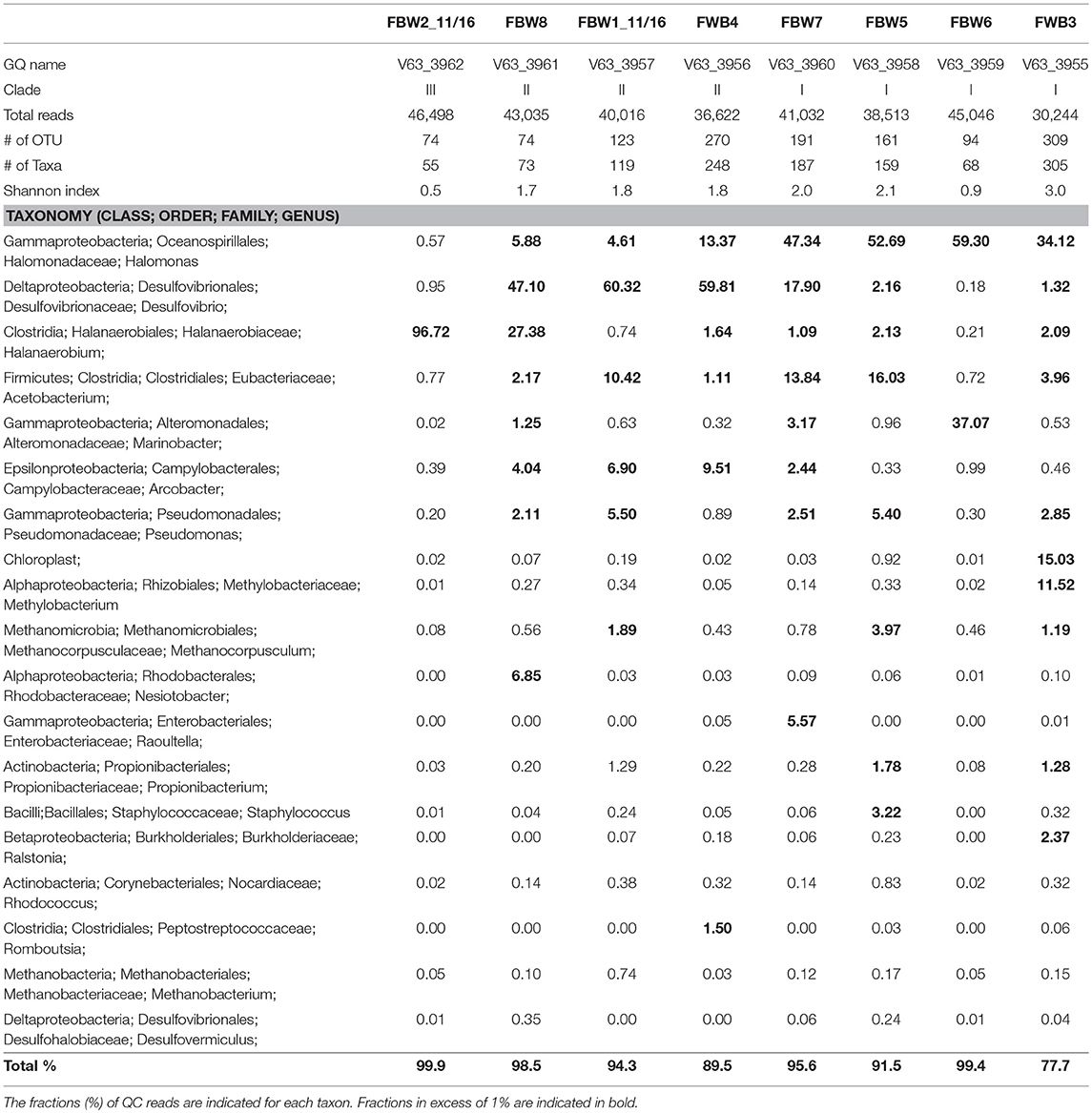
Table 2. Microbial community compositions of flow-back water samples from a Duvernay shale gas field received in November 2016.
Most C-ring samples had a higher microbial diversity (Shannon index of 2.3 to 3.6) than that of the 11/16 flow-back water samples, (Tables 2, 3). At the phylum/class level the microbial community compositions of the four C-ring storage tank water samples indicated the presence high proportions of Firmicutes, Deltaproteobacteria, Bacteroidetes, Thermotogae, and Methanosarcinales (Figure S1). At the genus level these were represented by Halanaerobium, Desulfovibrio, Anaerophaga, Geotoga, and Methanohalophilus (Table 3).
