Protein Serine/Threonine Phosphatase Type 2C of Leishmania mexicana
- 1Unidad de Investigación UNAM-INC, División de Investigación, Facultad de Medicina, Instituto Nacional de Cardiología Ignacio Chávez., Ciudad de México, Mexico
- 2Ciencias Experimentales, Escuela Nacional Colegio de Ciencias y Humanidades Plantel, Naucalpan, Mexico
- 3Departamento de Bioquímica y Biología Estructural, Instituto de Fisiología Celular, Universidad Nacional Autónoma de México, Ciudad de México, Mexico
- 4Departamento de Bioquímica, Centro de Investigación y Estudios Avanzados (CINVESTAV-IPN), Ciudad de México, Mexico
- 5 División de Ingeniería en Biotecnología, Universidad Politécnica del Valle de Toluca, Almoloya de Juárez, Mexico
- 6Escuela Superior de Medicina, Instituto Politécnico Nacional, Ciudad de México, Mexico
- 7Unidad de Investigación en Medicina Experimental, Facultad de Medicina, Universidad Nacional Autónoma de México, Hospital General de México, Ciudad de México, Mexico
Protein phosphorylation and dephosphorylation are increasingly recognized as important processes for regulating multiple physiological mechanisms. Phosphorylation is carried out by protein kinases and dephosphorylation by protein phosphatases. Phosphoprotein phosphatases (PPPs), one of three families of protein serine/threonine phosphatases, have great structural diversity and are involved in regulating many cell functions. PP2C, a type of PPP, is found in Leishmania, a dimorphic protozoan parasite and the causal agent of leishmaniasis. The aim of this study was to clone, purify, biochemically characterize and quantify the expression of PP2C in Leishmania mexicana (LmxPP2C). Recombinant LmxPP2C dephosphorylated a specific threonine (with optimal activity at pH 8) in the presence of the manganese divalent cation (Mn+2). LmxPP2C activity was inhibited by sanguinarine (a specific inhibitor) but was unaffected by protein tyrosine phosphatase inhibitors. Western blot analysis indicated that anti-LmxPP2C antibodies recognized a molecule of 45.2 kDa. Transmission electron microscopy with immunodetection localized LmxPP2C in the flagellar pocket and flagellum of promastigotes but showed poor staining in amastigotes. Interestingly, LmxPP2C belongs to the ortholog group OG6_142542, which contains only protozoa of the family Trypanosomatidae. This suggests a specific function of the enzyme in the flagellar pocket of these microorganisms.
Introduction
Phosphorylation is the process of adding a phosphate group to a molecule and dephosphorylation the process of removing the same. Addition of phosphates is carried out by protein kinases and removal by protein phosphatases, generally occurring on tyrosine, serine and threonine residues (Cohen, 2002; Wang et al., 2008). There are protein tyrosine phosphatases and protein serine/threonine phosphatases.
The latter type has three families of molecules, one of them being phosphoprotein phosphatases (PPPs), which have great structural and functional diversity. Apart from being responsible for removing the phosphate group from molecules, they regulate a variety of cell functions. The PPP family consists of PP1, PP2A and PP2B phosphatases as well as metal- (Mg2+- or Mn2+-) dependent protein phosphatases (PPMs). Among PPMs are protein phosphatase type 2C (PP2C) and pyruvate dehydrogenase phosphatase (PDP) (Cohen, 2002; Wang et al., 2008; Virshup and Shenolikar, 2009). Of the 199 phosphatases in the human phosphatome, only 13 are PPMs, but these have a wide range of activity, from regulation of the cell cycle to metabolism and apoptosis (Das et al., 1996; Andreeva and Kutuzov, 2004; Schweighofer et al., 2004; Lammers and Lavi, 2007; Moorhead et al., 2007).
The evidence of PP2Cs in archaea, bacteria, fungi, plants, animals and parasites shows an early evolutionary emergence. Since they evolved from prokaryotes to multicellular eukaryotes, PP2C sequences are very diversified (Fuchs et al., 2013). PP2C enzymes have been cloned and characterized in Leishmania chagasi (LcPP2C), Leishmania major (LmPP2C) and Leishmania donovani (LdPP2C). To our knowledge, however, there is no information in the literature about the function of this enzyme, either in the parasite or cells of the host (Burns et al., 1993; Escalona-Montaño et al., 2017; Jakkula et al., 2018).
The genome of L. major was discovered in 2005 (Ivens et al., 2005), and only 2 years later the first analysis of the catalytic domains of protein phosphatases in trypanosomatids was published. A total of 88 protein phosphatases were identified in L. major, including 58 serine/threonine phosphatases. Of the latter, 30 are in the PPP family, 13 dephosphorylate the carboxy-terminal domain of RNA polymerase II (F-cell production eukaryotic-like phosphatases), and 15 are PP2C eukaryotic-like phosphatases (Brenchley et al., 2007). The function of these PP2Cs is still unknown.
Our group recently reported the cloning, purification, biochemical characterization and expression level of a protein of L. major, concluding that it is a PP2C of threonine/serine protein phosphatases. Additionally, we found PP2C in different Leishmania species and described its ultrastructural location in promastigotes, mainly along the flagella (Escalona-Montaño et al., 2017).
Leishmania is a dimorphic protozoan parasite and the causal agent of leishmaniasis, a set of neglected tropical diseases (NTDs). According to epidemiological studies, 12 million people are infected worldwide, with 2 million new cases each year (Alvar et al., 2012). Approximately 350 million people are currently at risk of contracting leishmaniasis, mostly in developing countries (Aguirre-Garcia et al., 2018). Leishmania mexicana, endemic to the south-eastern region of Mexico, induces various clinical forms of cutaneous leishmaniasis, classified as localized and diffuse leishmaniasis.
Since leishmaniasis is a significant public health problem in México, it is important to determine which enzymes of L. mexicana participate in the disease-related signaling pathways. Those showing such activity could possibly be used as a drug target, an immunomodulatory molecule, or a diagnostic marker. The aim of the present study was to clone, purify, biochemically characterize, the PP2C in L. mexicana. PP2Cs were also identified in other Leishmania species. Based on immunodetection with a transmission electron microscope (TEM), the enzymes were localized in the flagellum and flagellar pocket of promastigotes and amastigotes of L. mexicana.
Materials and Methods
Culture of L. mexicana Promastigotes
Promastigotes of L. (V.) panamensis MHOM/PA/71/LS94, L. (V.) brazilienzis MHOM/BR/75/M2903, L. (L.) venezuelensis MHOM/VE/80/PMH3, L. (V.) donovani, L. (L.) amazonensis IFLA/BR/67/PH8, and L. (L.) mexicana MNYC/BZ/62/M379 (a generous gift from Dr. Nancy Sarabia, Centro Internacional de Entrenamiento e Investigación Médica in Calí, Colombia (CIDEIM)) were cultivated at 26°C in RPMI 1640 medium containing penicillin (100 units/mL), streptomycin (100μg/mL), and 10% fetal bovine serum. All reagents were purchased from Gibco (Invitrogen Corp, Carlsbad, CA, USA). The cultured promastigotes were washed twice with phosphate-buffered saline (PBS) at pH 7.5 and centrifuged for 10 min at 2,500 × g at room temperature (RT).
Extraction of DNA and Polymerase Chain Reaction Amplification of the Gene for PP2C of L. mexicana
Total DNA was extracted from L. mexicana promastigotes with TRIZOL. Polymerase chain reaction (PCR) was performed with 1U Taq polymerase (Invitrogen). The optimal magnesium concentration was established by means of testing concentrations in the range of 0-4 mM. Two oligonucleotides were constructed based on alignments of known sequences of PP2C. The sequences for the sense and the antisense oligonucleotides are 5 ′ATGGGSATTCCWCTTCCSA 3 ′ and 5 ′ CCTATTTTCTTGCATTGTTCACT 3 ′, respectively (where S = C/G and W= A/T). Genomic DNA was used as a template under the following conditions: 1 cycle of 5 min at 94°C; 40 cycles of 30 s at 94°C, 1 cycle of 30 s at 51.4°C, 1 cycle of 1 min at 72°C, and 1 cycle of 10 min at 72°C.
Cloning of the Gene and Sequence Analysis
The amplified gene was ligated to the Topo TA Vector with a cloning kit (Thermo Fisher Scientific) and competent E. coli cells (DH5 alpha strain) were transformed. The gene was completely sequenced.