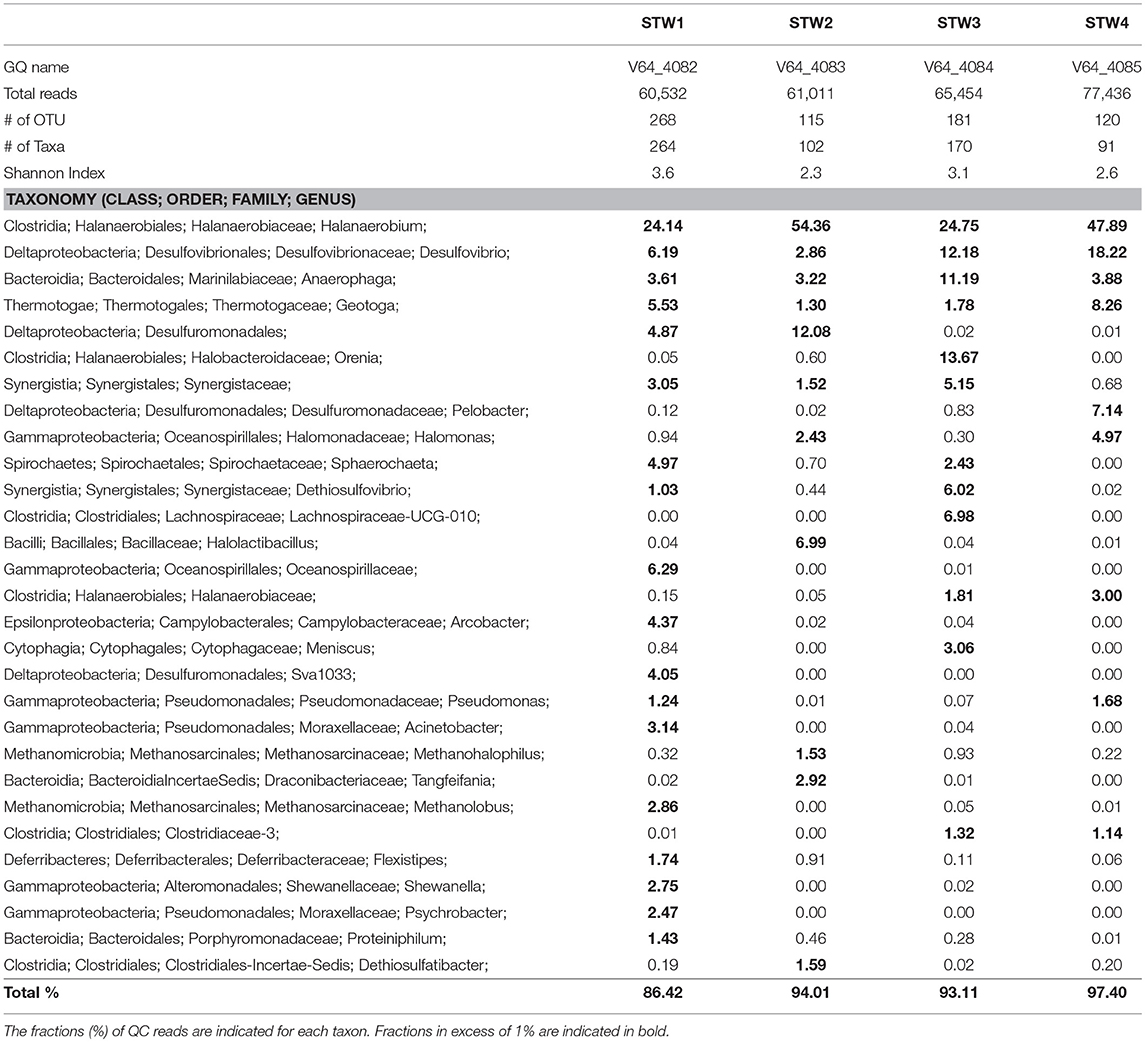
Table 3. Microbial community compositions of C-ring storage tank water samples from a Duvernay shale gas field received in January 2017.
Corrosivity of Field Samples
Corrosion rates of carbon steel BBs incubated with Duvernay flow-back water samples received in November 2016, and storage tank samples received in January 2017 are listed in Table 4. BBs incubated with FBW6, FBW2_11/16 or FBW8 had high corrosion rates of 0.40–0.44 mm/year (Table 4). C-ring STW samples had intermediate to high corrosion rates (Table 4: 0.10–0.35 mm/year). BBs incubated with STW2 had the highest general corrosion rate of 0.35 ± 0.05 mm/year, followed by those incubated with STW4 (0.20 ± 0.07 mm/year). SEM images of the BBs incubated with these two samples indicated pitting corrosion of the steel surface (Figures 2H,I,K,L, respectively). BBs incubated with FWB4 or FBW1_11/16 had intermediate corrosion rates of 009–0.10 mm/year (Table 4). BBs incubated with FWB3, FBW5, or FBW7 had low corrosion rates of 0.03–0.05 mm/year, which were only slightly higher than the control rate of 0.00 mm/year (Table 4). SEM images of BBs incubated with FBW5 (0.05 mm/year) showed a smooth surface similar to that of the control with no significant damage (Figure 2). On the contrary, BBs incubated with FBW2_11/16 and FBW8, which had high corrosion rates showed significant corrosion damage of the steel surface (Figure 2). Interestingly, numerous pits were observed on the surface of BBs incubated with FBW2_11/16 (Figures 2C,F), but not on that of BBs incubated with FBW8 (Figures 2G,J). The corrosion rates of BBs incubated with 11/16 flow-back water samples (Table 4) did not correlate with the fraction of the SRB Desulfovibrio in these samples (Table 3). Re-incubation of 5 BBs with FBW6, which was either used directly or was filtered using an 0.2 μm Millipore nylon membrane (VWR International, Edmonton, AB) gave general corrosion rates of 0.55 ± 0.06 and 0.57 ± 0.02 mm/year, respectively (data not shown). These results indicate that MIC may not be a major contributor to the high corrosion rates observed for some of these shale gas samples, chemical corrosion is more likely (Sharma et al., 2017).
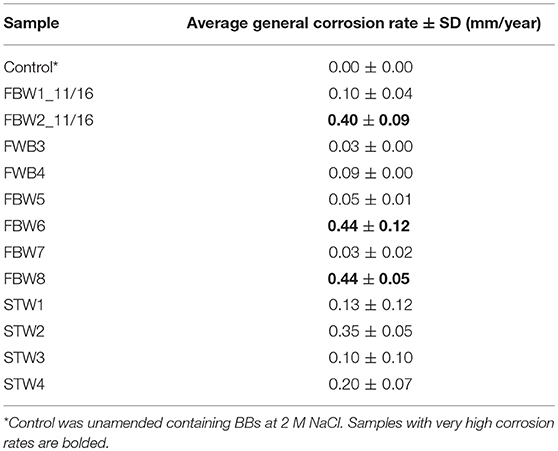
Table 4. Average general corrosion rates of unfiltered flowback water and C-ring samples incubated with five carbon steel BBs each.