Expression of the PP2C of L. mexicana
The gene was amplified with oligonucleotides containing NdeI and HindIII restriction sites 5’ GGGAATTCCATATGGGCATTCC 3’ and 5 ‘CCCAAGCTTTCACTGCGTCTG 3’, respectively. The released gene was treated with NdeI and Hind III and then ligated to the pET-23b vector (Novagen) expression plasmid, which encodes for proteins that have a carboxyl-terminal His6-Tag. The expression of the PP2C of L. mexicana was quantified in the Rosetta-gami B (DE3) strain of E. coli cells (Novagen), as previously described with some modifications. Expression was induced in bacterial cultures at A 600nm= 0.8 by adding 1.0 mM isopropyl-beta-D-thiogalactoside (IPTG) and culturing the cells for an additional 3 h. Upon completion of this time, the cells were harvested by centrifugation and processed immediately or frozen at -70 to await further use (Escalona-Montaño et al., 2017)
Purification of Recombinant PP2C of L. mexicana
The LmxPP2C protein was purified as reported [11], with some modifications (patent in Mexico, MX/a/2020/011271). Cell cultures were centrifuged in volumes of 100 mL and suspended in 50 mL lysis buffer (50 mM TrisHCl at pH 8, 300 mM NaCl, 1 mM benzamidine, 100µM leupeptin, 2 µg/mL aprotinin and 5 mM imidazole). The suspension was sonicated on a ModelVCX 650 Ultrasonic processor (Ultrasonics, Inc.) five times at 30% amplitude for 60 s intervals, with resting periods of 1 min between each interval. The homogenate was centrifuged at 21,000 × g for one hour at 4°C to obtain the supernatant containing the soluble protein. The supernatant was loaded onto an Ni-charged column previously equilibrated with binding buffer (50 mM Tris-HCl at pH 8, 300 mM NaCl and 5 mM imidazole). The recombinant protein (LmxPP2C) was purified and eluted with an elution buffer (50 mM Tris-HCl at pH 8, 300 mM NaCl and 50-500 mM imidazole) and the protein concentration was quantified by the Bradford method (Bradford et al., 1976).
Production of Polyclonal Antiserum
Anti-LmxPP2C antibodies were generated in rabbits, following the procedure established by Montfort et al. (1994). Briefly, rabbits were injected intramuscularly with 135 µg of recombinant LmxPP2C emulsified in complete Freund’s adjuvant, and the same procedure was repeated 2 weeks later without adjuvant. The immunization was based on two weekly intramuscular injections, after which the animals were bled and antiserum was separated by centrifugation and stored at -20°C. Rabbits were housed at the animal facility of the Research Unit of Experimental Medicine of the Medicine Faculty, UNAM, and handled in accordance with the National Ethical Guidelines for Animal Health NOM-062-ZOO-1999 and the guidelines recommended for animal care by the institutional Ethics in Research Committee.
Phosphatase Activity Assays
p-NPP substrate. Acid phosphatase activity was determined as described by Dissing et al. (1979). Recombinant LmxPP2C (25 µg) was incubated in the following buffers at 37°C for 60 min: sodium acetate buffer (pH 4.5-5.5), 50 mM MES (pH 6.0 and 6.5), 50 mM HEPES (pH 7.0, 7.5 and 8.0), 50 mM TRIS (pH 8.5-9), or 50 mM CAPS (pH 10-11) added to 10 mM MgCl2 and 10 mM p-nitrophenyl phosphate [p-NPP] in a final volume of 100 μL. Upon completion of this time, the reaction was stopped with 20 μL of 2 N NaOH. Divalent cations (10 mM MgCl2 or MnCl2) in 50mM HEPES (pH 8) were added and the absorbance was read at 405 nm on a microtiter plate reader, as previously reported with some modifications (Escalona-Montaño et al., 2017).
Phosphopeptides. Tyrosine and serine/threonine phosphatase activity was assessed by employing a non-radioactive tyrosine phosphatase assay system (Promega). The release of inorganic phosphate (Pi) was monitored by measuring the absorbance of the molybdate-malachite green-phosphate complex. Briefly, 1.2 µg of recombinant LmxPP2C was incubated in a total volume of 100 µL of assay buffer containing 50 mM HEPES at pH 8.0 plus 10 mM MgCl2. The reaction was started by adding 50 µM Tyr phosphopeptide-1 substrate [END (pY) INASL] and 50 µM Thr [RRA (pT)VA]. After 30 min at RT, the reaction was stopped with 50 µL molybdate dye/additive mixture. The optical density of the samples was read at 630 nm, using a curve of phosphate density as the standard (Escalona-Montaño et al., 2017).
Effect of Phosphatase Inhibitors on Recombinant LmxPP2C
The activity of recombinant LmxPP2C (1.2 µg) was evaluated in the presence of specific protein tyrosine phosphatase (PTP) inhibitors: 200 µM sodium orthovanadate, 100 µM sodium tungstate and 50 µM sodium pervanadate. Additionally, serine/threonine phosphatase inhibitors were tested: 5 nM calyculin, 1 µM okadaic acid and 20 µM sanguinarine, the latter a specific inhibitor of PP2C. For the inhibition assays, 100 µL of the reaction mixture was pre-incubated at RT for 15 min before adding the p-NPP substrate (all reagents from Sigma-Aldrich, St. Louis, MO, USA). Subsequently, the solution was incubated at 37°C for 60 min and then the reaction was stopped with 20 µL of 2 N NaOH and the absorbance was read at 405 nm on a microtiter plate reader.
SDS-PAGE and Western Blot
The His-Tag motif was found in recombinant LmxPP2C. The total extract (TE) of bacteria was analyzed by means of sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS-PAGE) in 10% acrylamide gels, and then electrotransferred to nitrocellulose membranes. The blots were incubated with a polyclonal HRP anti-His6-Tag antibody at a 1:1000 dilution in TBS-T (200 mM Tris-HCl, 150mM NaCl and 0.005% Tween-20) and washed four times every 10 min with TBS-T. The bands were detected with enhanced chemiluminescent substrate (Super-Signal West Pico Chemiluminescent Substrate, Pierce, Rockford IL, USA), according to the manufacturer’s instructions.
Identification of PP2C in Total Extracts of Different Leishmania Species
Promastigotes of L. (V.) panamensis MHOM/PA/71/LS94, L. (V.) brazilienzis MHOM/BR/75/M2903, L. (L.) venezuelensis MHOM/VE/80/PMH3, L. (V.) donovani, L. (L.) amazonensis IFLA/BR/67/PH8, and L. (L.) mexicana MNYC/BZ/62/M379 were harvested after centrifugation at 2,000 × g for 10 min and washing with PBS three times. The pellet containing the parasites was suspended in cold lysis buffer (10 mM imidazole at pH 7.2, 2 μg mL-1 leupeptin, 10 μg mL-1 aprotinin and 2mM benzamidine) and sonicated to obtain the TE. Ten μg of TE from each Leishmania species and 1 µg of LmxPP2C were evaluated with electrophoresis (SDS-PAGE) in 10% acrylamide gels and then electrotransferred onto membranes of low fluorescence. The membranes were blocked with Li-Cor blocking buffer (Lincoln, NE, USA) at RT for 30 min and incubated with antibodies against LmxPP2C using a dilution of 1:1000 in 1% Tween Li-Cor blocking buffer overnight at 4°C. The membranes were washed with PBS-Tween and incubated with a secondary antibody, IRDye 680LT goat anti-rabbit IgG (926-68021, LI-COR Biosciences), at a dilution 1/10000 for 1 h under gentle shaking. The membranes were washed three times with PBS 1X and the proteins were examined on an Odyssey Infrared Imaging system (Li-Cor, Lincoln, NE, USA), according to the manufacturer´s instructions.
Phylogenetic Analysis of the Ortholog Group of LmxPP2C
The ortholog group of L. mexicana PP2C (LmxM.25.0750) was determined with OrthoMCL DB (Li et al., 2003). The phylogenetic tree and branch support of all aligned sequences of PP2C orthologs were constructed with MEGA 6 (multiple sequence alignment) (Tamura et al., 2013). The maximum likelihood method was carried out with an LG model and 5 categories of gamma distribution with invariant sites to find the best-fitting model among those tested. Bootstrap values were based on 100 replicates to estimate support for the nodes of the maximum likelihood tree. The final bootstrapped dendogram was visualized on the Interactive Tree of Life (iTOL) v3 (Letunic and Bork, 2016).
Multiple Sequence Alignment
L. mexicana and L. major PP2C sequences (LmxM.25.0750 and LmF.25.0750, respectively) were retrieved from the TriTrypDB database (Aslett et al., 2010). The human sequence NP_066283.1 (PPM1A) was retrieved from the NCBI reference sequence and was used as a seed for multiple sequence analysis. Full-length PP2C protein sequences were aligned on MAFFT v7 (Katoh and Standley, 2013) with L-INS-i (a very slow iterative refinement method recommended for <200 sequences with one conserved domain and long gaps). The final alignment was edited by hand considering the 11 motifs described by Bork et al. (1996), utilizing the alignment explorer of MEGA 6 software (Tamura et al., 2013). Alignment confidence was assessed with the Guidance2 server (Sela et al., 2015).
Prediction and Structural Alignment of the 3D Model for Leishmania mexicana PP2C
The full-length PP2C amino acid sequences from L. mexicana and L. major were retrieved from the TriTrypDB database with the gene IDs LmxF.25.0750 and LmF.25.0750, respectively. With each sequence, a 3D model was predicted on the I-TASSER server (Zhang, 2008). The final models were selected on I-TASSER, clustering all the decoys on the SPICKER program based on with their pair-wise structure similarity. Up to five models are returned by the program, corresponding to the five largest clusters of structures. The confidence of each model is quantitatively measured by a C-score, calculated in accordance with the significance of threading template alignments and the convergence parameters of the structure assembly simulations. The C-score is typically in the range of [-5, 2], where a C-score of a higher value indicates a model with a greater degree of confidence (Zhang, 2008). The Protein Data Bank (PDB) files of the predicted models were downloaded and the structural alignment to match all structures was performed in the UCSF Chimera program (Pettersen et al., 2004). The crystal structure of human 1a6q was downloaded from the PDB data base.
Distribution of PP2C in Promastigotes and Amastigotes of L. mexicana by Immune Electron Microscopy
The distribution of PP2C in promastigotes and amastigotes of L. mexicana was determined by immune electron microscopy (IEM), as previously reported (Escalona-Montaño et al., 2016). Briefly, promastigotes and amastigotes of L. mexicana were washed with PBS and fixed with 4% paraformaldehyde and 0.1% glutaraldehyde in PBS at RT for 1 h. Parasites were gradually dehydrated in ethanol, embedded in LR White resin (London Resin, Polysciences, Inc., Warrington, PA, USA), and then left overnight to polymerize in gelatin capsules under UV light at 4°C. Thin sections were cut in an Ultracut E ultra-microtome (Reichert Jung, Austria), mounted on Formvar-covered nickel grids, and incubated with a rabbit antibody against LmxPP2C diluted in PBS-T overnight at 4°C. Grids were washed with PBS-T and incubated with a goat anti-rabbit polyclonal antibody coupled to 10 nm gold particles at RT for 2 h (Zymed, Thermo Scientific, PA, USA). After thorough washings in PBS and distilled water, sections were stained with 2% uranyl acetate and a saturated solution of lead citrate before being viewed on a TEM (JEOL 1400x, JEOL Ltd, Japan). As the negative control, sections were incubated with pre-immune rabbit serum diluted in PBS-T and then with the secondary antibody coupled to gold particles. To recognize the subcellular structures detected by the antibodies, promastigotes and amastigotes were processed in parallel to preserve their ultrastructure, as previously described [28]. Briefly, parasites were fixed in 2.5% glutaraldehyde for 1 h, rinsed with PBS, post-fixed in 1% OsO4 at 4°C, rinsed again, gradually dehydrated in ethanol, and finally embedded in Spurr resin. Thin sections were cut on an Ultracut E ultramicrotome and stained with uranyl acetate and lead citrate. Samples were examined on a TEM at 80 keV. Digital images were taken and processed on Adobe Photoshop software (USA).
Statistical Analysis
All data is expressed as the mean ± SEM (standard error of the mean). The graph portrays the mean value of three independent assays.
Results
Cloning and Purification of PP2C of L. mexicana
The amplified LmxPP2C gene was cloned in the pET-23b plasmid to verify that the E. coli strain encoded a protein with the characteristics of PP2C phosphatase. The bacterial strain was induced and underwent cell fractionation, from which the TE, cytosolic fraction (CF) and the membrane fraction (MF) were obtained. The three extracts were evaluated on SDS-PAGE gel stained with Coomassie blue (Figure 1A). Various proteins of diverse molecular weights can be observed in lanes 2 (TE), 3 (CF) and 4 (MF), including an enriched molecule of 45.2 kDa in lanes 2 and 3 but not in lane 4. Hence, the protein phosphatase was found in the CF.
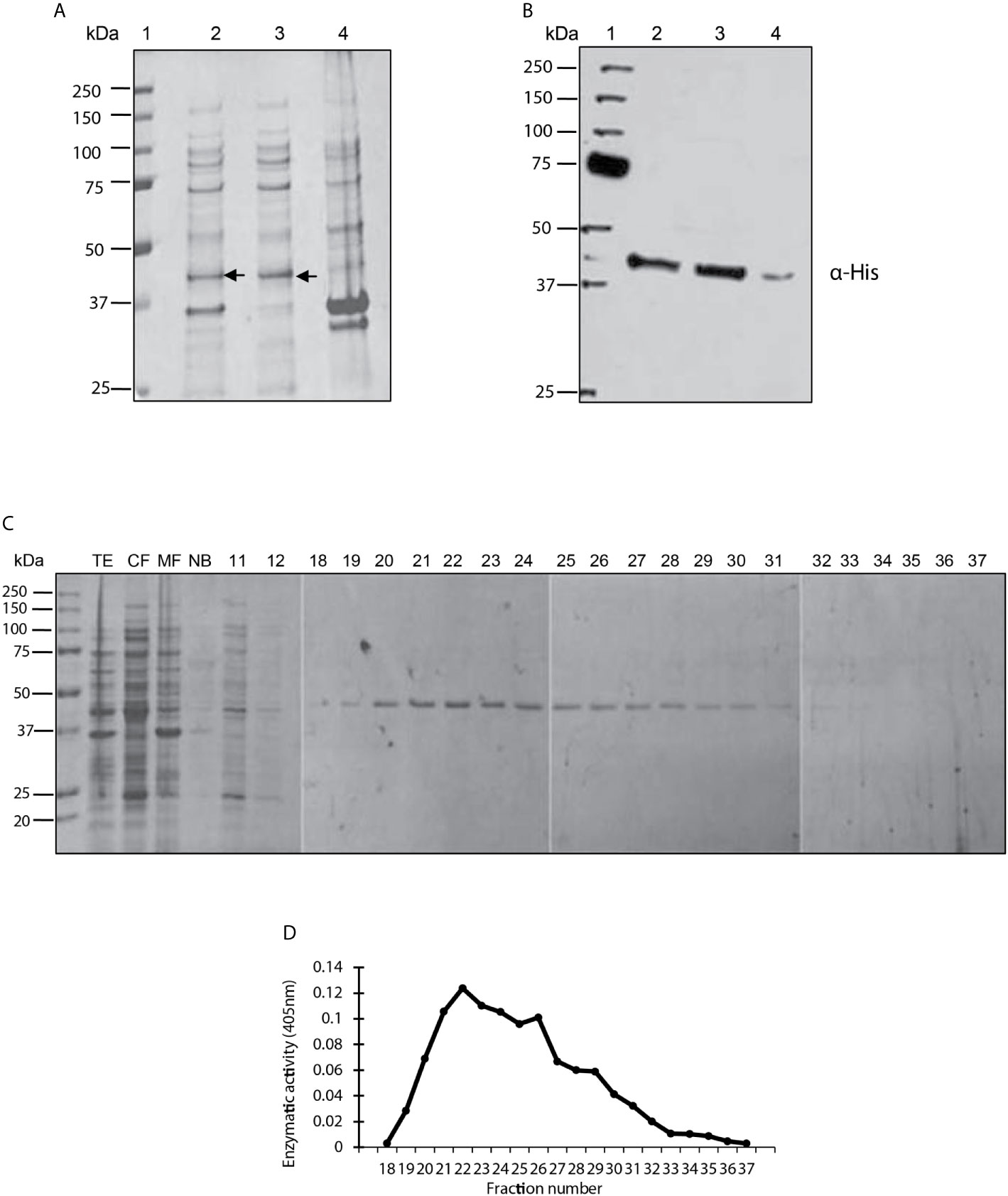
Figure 1 Purification, identification and analysis of enzymatic activity of the PP2C of Leishmania mexicana. (A) Expression of PP2C of L. mexicana in different subcellular fractions of Escherichia coli cells. Molecular weight marker (line 1); total extract prepared from the E. coli codon plus, induced with IPTG (line 2); cytosolic fraction (line 3); and membrane fraction (line 4). The three extracts were evaluated on SDS-PAGE gel stained with Coomassie blue. (B) Western blot analysis of the same sample used in molecular weight marker (line 1); total extract prepared from the E. coli codon, induced with IPTG (line 2); cytosolic fraction (line 3); and membrane fraction (line 4). The C-terminal His-Tag was detected with a monoclonal antibody. (C) Purification steps, utilizing SDS-PAGE gel stained with Coomassie blue dye. The cytosolic fraction (CF) was applied to a Ni2+–NTA resin and the bound material eluted with an imidazole gradient. TE, CF, MF and not bound (NB); 11 and 12 correspond to the washes; 18-37 are the eluents from which the purified protein was obtained. (D) The phosphatase activity of the eluents 18-27 was examined by using p-NPP as the substrate.