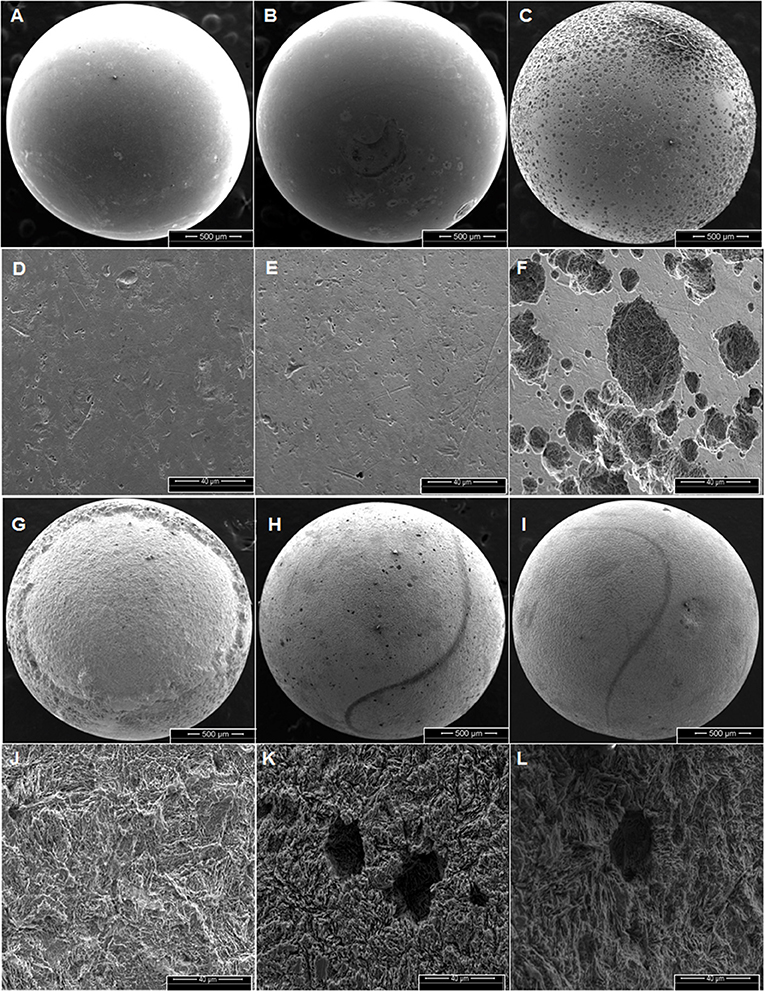
Figure 2. Scanning electron microscope (SEM) images of BBs from incubations with the control (AD), FBW5 (B,E), FBW2_11/16 (C,F), FBW8 (G,J), STW2 (H,K), and STW4 (I,L) at the end of incubation. Scales are indicated at the bottom of each image. The general corrosion rates observed in these incubations were (A,D) 0.0 ± 0.0, (B,E) 0.01 ± 0.01, (C,F) 0.40 ± 0.09, (G,J) 0.44 ± 0.05, (H,K) 0.35 ± 0.05, and (I,L) 0.20 ± 0.07 mm/year, respectively.
Methanogenesis With Methylated Amines at High Salinity
To test whether halophilic methylotrophic methanogens in shale samples can metabolize methylamines to methane and ammonium Bakken shale oil samples were selected as inocula. These samples had higher fractions of halophilic methanogens than found in Duvernay shale gas field samples. Medium with 2 M NaCl and 20 mM TMA were inoculated with 10% (v/v) of field sample. This resulted in the production of 30–60 mM of methane and 20–25 mM in 3 of 7 tested field samples, namely 10TW_11/13, 2PW_08/15, and 9FW_08/15 (Figure 3). Microbial community analyses of these primary enrichments showed a dominance of Methanohalophilus and Halanaerobium. A high proportion of Sphingobacteriales was also detected (Table S2).
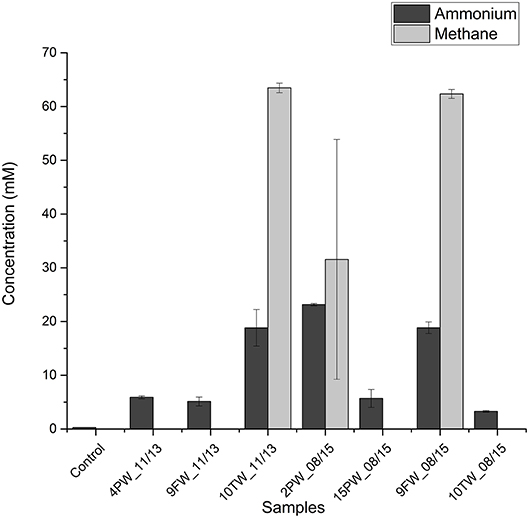
Figure 3. Bakken field samples inoculated in medium with 20 mM trimethylamine at 2.0 M NaCl. Concentrations of ammonium in the medium and methane in the headspace ± SD (duplicates) are shown for day 0 and day 60.
The primary enrichments of Bakken 10TW_11/13, 2PW_08/15 and 9FW_08/15 in medium with TMA were then used to inoculate CSBK medium with 2.5 M NaCl and one of the following 20 mM MA, 20 mM DMA, 20 mM TMA, or 10 mM GB. Secondary enrichments inoculated with the TMA-enrichment of 10TW_11/13 showed methane formation with MA, TMA, and GB, but not with DMA (Figure 4). The increase in concentrations in these enrichments from 1 to 35 days (Figure 4) was 9.6 mM for MA, 6.5 mM for DMA, 8.7 mM for TMA, and 3 mM for GB. Secondary enrichments inoculated with the TMA-enrichment of 2PW_08/15 showed methane formation with MA, DMA, and TMA, but not with GB (Figure 4). The increase in concentrations in these enrichments from 1 to 35 days (Figure 4) was 17.9 mM for MA, 18.2 mM for DMA, 15 mM for TMA, and 3.8 mM for GB. Lastly, secondary enrichments inoculated with the TMA-enrichment of 9FW_08/15 showed methane formation with all four substrates with the least methane being formed with GB (Figure 4). The increase in concentrations in these enrichments from 1 to 35 days (Figure 4) was 14.4 mM for MA, 14.4 mM for DMA, 9.5 mM for TMA, and 2.8 mM for GB. Hence, the enrichments with 10TW_11/13 were less active than those with 9FW_08/15 and 2PW_08/15 in both methane and ammonium production. The average methane concentrations formed with MA, DMA, TMA, and GB for all three samples were 49.7 ± 13.2 mM, 32.5 ± 28.3 mM, 41.3 ± 28.0 mM, and 12.5 ± 21.4 mM (n = 6), respectively. The average ammonium concentrations formed with MA, DMA, TMA, and GB for all three samples were 18.1 ± 4.4, 13.0± 6.4, 12.6 ± 4.1, and 9.2 ± 0.9 mM (n = 6), respectively. Overall, methane and ammonium production correlated positively with each other (r2 = 0.82). The least methane and ammonium were formed with GB.