A Western blot was performed with an anti-histidine antibody to determine whether the recombinant LmxPP2C protein has a sequence of 6 histidines at its extreme carboxyl terminal (Figure 1B). The antibody recognized a molecule of approximately 45.2 kDa in the TE (lane 2) and CF (lane 3). A very light phosphorescence is displayed in the MF (lane 4). The results corroborated the presence of histidines in the recombinant protein.
The recombinant protein was purified under nondenaturing conditions with Ni2+–NTA resin. Elution was carried out with a linear gradient of imidazole and the eluted fractions were analyzed by electrophoresis (SDS-PAGE), revealing a single protein with an approximate molecular weight of 45.2 kDa (Figure 1C). Phosphatase activity was evidenced in fractions 22-25 by a broad peak (Figure 1D).
Identification of PP2C in Different Leishmania Species
The PP2C protein was detected with antibodies against recombinant LmxPP2C (anti-LmxPP2C) applied to TE promastigotes of various Leishmania species (Figure 2): L. (V.) panamensis MHOM/PA/71/LS94 (lane 1), L. (V.) brazilienzis MHOM/BR/75/M2903 (lane 2), L. (L.) venezuelensis MHOM/VE/80/PMH3 (lane 3), L. (V.) donovani (lane 4), L. (L.) amazonensis IFLA/BR/67/PH8 (lane 5), (L.) mexicana MNYC/BZ/62/M379 (lane 6), and LmxPP2C (as the control, lane 7). In all Leishmania species (Figure 2, lanes 1-6), the protein was of 45.2 kDa, having the same molecular mass as the recombinant protein LmxPP2C (Figure 2, lane 7). Tubulin served as the loading control (top panel).
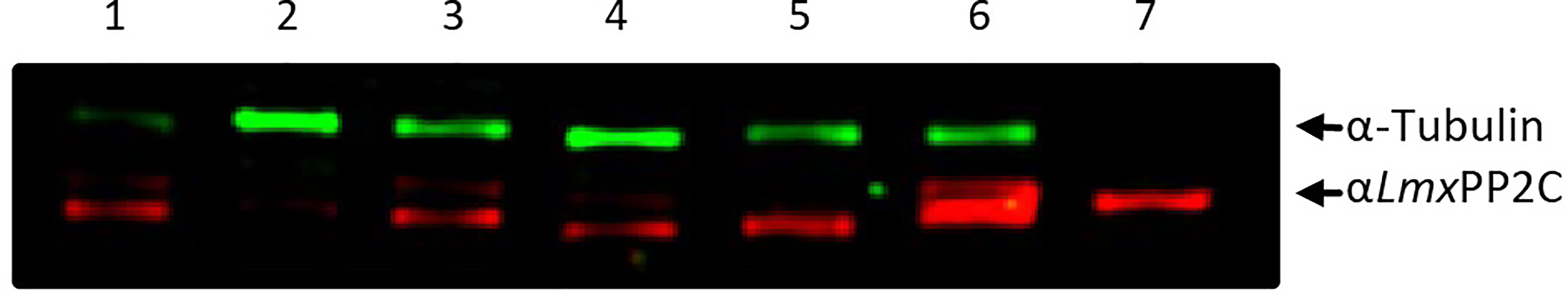
Figure 2 Identification of PP2C in the total extract of promastigotes of different species of Leishmania. Immunodetection of the PP2C protein in the total extract (TE) of the promastigotes of L. (V.) panamensis MHOM/PA/71/LS94 (lane 1), L. (V.) brazilienzis MHOM/BR/75/M2903 (lane 2), L. (L.) venezuelensis MHOM/VE/80/PMH3 (lane 3), L. (V.) donovani (lane 4), L. (L.) amazonensis IFLA/BR/67/PH8 (lane 5), and L. (L.) mexicana MNYC/BZ/62/M379 (lane 6). LmxPP2C (lane 7) served as the control and tubulin as a loading control.
Biochemical Characterization of PP2C of Leishmania mexicana
Cation dependence. Phosphatase activity was measured in the absence of divalent metal ions and in the presence of MnCl2, MgCl2 and CaCl2 (Figure 3A) with the generic phosphatase substrate p-NPP. There was a high level of hydrolysis of p-NPP induced by 10 mM of MnCl2 and a low level by the divalent cations CaCl2 and MgCl2. No phosphatase activity was observed in the control sample without cations.
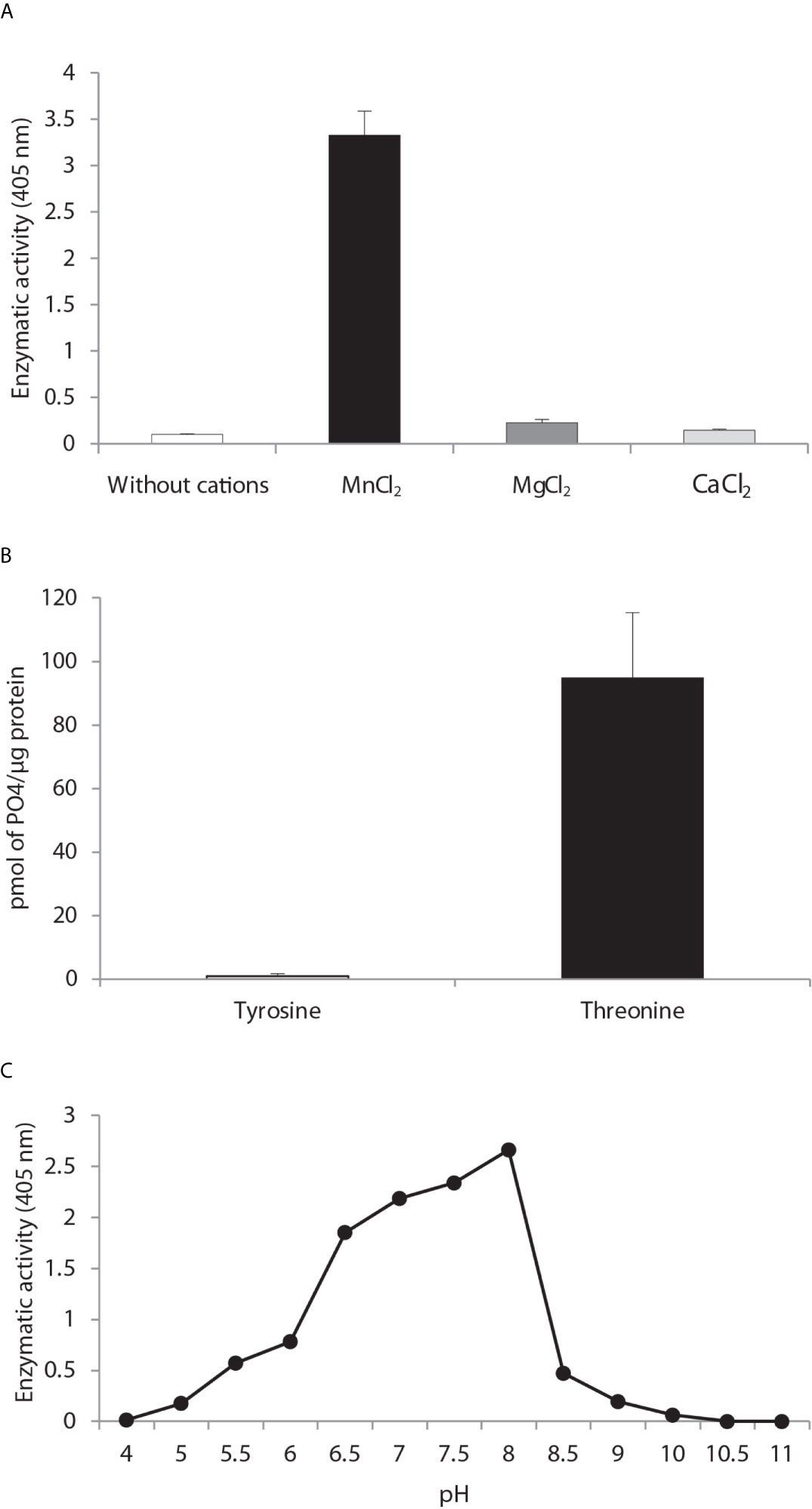
Figure 3 Biochemical characterization of PP2C of Leishmania mexicana. (A) The effect of MgCl2, MnCl2 and CaCl2 on the activity of PP2C of L. mexicana (LmxPP2C) was measured under standard conditions with 10 mM p-NPP as the substrate. (B) Hydrolysis of [END (pY) INASL] and Thr [RRA (pT) VA] by LmxPP2C. (C) Determination of the optimal pH for the activity of LmxPP2C, which was incubated with the appropriate buffers at different pH levels (4–11) in the presence of 10 mM MgCl2, with p-NPP as the substrate. Bars depict the mean percentage of relative activity of three independent assays.