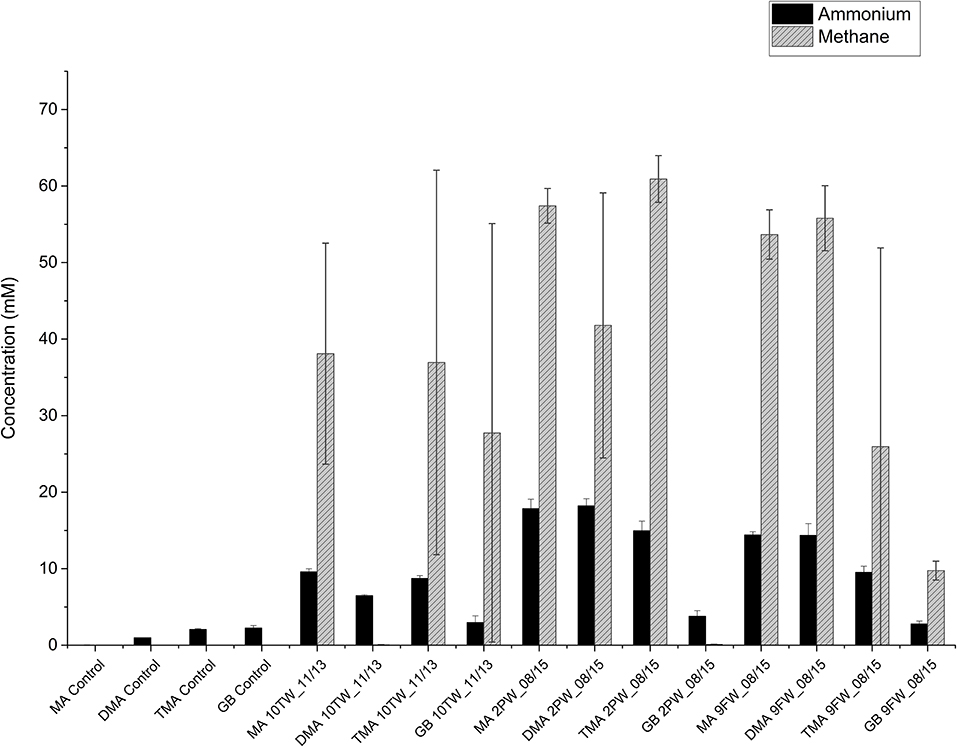
Figure 4. Ammonium and methane production in secondary enrichments of field samples in medium with 20 mM of methylamine (MA), dimethylamine (DMA), trimethylamine (TMA), or 10 mM of glycine betaine (GB) after 35 days of duplicate incubations (± SD). Control incubations without inoculum are also shown and gave no methane.
Dominant taxa of all incubations were Methanohalophilus and Halanaerobium (Table S3). Interestingly, high proportions of Sphingobacteriales were also detected in the enrichments with the exception of 10TW_11/13 (Table S3). Members of the Sphingobacteriales are involved with fermentation and osmotic solute production (Krieg et al., 2009), which could support the growth of Halanaerobium and Methanohalophilus in these enrichments.
Discussion
Waters produced from a Duvernay shale gas field had high ammonium concentrations. These high ammonium concentrations are of interest and have been reported in other shale systems (Harkness et al., 2015). In produced waters from a Bakken shale oil field the ammonium concentrations were linearly correlated with salinity (An et al., 2017), suggesting that the ammonium was contributed by the shale. However, in the Duvernay shale gas samples studied here the higher ammonium concentration in early, less saline flow-back waters suggests a different origin of at least some of this ammonium. Early and late flow-back waters converged in C-ring storage tanks, which had an intermediate salinity of 1.5 Meq of NaCl, a pH of 6.3, 0.64 mM sulfate, 0.15 mM sulfide, 9.2 mM ammonium, and 5.5 mM acetate (Table 1). The lower concentrations of sulfate and elevated concentrations of hydrogen sulfide, as compared to flow-back waters, indicated active sulfate reduction.
The microbial community compositions of the four types of samples reflected the water chemistry data. Source water samples were dominated by freshwater microorganisms (Table S1), such as Limnohabitans capable of growth in both oxic and anoxic conditions with high sensitivity to acidity and salinity (Kasalický et al., 2013). Early flow-back waters had Thermus as a dominant community member, which is thermophilic (Mesbah and Wiegel, 2012), as well as Anoxybacillus, which is thermophilic and anaerobic (Urbieta et al., 2015). However, early flow-back water samples also had minor fractions of Flavobacterium, which is often found in fresh water environments (Ahn et al., 2016) and which was a dominant community member in the source waters (Table S1). These changes indicated transition from an oxic fresh water into a warmer, more anoxic reservoir environment.