Substrate specificity. Substrate specificity was tested by assays with peptides phosphorylated in tyrosine and threonine residues. LmxPP2C dephosphorylated the threonine but not tyrosine substrate (Figure 3B).
Optimal pH. The pH sensitivity of LmxPP2C was assessed by utilizing p-NPP in distinct buffers to determine the optimal conditions for phosphatase activity, which proved to be pH 8 (Figure 3C).
Effect of Some Phosphatase Inhibitors on the Activity of PP2C of Leishmania mexicana
The effect of potential inhibitors on L. mexicana phosphatase was evaluated on Thr [RRA (pT)VA]. The purified phosphatase was incubated with various inhibitors that target different classes in the serine/threonine protein phosphatase and tyrosine phosphatase families.
Calyculin and okadaic acid, inhibitors of serine/threonine phosphatases, produced 18 and 14% inhibition, respectively, on LmxPP2C. Sanguinarine (a specific inhibitor of PP2C activity) generated an 81% inhibition of LmxPP2C (Figure 4A). The phosphatase inhibitors (sodium orthovanadate, tungstate and pervanadate) did not have any significant effect (Figure 4B).
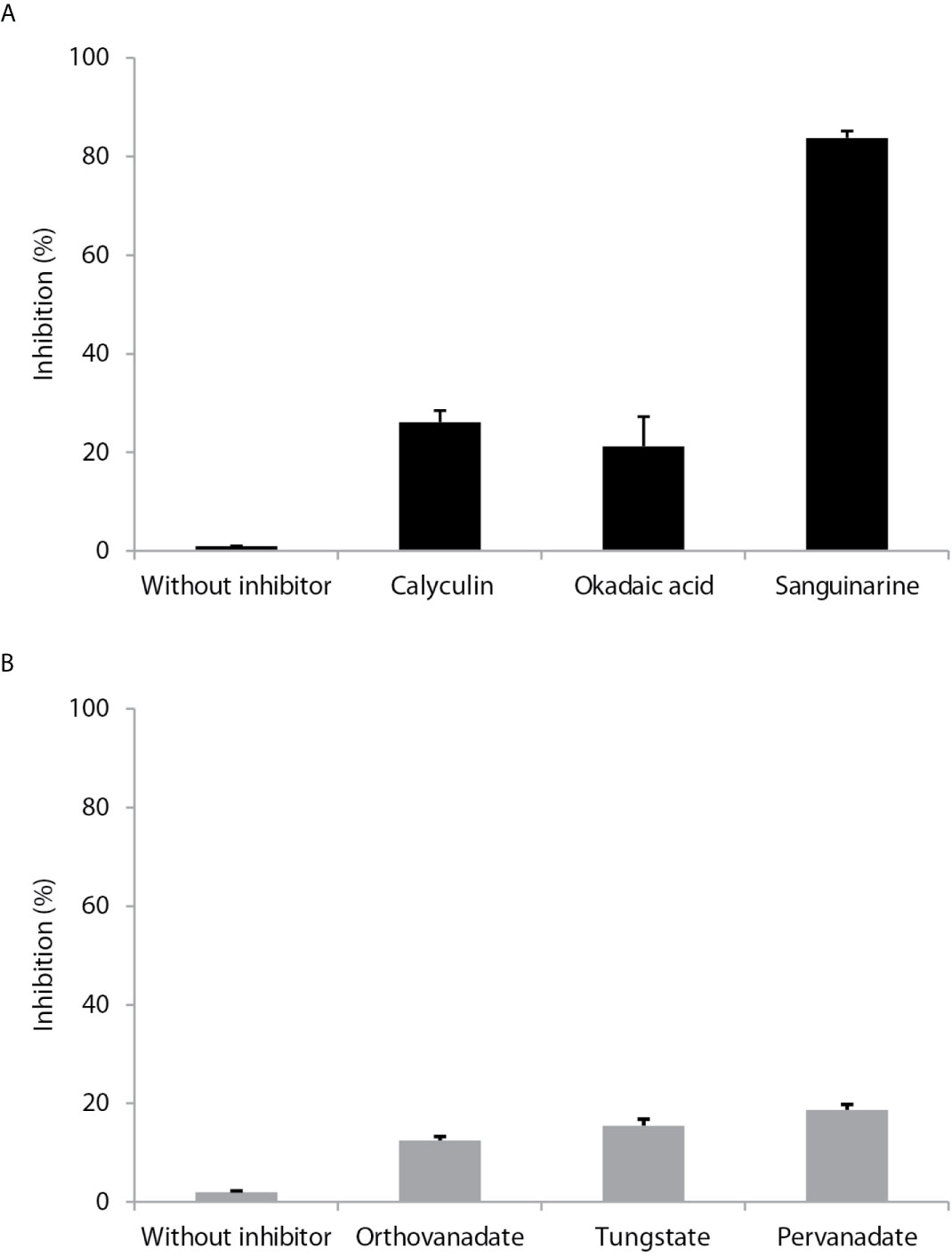
Figure 4 Effect of different inhibitors on PP2C of Leishmania mexicana. (A) Inhibitors (calyculin and okadaic acid) of serine threonine phosphatases and an inhibitor (sanguinarine) of the PPM family were incubated with PP2C of L. mexicana (LmxPP2C), and then phosphatase activity was assessed with p-NPP as the substrate. (B) Inhibitors of PTPs (orthovanadate, tungstate and pervanadate) were incubated with LmxPP2C and then phosphatase activity was evaluated with p-NPP as the substrate. Bars represent the mean percentage of relative activity of three independent assays.
Multiple Sequence Alignment
To examine the conserved sequence motifs for L. mexicana, L. major and human PP2Cs, a multiple sequence alignment was performed on the MAFFT program. The colored boxes display the conserved motifs while the red triangles show the conservation of the four aspartate residues that are crucial in the catalytic activity. LmxPP2C has a 98.77% sequence similarity with LmPP2C and 44.10% with human PP2C, indicating a different role of these enzymes in Leishmania spp. versus humans (Figure 5A).
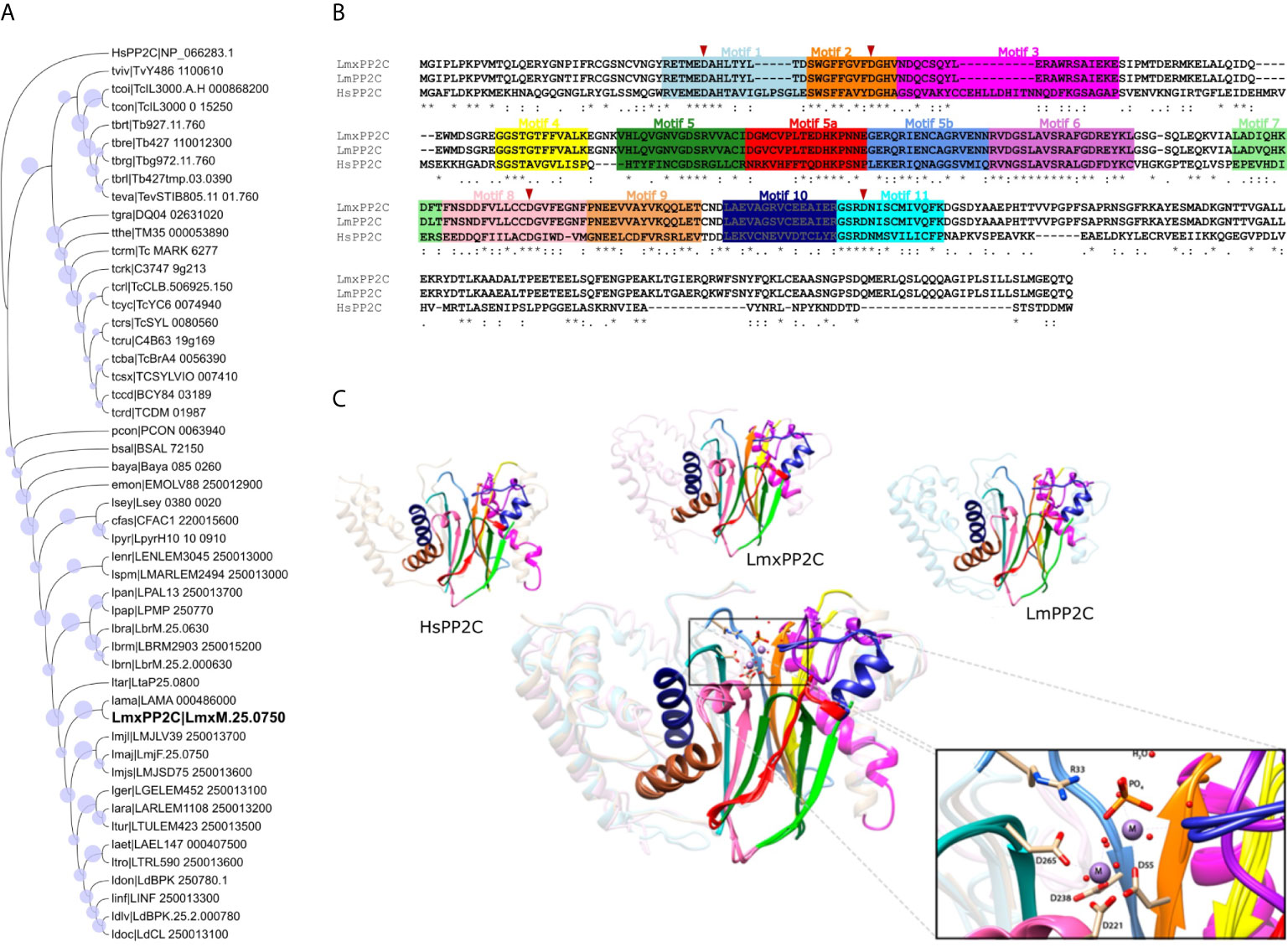
Figure 5 Phylogenetic tree, multiple sequence alignment and structural alignment of Leishmania PP2C. (A) Analysis of the phylogenetic tree of Leishmania PP2C, finding that they belong to the ortholog group OG6_142542 (using the maximum likelihood method based on the LG+G+I model). The analysis involved 50 amino acid sequences. Bootstrap values for each node are shown as a circle. (B) Multiple sequence alignment of L. mexicana, L. major and human PP2C (LmxM.25.0750, LmF.25.0750 and NP_066283.1, respectively). The 11 conserved motifs are indicated by colored boxes with their corresponding motif numbers in the upper part of the figure. The four aspartates implicated in the catalytic mechanism are conserved in each sequence (red arrowhead). (C) Structural alignment of the predicted LmxPP2C, LmPP2C and HsPP2C (1A6Q) carried out on the USCF Chimera program. The 11 conserved motifs of the PP2C catalytic domain are depicted in cartoon representation with distinct colors for each conserved motif. The amino acids participating in the catalysis are portrayed as sticks, while the rest of the protein is illustrated in the background. The structural alignment demonstrates conservation between PP2C sequences.