Microbial communities in late flow-back waters lacked fresh water taxa and had increased proportions of halophilic and halotolerant taxa, including Halomonas, Desulfovibrio, and Halanaerobium, reflecting the increased salinity of these waters (Table 2). High fractions of the SRB Desulfovibrio (Table 2: 1.3–60%) indicated a potential for microbial formation of H2S, referred to as souring (Voordouw et al., 1990; Finster and Kjeldsen, 2010). This taxon was also observed in high salinity shale oil samples (An et al., 2017). High fractions of Halomonas indicate a potential of nitrate-mediated souring control, as Halomonas reduces nitrate to nitrite, which is a strong SRB inhibitor (Vreeland et al., 1980; An et al., 2017). Other taxa, such as Halanaerobium, Acetobacterium, and Marinobacter, are capable of anaerobic, fermentative metabolism at high salinity (Daly et al., 2016; Liang et al., 2016). Microbial community compositions in C-ring storage facilities were also dominated by halophiles with high fractions of Halanaerobium, Desulfovibrio, Anaerophaga, and Geotoga (Table 3). Both STW1 and STW2 had high fractions of Desulfuromonadales and Synergistaceae, whereas STW1 also had a high fraction of Arcobacter (Table 3). These taxa may act in oxygen-mediated sulfide oxidation and sulfur reduction, which is expected in an air-exposed storage facility containing aqueous sulfide. STW3 had high fractions of Orenia, whereas Halomonas was present in high fractions in both STW2 and STW4 (Table 3). These taxa are capable of fermentation and iron-reduction (Denger et al., 2002; Feio et al., 2004; Greene et al., 2009; Liang et al., 2016), reflective of the storage tank conditions. STW3 and STW4 had higher fractions of Desulfovibrio compared to STW1 and STW2, which may reflect the increased aqueous H2S concentration in these samples (Table 1).
Overall, the microbial community compositions of the samples reflected the transition from fresh water (0 M NaCl) to sea water (0.5 M NaCl) to high salinity reservoir (2–3 M NaCl) properties, as has been described for other shale oil (An et al., 2017) and shale gas reservoirs (Wuchter et al., 2013; Cluff et al., 2014; Gaspar et al., 2014; Daly et al., 2016; Borton et al., 2018). The presence of high fractions of Desulfovibrio in late flow-back waters and C-ring storage facility waters indicates potential for souring and associated corrosion. However, the high corrosion rates observed upon incubation of carbon steel BBs with some of the samples were not correlated with a particular community composition, i.e., communities in samples with high corrosion rates were found in all of the three clades of Figure 1. Hence, despite the demonstrated presence of taxa implicated in corrosion including Halanaerobium (Ravot et al., 2006; Liang et al., 2016; Lipus et al., 2017), Desulfovibrio (Enning and Garrelfs, 2014; Mand et al., 2014), and Acetobacterium (Ravot et al., 2006; Mand et al., 2014), MIC may be only a minor contributor to the observed corrosion. The observation that passage of sample FBW6 through an 0.2 μm filter had little effect on the corrosion rate and that three separate measurements gave very similar results (0.44 ± 0.12 mm/year unfiltered, 0.55 ± 0.06 mm/year unfiltered, and 0.57 ± 0.02 mm/year filtered) indicates chemical corrosion. Similar studies on the corrosivity of Duvernay shale gas samples from another field implicated the biocide used in the high corrosion rate of 1.25 mm/year observed for one out of 16 samples (Sharma et al., 2017). In the present study we cannot comment on which chemical(s) may have been contributing to the observed high corrosion rates in some of the samples.
Hydrogenotrophic and acetotrophic methanogens in oil fields are involved in the anaerobic metabolism of oil components to methane and CO2 (Grigoryan and Voordouw, 2008; Jones et al., 2008; Berdugo-Clavijo and Gieg, 2014), as well as in corrosion of carbon steel (Enning and Garrelfs, 2014). However, methylotrophic methanogens may be major contributors to biogenic methane formation in subsurface environments that are rich in low-maturity kerogens (Strapoc et al., 2011) such as in shale oil and shale gas reservoirs. Methylotrophic methanogens such as Methanolobus, Methanosarcina, and Methanohalophilus are often found in shale oil and shale gas fields (Michimaru et al., 2009; Strapoc et al., 2011; Cluff et al., 2014; Katayama et al., 2014; Shimizu et al., 2015; Daly et al., 2016). Methanohalophilus and Halanaerobium form a syntrophy in which GB, used as an osmolyte by Methanohalophilus is metabolized by Halanaerobium to trimethylamine, which is then metabolized by Methanohalophilus to methane and ammonium (Daly et al., 2016; Borton et al., 2018). Microbial communities in samples from the Bakken shale oil field (An et al., 2017) had high proportions of Halanaerobium and Methanohalophilus, which were also found in many different shale gas formations (Mouser et al., 2016), including in samples from the Duvernay formations studied here (Tables 2, 3).
Several primary enrichments of methanogens using Bakken field samples inoculated with 20 mM TMA at 2.0 M NaCl showed on average formation of 52 mM methane and 22.5 mM ammonium (Figure 3). This agrees with the stoichiometry of methylotrophic methanogenesis of TMA (Table S4). Microbial community analyses of the primary TMA enrichments showed high proportions of Methanohalophilus, which thrived in the medium provided, and lower but still high fractions of Halanaerobium, which may have fed on GB or other metabolites produced by Methanohalophilus (Table S2) (Tsai et al., 1995; Cayol et al., 2002; Oren, 2008). Secondary enrichments with different methylated amines (MAs) or GB showed on average higher production of methane and ammonium with the MAs than with GB (Figure 4). The microbial community compositions of these secondary enrichments were again dominated by Methanohalophilus and Halanaerobium for all three of the samples tested (Table S4; Equations 1–3). Formation of methane and ammonium from GB therefore likely involved syntrophy of Halanaerobium and Methanohalophilus (Table S4, Equations 3, 4; Daly et al., 2016; Borton et al., 2018). Interestingly, members of the Sphingobacteriales can be closely associated with Halanaerobium and Methanohalophilus, possibly involved in the carbon cycle at extreme conditions though little are known about the metabolic pathways involved (Wang et al., 2017).
Methylotrophic methanogenesis can only produce ammonium concentrations of 10 to 30 mM, as observed in the Duvernay shale gas field samples (Table 1), if the MA and/or GB substrates are present at such high concentrations (Figures 3, 4). However, the GB concentration in shale gas samples was estimated to be only 8 μM (Borton et al., 2018), which is too little. Instead, ammonium may have been contributed by the shale formation. If MAs are present in flow-back waters or in shale oil or gas, then these can be converted to methane and ammonium by halophilic methylotrophic methanogens such as Methanohalophilus, which are found in shale gas and shale oil reservoirs.