Phylogenetic Tree Analysis of the Ortholog Group of Leishmania PP2C
LmxPP2C belongs to the ortholog group OG6_142542, which contains 49 species belonging to the family Trypanosomatidae. To investigate the phylogenetic relationship of all PP2C protein sequences in this ortholog group with human PP2C (NP_066283.1, OG6_100270), a phylogenetic tree was constructed based on the alignments of PP2C domains using the maximum likelihood method. According to the phylogenetic analysis, these 49 PP2C sequences are grouped by taxa. Since human PP2C is found in a distinct ortholog group, it probably has a different function than that of the enzymes of Leishmania and other Trypanosomatidae species (Figure 5B).
Prediction and Structural Alignment of the 3D Model of PP2C of Leishmania mexicana
To identify structural similarities and differences between LmxPP2C, LmPP2C and human PP2C, firstly a homology prediction was made using the I-TASSER server and then the structural alignment was visualized in the UCSF-Chimera program. The eleven motifs of PP2C in the three species are shown in colors, as was done in the multiple sequence alignment. The catalytic PP2C domain is the typical β-sheet sandwich surrounded by α-helices. In the case of L. mexicana, the catalytic site of PP2C is localized in a cleft between two central β sheets formed by aspartate residues D38, D55, D221 and D265 as well as arginine R33. The structure alignment of these three species indicates that they are homologous, containing conserved elements of the PP2C family with a few changes in the motif 3 secondary structure and the C-terminal length (Figure 5C).
Ultrastructural Localization of PP2C in Promastigotes and Amastigotes of L. mexicana
Distribution of PP2C in Promastigotes and Amastigotes of L. mexicana
PP2C was observed in promastigotes of L. mexicana by IEM (based on an accumulation of the gold label), localized in the flagellar pocket and particularly in the corresponding vesicles (Figures 6A(a, b), arrow). The label was also detected along the flagellum (Figure 6A, image c). In order to recognize the precise location of the labeling with gold particles, ultrastructural micrographs of promastigotes and of the flagellar pocket are herein included (Figure 6A, Images d and e). Spheroid structures in the flagellar pocket were labeled by the antibody against LmxPP2C and are easily seen in the micrograph of the respective control of ultrastructure as electron-dense vesicles (Figure 6A, images b and e respectively, arrow). Samples incubated with pre-immune serum displayed no IEM labelling of the parasites (data not shown).
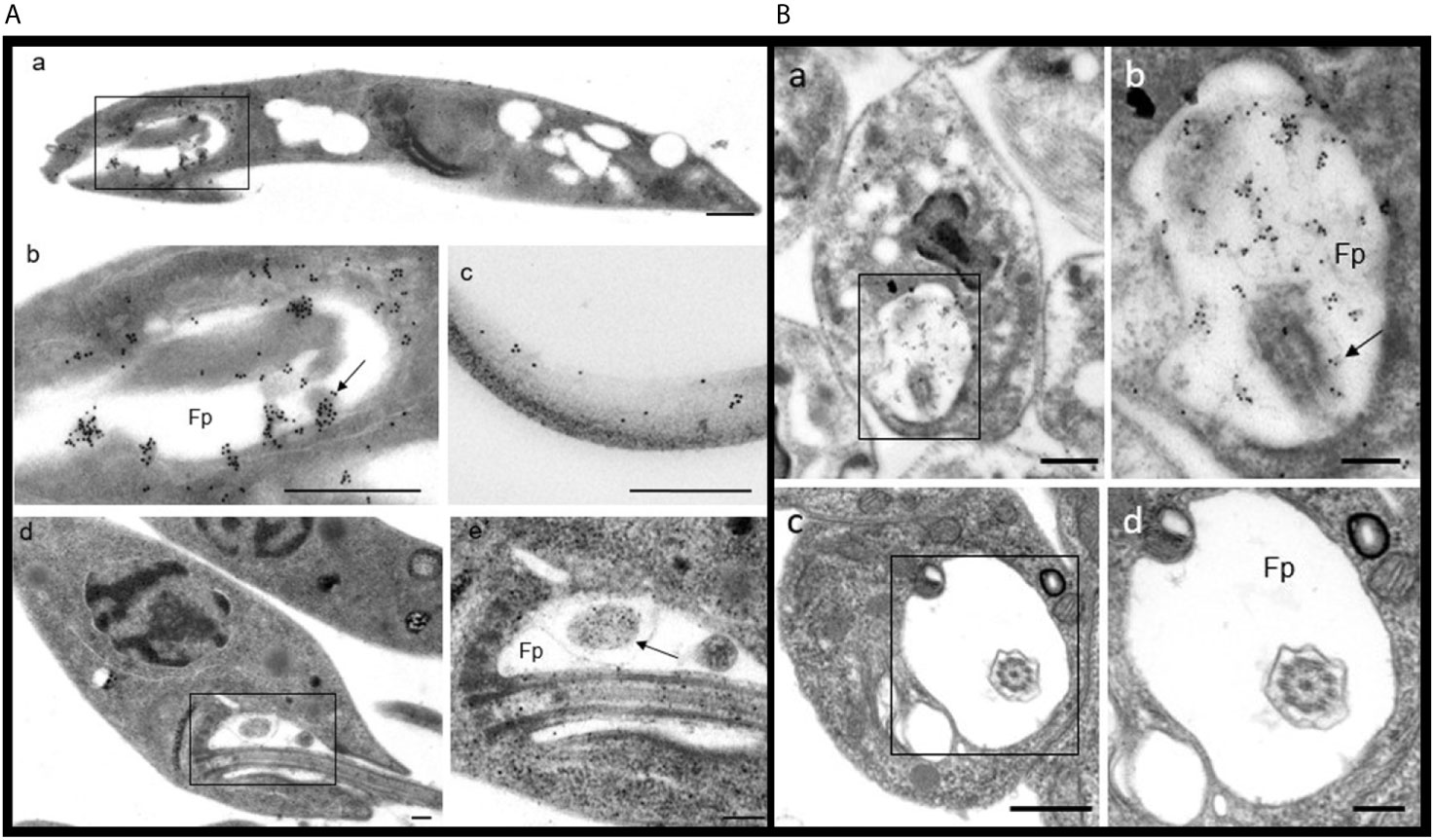
Figure 6 Distribution of PP2C in promastigotes and amastigotes of L. mexicana. (A) Immune electron microscopy of LmxPP2C in promastigotes. The micrographs in A are of low (Images a and d) and high magnification (images b, c and e) of promastigotes. Image (b) corresponds to a magnification of the inset in (a). Micrographs (d) and (e) correspond to the control for the ultrastructure of a promastigote. The image (e) represents a magnification of the inset in (a). Scale bars = 500 nm. (B) Immune electron microscopy of LmxPP2C in amastigotes. The micrographs are of low (images a and c) and high magnification (b and d) of amastigotes. Image (b) portrays a magnification of the inset in (a). Images (c) and (d) correspond to the control for ultrastructure of an amastigote. Fp, flagellar pocket. Scale bars in a and c = 500 nm; scale bars in b and d = 200 nm.
The distribution of PP2C in amastigotes of L. mexicana was viewed by IEM, finding a specific location of the immune gold label in the flagellar pocket (Figure 6B, images a and b). The flagellum located in the flagellar pocket (Image b, arrow) was poorly marked with gold particles. In contrast, the electron-dense vesicles and the fuzzy material located in the flagellar pocket were heavily marked by the immune gold label (Image b). There was scarce labeling in the cytoplasm (data not shown). The precise location of the gold labelling was determined based on the amplified images of the respective control of ultrastructure of amastigotes and the flagellar pocket (Figure 6B, Images c and d). Thin sections incubated with pre-immune serum did not exhibit any labelling of the parasites (data not shown).
Discussion
Protein phosphatases have served as a target for the design of anti-cancer drugs (Le Guezennec and Bulavin, 2010). The PP2C delta phosphatase (Wip) plays an important role in homeostasis as well as in the pathogenesis of several human diseases. Wip1 is considered a negative regulator of the p53 oncogene pathway in tumor lines (Goloudina et al., 2016). PP2Cs are monomeric enzymes that require metal cations (Mg+2 or Mn+2) for their enzymatic activity. They are comprised of a catalytic domain with 11 conserved motifs containing aspartic acid. Their catalytic activity is highly regulated and may have enormous therapeutic potential (Baskaran and Velmurugan, 2018).
In parasites, protein phosphatases of serine/threonine and tyrosine have been cloned and characterized. Our group has characterized protein phosphatases in protozoan parasites such as L. major and Cryptosporidium parvum (Escalona-Montaño et al., 2017; Gómez-Sandoval et al., 2020). In the current contribution, the recombinant protein PP2C of L. mexicana was cloned and purified.
PP2C protein phosphatases have been characterized in Leishmania, including the PP2C of L. chagasi (LcPP2C), L. major (LmPP2C) and L. donovani (LdPP2C), (Burns et al., 1993; Escalona-Montaño et al., 2017; Jakkula et al., 2018). These enzymes share very similar biochemical characteristics. For example, purified LdPP2C is enzymatically active, and according to an inhibition study, sanguinarine likely acts as a non-competitive inhibitor (at pH 3.5-8.5). The molecular weight of the purified protein was 45 kDa (Jakkula et al., 2018). LcPP2C was reported to have a molecular weight of 42 kDa in both the infective promastigotes and the tissue amastigotes of L. chagasi and L. amazonensis.
To verify the relationship between LcPP2C and mammalian PP2Cs observed in predicted amino acid sequence comparisons, enzymatically active LcPP2C was expressed. Purified LcPP2C readily dephosphorylated [32P] casein in an Mg+2-dependent and okadaic acid-independent manner (Burns et al., 1993). In L. major, there was a protein of 44.9 kDa with PP2C activity, the latter being dependent on divalent cations (Mg+2 and Mn+2) and found to be optimal at a pH of 8.5 when using phosphothreonine as the substrate. Sanguinarine inhibited the activity of recombinant LmPP2C (Escalona-Montaño et al., 2017).
The present results reveal that recombinant PP2C of L. mexicana has a molecular weight of 45.2 kDa, is Mn+2-dependent, and has optimal activity at pH 8 with p-NPP as the substrate. It preferentially dephosphorylated threonine (versus tyrosine), an activity inhibited by sanguinarine. The Mn+2-dependency has been described for other microorganisms, such as a Chlamydia spp. (Claywell et al., 2018).
The similarity of PP2C between distinct species of Leishmania makes this enzyme an important subject of research for the development of diagnostic tools, vaccine immunogens, and drugs for treating leishmaniasis. However, the function of PP2C enzymes in the Leishmania species is still unknown. LdPP2C affects the host innate immune response by upregulating pro-inflammatory cytokines (TNF-α and IL-6) as well as nitric oxide (Jakkula et al., 2018), while LcPP2C elicits a strong proliferative response of T cells in patients with leishmaniasis (Burns et al., 1993).
In both the promastigote and amastigote forms, there is an invagination of the plasma membrane at the base of the flagellum (the flagellar pocket). This key feature of trypanosomatid cells is central to several vital processes, including differentiation, endo/exocytosis, flagellum assembly, and the definition of surface membrane boundaries. Such processes are critical for the cell biology underlying the life cycle of Leishmania (Sunter et al., 2018). The location of PP2C in the flagellum of L. major suggests a potential regulatory activity in this organelle. Nevertheless, the functional role of LmPP2C has yet to be established (Escalona-Montaño et al., 2017). In other trypanosomatid parasites, such as Trypanosoma rangeli, a PTP was associated with the parasite flagellum, although its function is unknown (Prestes et al., 2012). In most of these parasites, the PP2C enzymes are mainly located in the cytosol. During invasion, the T. gondii parasite secretes PP2C from the rhoptries to deliver it to the host cell and target the respective nucleus (Gilbert et al., 2007).
In L. mexicana, the presence of LmxPP2C in the flagellar pocket of the parasite indicates that the enzyme is probably discharged from Golgi-derived multivesicular bodies in the same location, which would explain why the labelling was found in the vesicles of promastigotes and amastigotes. Hence, phosphorylation is likely carried out by yet unidentified kinases in the Golgi apparatus involved in a reciprocal regulatory relationship with LmxPP2C.
Since PP2C is located in the flagellar pocket of promastigotes (an important organelle in Leishmania), it represents a possible drug target or diagnostic marker of leishmaniasis. Comparative analysis of gene expression across promastigote and amastigote forms revealed a total of 3,832 genes differentially expressed between promastigotes and intracellular amastigotes. During differentiation to amastigotes, a large proportion of the downregulated genes are linked to the function of the motile flagellum, while the upregulated genes encode cell surface proteins, transporters and peptidases, as well as many other proteins yet to be defined (the undefined genes comprise 293 of the 936 novel genes) (Fiebig et al., 2015). The LmxM.25.0750 gene has higher expression in promastigotes than amastigotes, which is correlated with its location in the flagellar pocket. Given that LmxPP2C belongs to the ortholog group OG6_142542 (containing only protozoa of the Trypanosomatidae family), it probably has a specific role in the flagellar pocket of these microorganisms.
The finding of a recombinant LmxPP2C protein opens a new field of research on its use as a drug target, an immunomodulatory molecule, and/or a diagnostic marker for leishmaniasis. Further research is needed on the function of this protein in the parasite, since it may be a key molecule in L. mexicana and in the pathogenesis of leishmaniasis.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The protocol for the experiments carried out with animals in this study was reviewed and approved by the Ethics in Research Committee and the Internal Committee on the Care and Use of Lab Animals (CICUAL, according to the name of the committee in Spanish), both of the Medicine Faculty of the Universidad Nacional Autónoma de México. The fundamental purpose of these committees is to evaluate and if appropriate authorize the use of protocols for the care and use of lab animals in research or teaching, with the aim of regulating such practices so that they conform to the Mexican norms established in the law known as NOM-062-ZOO-1999.
Author Contributions
The purification of LmxPP2C was performed by AE-M. The experimental characterization of PP2C activity and the Western blot assay were carried out by MZ-F. The characterization of LmxPP2C was supervised and validated by IB in her lab. The gene cloning experiments were designed by RP-M, who supervised and validated them in her lab. The gene encoding LmxPP2C was cloned and overexpressed by NC. LmxPP2C was localized in promastigotes and amastigotes with a transmission electron microscope (TEM) by RM-F. The parasites used for localizing LmxPP2C with a TEM were provided by IB. The in silico model of PP2C was constructed by JG-S. The polyclonal antibodies against LmxPP2C were generated by AG-C. The statistical analysis and the creation of the figures was done by AR-B. The analysis of the results involved all authors under the direction of IB and MA-G. The conceptualization of the study and funding acquisition for the experimental work was the responsibility of MA-G, and the manuscript was structured by MA-G. The original draft was prepared by AE-M and MA-G, and the writing was reviewed and edited by AE-M, JG-S, LG-K and MA-G. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Secretaría de Educación Pública - Consejo Nacional de Ciencia y Tecnología (SEP-CONACyT, Mexico), grant number 284018, and partially sponsored by the DGAPA-PAPIIT, grant number IN218619, given to MA-G.