In conclusion, analyses of samples from a Duvernay shale gas field suggested that high ammonium concentrations in initial flow back waters may be partially attributed to the fracturing process, e.g., by biotic conversion of methylamines used during the fracturing process. Osmolytes, such as GB, which stabilize a significant portion of the halophilic microbial community can be converted to methane and ammonium through syntrophic metabolism by Halanaerobium and Methanohalophilus (Daly et al., 2016; Borton et al., 2018). However, these are present at concentrations that are too low to contribute to these very high ammonium concentrations. The high corrosion rates observed for some field samples were not correlated with the presence of Desulfovibrio or of Halanaerobium as suggested elsewhere (Liang et al., 2016; Lipus et al., 2017). The high ammonium concentrations in late highly saline flow back waters most likely originated from the reservoir.
Author Contributions
BA did the experimental setup, data collection, interpretation, execution of the experiments, drafting, and revision of the manuscript. YS conducted the water chemistry analyses of the field samples. JV performed the DNA isolations. GV provided funding, supervision, conception of the work, and helped write the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by an NSERC Industrial Research Chair Award to GV, which was also supported by BP America Production Co., Baker Hughes Canada, Computer Modeling Group Limited, ConocoPhillips Company, Dow Microbial Control, Enbridge Inc., Enerplus Corporation, Intertek Commercial Microbiology, Oil Search (PNG) Limited, Shell Global Solutions International, Suncor Energy Inc., and Yara Norge AS, as well as by Alberta Innovates. We thank Matthew Fay, Shell Canada, for providing field information and sample collection and discussion.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenrg.2019.00023/full#supplementary-material
References
Ahn, T.-S., Kim, H., Kim, T.-S., Kim, S. B., Han, J.-H., Kang, H., et al. (2016). Flavobacterium paronense sp. nov., isolated from freshwater of an artificial vegetated island. Int. J. Syst. Evol. Microbiol. 66, 365–370. doi: 10.1099/ijsem.0.000727
Aminot, A., Kirkwood, D. S., and Kérouel, R. (1997). Determination of ammonia in seawater by the indophenol-blue method: evaluation of the ICES NUTS I/C 5 questionnaire. Mar. Chem. 56, 59–75. doi: 10.1016/S0304-4203(96)00080-1
An, B. A., Shen, Y., and Voordouw, G. (2017). Control of sulfide production in high salinity bakken shale oil reservoirs by halophilic bacteria reducing nitrate to nitrite. Front. Microbiol. 8:1164. doi: 10.3389/fmicb.2017.01164
Berdugo-Clavijo, C., and Gieg, L. M. (2014). Conversion of crude oil to methane by a microbial consortium enriched from oil reservoir production waters. Front. Microbiol. 5:197. doi: 10.3389/fmicb.2014.00197
Boone, D. R., Mathrani, I. M., Liu, Y., Menaia, J. A. G. F., Mah, R. A., and Boone, J. E. (1993). Isolation and characterization of Methanohalophilus portucalensis sp. nov. and DNA reassociation study of the genus methanohalophilus. Int. J. Syst. Bacteriol. 43, 430–437. doi: 10.1099/00207713-43-3-430
Borton, M. A., Hoyt, D. W., Roux, S., Daly, R. A., Welch, S. A., Nicora, C. D., et al. (2018). Coupled laboratory and field investigations resolve microbial interactions that underpin persistence in hydraulically fractured shales. Proc. Natl. Acad. Sci. U.S.A. 115, E6585–E6594. doi: 10.1073/pnas.1800155115
Bowers, K. J., and Wiegel, J. (2011). Temperature and pH optima of extremely halophilic archaea: a mini-review. Extremophiles 15, 119–128. doi: 10.1007/s00792-010-0347-y
Brittingham, M. C., Maloney, K. O., Farag, A. M., Harper, D. D., and Bowen, Z. H. (2014). Ecological risks of shale oil and gas development to wildlife, aquatic resources and their habitats. Environ. Sci. Technol. 48, 11034–11047. doi: 10.1021/es5020482
Cayol, J. L., Fardeau, M. L., Garcia, J. L., and Ollivier, B. (2002). Evidence of interspecies hydrogen transfer from glycerol in saline environments. Extremophiles 6, 131–134. doi: 10.1007/s007920100229
Cluff, M. A., Hartsock, A., MacRae, J. D., Carter, K., and Mouser, P. J. (2014). Temporal changes in microbial ecology and geochemistry in produced water from hydraulically fractured Marcellus shale gas wells. Environ. Sci. Technol. 48, 6508–6517. doi: 10.1021/es501173p
Daly, R. A., Borton, M. A., Wilkins, M. J., Hoyt, D. W., Kountz, D. J., Wolfe, R. A., et al. (2016). Microbial metabolisms in a 2.5-km-deep ecosystem created by hydraulic fracturing in shales. Nat. Microbiol. 1:16146. doi: 10.1038/nmicrobiol.2016.146
Denger, K., Ludwig, W., Warthmann, R., and Schink, B. (2002). Anaerophaga thermohalophila gen. nov., sp. nov., a moderately thermohalophilic, strictly anaerobic fermentative bacterium. Int. J. Syst. Evol. Microbiol. 52, 173–178. doi: 10.1099/00207713-52-1-173
Dong, X., Kleiner, M., Sharp, C. E., Thorson, E., Li, C., Liu, D., et al. (2017). Fast and simple analysis of miseq amplicon sequencing data with metaamp. Front. Microbiol. 8:1461. doi: 10.3389/fmicb.2017.01461
Enning, D., and Garrelfs, J. (2014). Corrosion of iron by sulfate-reducing bacteria: new views of an old problem. Appl. Environ. Microbiol. 80, 1226–1236. doi: 10.1128/AEM.02848-13
Feio, M. J., Zinkevich, V., Beech, I. B., Llobet-Brossa, E., Eaton, P., Schmitt, J., et al. (2004). Desulfovibrio alaskensis sp. nov., a sulphate-reducing bacterium from a soured oil reservoir. Int. J. Syst. Evol. Microbiol. 54, 1747–1752. doi: 10.1099/ijs.0.63118-0
Fida, T. T., Chen, C., Okpala, G., and Voordouw, G. (2016). Implications of limited thermophilicity of nitrite reduction for control of sulfide production in oil reservoirs. Appl. Environ. Microbiol. 82, 4190–4199. doi: 10.1128/AEM.00599-16
Finster, K. W., and Kjeldsen, K. U. (2010). Desulfovibrio oceani subsp. oceani sp. nov., subsp. nov. and Desulfovibrio oceani subsp. galateae subsp. nov., novel sulfate-reducing bacteria isolated from the oxygen minimum zone off the coast of Peru. Antonie van Leeuwenhoek 97, 221–229. doi: 10.1007/s10482-009-9403-y
Gaspar, J., Mathieu, J., Yang, Y., Tomson, R., Leyris, J. D., Gregory, K. B., et al. (2014). Microbial dynamics and control in shale gas production. Environ. Sci. Technol. Lett. 1, 465–473. doi: 10.1021/ez5003242
Greene, A. C., Patel, B. K. C., and Yacob, S. (2009). Geoalkalibacter subterraneus sp. nov., an anaerobic Fe(III)- and Mn(IV)-reducing bacterium from a petroleum reservoir, and emended descriptions of the family Desulfuromonadaceae and the genus Geoalkalibacter. Int. J. Syst. Evolu. Microbiol. 59, 781–785. doi: 10.1099/ijs.0.001537-0
Grigoryan, A., and Voordouw, G. (2008). Microbiology to help solve our energy needs: methanogenesis from oil and the impact of nitrate on the oil-field sulfur cycle. Ann. N.Y. Acad. Sci. 1125, 345–352. doi: 10.1196/annals.1419.004
Harkness, J. S., Dwyer, G. S., Warner, N. R., Parker, K. M., Mitch, W. A., and Vengosh, A. (2015). Iodide, bromide, and ammonium in hydraulic fracturing and oil and gas wastewaters: environmental implications. Environ. Sci. Technol. 49, 1955–1963. doi: 10.1021/es504654n
Horton, D., and Jones, A. (1998). US Patent 5771971: Clay stabilizing agent and a method of use in subterranean formation to inhibit clay swelling.
Jones, D. M., Head, I. M., Gray, N. D., Adams, J. J., Rowan, A. K., Aitken, C. M., et al. (2008). Crude-oil biodegradation via methanogenesis in subsurface petroleum reservoirs. Nature 451, 176–180. doi: 10.1038/nature06484
Kasalický, V., Jezbera, J., Hahn, M. W., and Šimek, K. (2013). The diversity of the Limnohabitans Genus, an important group of freshwater Bacterioplankton, by characterization of 35 isolated strains. PLoS ONE 8:e58209. doi: 10.1371/journal.pone.0058209
Katayama, T., Yoshioka, H., Mochimaru, H., Meng, X. Y., Muramoto, Y., Usami, J., et al. (2014). Methanohalophilus levihalophilus sp. nov., a slightly halophilic, methylotrophic methanogen isolated from natural gas-bearing deep aquifers, and emended description of the genus Methanohalophilus. Int. J. Syst. Evolu. Microbiol. 64, 2089–2093. doi: 10.1099/ijs.0.063677-0
King, G. M. (1984). Metabolism of trimethylamine, choline, and glycine betaine by sulfate-reducing and methanogenic bacteria in marine sediments. Appli. Environ. Microbiol. 48, 719–725.
Krieg, N. R., Staley, J. T., Brown, D. R., Hedlund, B. P., Paster, B. J., Ward, N. L., et al. (2009). “Phylum XIV. Bacteroidetes phyl. nov.,” Systematic Bacteriology, eds W. B. Whitman (New York, NY: Springer), 25–469.
Lai, S. J., and Lai, M. C. (2011). Characterization and regulation of the osmolyte betaine synthesizing enzymes GSMT and SDMT from halophilic methanogen Methanohalophilus portucalensis. PLoS ONE 6:e25090. doi: 10.1371/journal.pone.0025090
Liang, R., Davidova, I. A., Marks, C. R., Stamps, B. W., Harriman, B. H., Stevenson, B. S., et al. (2016). Metabolic capability of a predominant Halanaerobium sp. in hydraulically fractured gas wells and its implication in pipeline corrosion. Front. Microbiol. 7:988. doi: 10.3389/fmicb.2016.00988
Lipus, D., Vikram, A., Ross, D., Bain, D., Gulliver, D., Hammack, R., et al. (2017). Predominance and Metabolic Potential of Halanaerobium spp. in produced water from hydraulically fractured marcellus shale wells. Appl. Environ. Microbiol. 83:02659–16. doi: 10.1128/AEM.02659-16
Mand, J., Park, H. S., Jack, T. R., and Voordouw, G. (2014). The role of acetogens in microbially influenced corrosion of steel. Front. Microbiol. 5:268. doi: 10.3389/fmicb.2014.00268
Mesbah, N. M., and Wiegel, J. (2012). Life under multiple extreme conditions: diversity and physiology of the halophilic alkalithermophiles. Appl. Environ. Microbiol. 78, 4074–4082. doi: 10.1128/AEM.00050-12
Michimaru, H., Tamaki, H., Hanada, S., Imachi, H., Nakamura, K., Sakata, S., et al. (2009). Methanolobus profundi sp. nov., a methylotrophic methanogen isolated from deep subsurface sediments in a natural gas field. Int. J. Syst. Evol. Microbiol. 59, 714–718. doi: 10.1099/ijs.0.001677-0
Mouser, P. J., Borton, M., Darrah, T. H., Hartsock, A., and Wrighton, K. C. (2016). Hydraulic fracturing offers view of microbial life in the deep terrestrial subsurface. FEMS Microbiol. Ecol. 92, 1–18. doi: 10.1093/femsec/fiw166
Oren, A. (1990). Formation and breakdown of glycine betaine and trimethylamine in hypersaline environments. Antonie van Leeuwenhoek 58, 291–298. doi: 10.1007/BF00399342
Oren, A. (2001). The bioenergetic basis for the decrease in metabolic diversity at increasing salt concentrations: implications for the functioning of salt lake ecosystems. Hydrobiologia 466, 61–72. doi: 10.1023/A:1014557116838
Oren, A. (2008). Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Syst. 4:2. doi: 10.1186/1746-1448-4-2
Oren, A. (2013). Life at high salt concentrations, intracellular KCl concentrations, and acidic proteomes. Front. Microbiol. 4:315. doi: 10.3389/fmicb.2013.00315
Ravot, G., Magot, M., Ollivier, B., Patel, B. K., Ageron, E., Grimont, P. A., et al. (2006). Haloanaerobium congolense sp. nov., an anaerobic, moderately halophilic, thiosulfate- and sulfur-reducing bacterium from an African oil field. FEMS Microbiol. Lett. 147, 81–88. doi: 10.1111/j.1574-6968.1997.tb10224.x
Roberts, M. F., Galinski, E., Martin, D., Ciulla, R., Roberts, M., Roberts, M., et al. (2005). Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst. 1:5. doi: 10.1186/1746-1448-1-5
Sharma, M., An, D., Liu, T., Pinnock, T., Cheng, F., and Voordouw, G. (2017). Biocide-mediated corrosion of coiled tubing. PLoS ONE 12:e0181934. doi: 10.1371/journal.pone.0181934
Shimizu, S., Ueno, A., Naganuma, T., and Kaneko, K. (2015). Methanosarcina subterranea sp. nov., a methanogenic archaeon isolated from a deep subsurface diatomaceous shale formation. Int. J. Syst. Evolu. Microbiol. 65, 1167–1171. doi: 10.1099/ijs.0.000072
Shrestha, N., Chilkoor, G., Wilder, J., Gadhamshetty, V., and Stone, J. J. (2017). Potential water resource impacts of hydraulic fracturing from unconventional oil production in the Bakken shale. Water Res. 108, 1–24. doi: 10.1016/j.watres.2016.11.006
Stan-Lotter, H., and Fendrihan, S. (2012). Adaption of Microbial Life to Environmental Extremes-Novel Research Results and Application. NewYork, NY: Springer Wien.
Strapoc, D., Ashby, M., Wood, L., Levinson, R., and Huizinga, B. (2011). “Applied microbiology and molecular biology in oilfield systems,” Applied Microbiology and Molecular Biology in Oilfield Systems, eds C. Whitby and T. L. Skovhus (Dordrecht: Springer), 77–84.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Trüper, H. G., and Schlegel, H. G. (1964). Sulphur metabolism in Thiorhodaceae, I. Quantitative measurements on growing cells of Chromatium okenii. Antonie van Leeuwenhoek 30, 225–238. doi: 10.1007/BF02046728
Tsai, C. R., Garcia, J. L., Patel, B. K., Cayol, J. L., Baresi, L., and Mah, R. A. (1995). Haloanaerobium alcaliphilum sp. nov., an anaerobic moderate halophile from the sediments of Great Salt Lake, Utah. Int. J. Syst. Bacteriol. 45, 301–307. doi: 10.1099/00207713-45-2-301
Tucker, Y. T., Kotcon, J., and Mroz, T. (2015). Methanogenic archaea in marcellus shale: a possible mechanism for enhanced gas recovery in unconventional shale resources. Environ. Sci. Technol. 49, 7048–7055. doi: 10.1021/acs.est.5b00765
Urbieta, M. S., Donati, E. R., Chan, K.-G., Shahar, S., Sin, L. L., and Goh, K. M. (2015). Thermophiles in the genomic era: biodiversity, science, and applications. Biotechnol. Adv. 33, 633–647. doi: 10.1016/j.biotechadv.2015.04.007
Voordouw, G., Menon, P., Pinnock, T., Sharma, M., Shen, Y., Venturelli, A., et al. (2016). Use of homogeneously-sized carbon steel ball bearings to study microbially-influenced corrosion in oil field samples. Front. Microbiol. 7:351. doi: 10.3389/fmicb.2016.00351
Voordouw, G., Niviere, V., Ferris, F. G., Fedorak, P. M., and Westlake, D. W. (1990). Distribution of hydrogenase genes in Desulfovibrio spp. and their use in identification of species from the oil field environment. Appli. Environ. Microbiol. 56, 3748–54.
Vreeland, R. H., Litchfield, C. D., Martin, E. L., and Elliot, E. (1980). Halomonas elongata, a new genus and species of extremely salt-tolerant bacteria. Int. J. Syst. Bacteriol. 30, 485–495. doi: 10.1099/00207713-30-2-485
Wang, X., Cao, A., Zhao, G., Zhou, C., and Xu, R. (2017). Microbial community structure and diversity in a municipal solid waste landfill. Waste Manage. 66, 79–87. doi: 10.1016/j.wasman.2017.04.023
Keywords: methanogenesis, methylotrophic methanogenesis, halophilic, shale gas, sulfate reducing bacteria (SRB), fermentative bacteria, corrosion, ammonium
Citation: An BA, Shen Y, Voordouw J and Voordouw G (2019) Halophilic Methylotrophic Methanogens May Contribute to the High Ammonium Concentrations Found in Shale Oil and Shale Gas Reservoirs. Front. Energy Res. 7:23. doi: 10.3389/fenrg.2019.00023
Received: 01 October 2018; Accepted: 14 February 2019;
Published: 07 March 2019.
Edited by:
Claire Dumas, Institut National de la Recherche Agronomique (INRA), FranceReviewed by:
Qaisar Mahmood, COMSATS University Islamabad, PakistanMohanakrishna Gunda, Qatar University, Qatar
Jorge Gonzalez-Estrella, University of New Mexico, United States
Copyright © 2019 An, Shen, Voordouw and Voordouw. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Biwen Annie An, biwen.an@bam.de
 Biwen Annie An
Biwen Annie An Yin Shen
Yin Shen Johanna Voordouw2
Johanna Voordouw2  Gerrit Voordouw
Gerrit Voordouw