Conflict of Interest
The procedure presently used to purify and quantify the expression of PP2C in L. mexicana has been patented in Mexico (MX/a/2020/011271) by AE-M and MA-G.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Adriana Ruiz Remigio and Marco Gudiño Zayas (Unidad de Investigación en Medicina Experimental, Facultad de Medicina, UNAM) for technical assistance. Ofelia Pérez Olvera and Daniel Andrés Sánchez Almaraz for technical assistance in Unidad de Investigación UNAM-INC from the Facultad de Medicina, UNAM. We are thankful for technical support given by Mónica Mondragón from the Biochemistry Department and Electron Microscopy Facility-LANSE at CINVESTAV, Mexico. The authors are grateful to Bruce Allan Larsen for proofreading the manuscript.
References
Aguirre-Garcia, M. M., Escalona-Montaño, A. R., Wilkins-Rodríguez, A., Kobeh-Gutiérrez, L. (2018). “Chapter 4: Innate Immune Evasion Strategies,” in “Leishmaniases as Re-emerging Diseases,” (London, UK: Editorial InTechOpen), 978–1-78984-102-2.
Alvar, J., Vélez, I. D., Bern, C., Herrero, M., Desjeux, P., Cano, J., et al. (2012). Who Leishmaniasis Control the WHO Leishmaniasis Control Team Leishmaniasis Worldwide and Global Estimates of Its Incidence. PloS One 7, e35671.ssw. doi: 10.1371/journal.pone.0035671
Andreeva, A. V., Kutuzov, M. A. (2004). Widespread presence of “bacterial-like” PPP phosphatases in eukaryotes. BMC Evol. Biol. 4, 47. doi: 10.1186/1471-2148-4-47
Aslett, M., Aurrecoechea, C., Berriman, M., Brestelli, J., Brunk, B. P., Carrington, M., et al. (2010). TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 38 (Database issue), D457–D462. doi: 10.1093/nar/gkp851
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1006/abio.1976.9999
Baskaran, R., Velmurugan, B. K. (2018). Protein phosphatase 2A as therapeutic targets in various disease models. Life Sci. 210, 40–46. doi: 10.1016/j.lfs.2018.08.063
Bork, P., Brown, N. P., Hegyi, H., Schultz, J. (1996. The protein phosphatase 2C (PP2C) superfamily: detection of bacterial homologues. Protein Sci. 5(7), 1421–1425. doi: 10.1002/pro.5560050720
Brenchley, R., Tariq, H., McElhinney, H., Szöor, B., Huxley-Jones, J., Stevens, R., et al. (2007). The TriTryp phosphatome: analysis of the protein phosphatase catalytic domains. BMC Genomics 8, 434. doi: 10.1186/1471-2164-8-434
Burns, J. M. Jr., Parsons, M., Rosman, D. E., Reed, S. G. (1993). Molecular cloning and characterization of a 42-kDa protein phosphatase of Leishmania chagasi. J. Biol. Chem. 268 (23), 17155–17161. doi: 10.1016/S0021-9258(19)85316-4
Claywell, J. E., Matschke, L. M., Plunkett, K. N., Fisher, D. J. (2018). Inhibition of the Protein Phosphatase CppA Alters Development of Chlamydia trachomatis. J. Bacteriol. 200 (19), e00419–e00418. doi: 10.1128/JB.00419-18
Cohen, P. (2002). The origins of protein phosphorylation. Nat. Cell Biol. 4 (5), E127–E130. doi: 10.1038/ncb0502-e127
Das, A. K., Helps, N. R., Cohen, P. T., Barford, D. (1996). Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution. EMBO J. 15 (24), 6798–6809. doi: 10.1002/j.1460-2075.1996.tb01071.x
Dissing, J., Dahl, O., Svensmark, O. (1979). Phosphonic and arsonic acids as inhibitors of human red cell acid phosphatase and their use in affinity chromatography. Biochim. Biophys. Acta 569 (2), 159–176. doi: 10.1016/0005-2744(79)90051-2
Escalona-Montaño, A. R., Ortiz-Lozano, D. M., Rojas-Bernabé, A., Wilkins-Rodriguez, A., H. Torres-Guerrero, H., Mondragón-Flores, R., et al. (2016). Leishmania mexicana: promastigotes and amastigotes secrete protein phosphatases and this correlates with the production of inflammatory cytokines in macrophages. Parasitology 143, 1409–1420. doi: 10.1017/S0031182016000949ISSN:1469-8161
Escalona-Montaño, A. R., Pérez-Montfort, R., Cabrera, N., Mondragón-Flores, R., Vélez-Ramírez, D. E., Gómez-Sandoval, J. N., et al. (2017). Protein phosphatase PP2C in the flagellum of Leishmania major: cloning and characterization. Parasitol. Open. 3,e15, 1–9. doi: 10.1017/pao.2017.14
Fiebig, M., Kelly, S., Gluenz, E. (2015). Comparative Life Cycle Transcriptomics Revises Leishmania mexicana Genome Annotation and Links a Chromosome Duplication with Parasitism of Vertebrates. PloS Pathog. 11 (10), e1005186. doi: 10.1371/journal.ppat.1005186
Fuchs, S., Grill, E., Meskiene, I., Schweighofer, A. (2013). Type 2C protein phosphatases in plants. FEBS J. 280 (2), 681–693. doi: 10.1111/j.1742-4658.2012.08670
Gilbert, L. A., Ravindran, S., Turetzky, J. M., Boothroyd, J. C., Bradley, P. J. (2007). Toxoplasma gondii targets a protein phosphatase 2C to the nuclei of infected host cells. Eukaryot. Cell. 6 (1), 73–83. doi: 10.1128/EC.00309-06
Goloudina, A. R., Kochetkova, E. Y., Pospelova, T. V., Demidov, O. N. (2016). Wip1 phosphatase: between p53 and MAPK kinases pathways. Oncotarget 7 (21), 31563–31571. doi: 10.18632/oncotarget.7325
Gómez-Sandoval, J. N., Okhuysen, P., Mondragón-Flores, R., Escalona-Montaño, A. R., Aguirre-García, M. M. (2020). Cellular Identification and In Silico Characterization of Protein Phosphatase 2C (PP2C) of Cryptosporidium parvum. Acta Parasitol. 65 (3), 704–715. doi: 10.2478/s11686-020-00209
Ivens, A. C., Peacock, C. S., Worthey, E. A., Murphy, L., Aggarwal, G., Berriman, M., et al. (2005). The genome of the kinetoplastid parasite, Leishmania major. Science 309 (5733), 436–442. doi: 10.1126/science.1112680
Jakkula, P., Qureshi, R., Iqbal, A., Sagurthi, S. R., Qureshi, I. A. (2018). Leishmania donovani PP2C: Kinetics, structural attributes and in vitro immune response. Mol. Biochem. Parasitol. 223, 37–49. doi: 10.1016/j.molbiopara.2018.06.005
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 (4), 772–780. doi: 10.1093/molbev/mst010
Lammers, T., Lavi, S. (2007). Role of type 2C protein phosphatases in growth regulation and in cellular stress signaling. Crit. Rev. Biochem. Mol. Biol. 42 (6), 437–461. doi: 10.1080/10409230701693342
Le Guezennec, X., Bulavin, D. V. (2010). WIP1 phosphatase at the crossroads of cancer and aging. Trends Biochem. Sci. 35 (2), 109–114. doi: 10.1016/j.tibs.2009.09.005
Letunic, I., Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 8;44 (W1), W242–W245. doi: 10.1093/nar/gkw290
Li, L., Stoeckert, C. J. Jr., Roos, D. S., Ortho, M. C. L. (2003). identification of ortholog groups for eukaryotic genomes. Genome Res. 13 (9), 2178–2189. doi: 10.1101/gr.1224503
Montfort, I., Pérez-Tamayo, R., Pérez-Montfort, R., González Canto, A., Olivos, A. (1994). Purification and immunologic characterization of a 30-kDa cysteine proteinase of Entamoeba histolytica. Parasitol. Res. 80 (7), 607–613. doi: 10.1007/BF00933010
Moorhead, G. B., Trinkle-Mulcahy, L., Ulke-Lemée, A. (2007). Emerging roles of nuclear protein phosphatases. Nat. Rev. Mol. Cell Biol. 8 (3), 234–244. doi: 10.1038/nrm2126
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25 (13), 1605–1612. doi: 10.1002/jcc.20084
Prestes, E. B., Bayer-Santos, E., Hermes Stoco, P., Sincero, T. C., Wagner, G., Umaki, A., et al. (2012). Trypanosoma rangeli protein tyrosine phosphatase is associated with the parasite’s flagellum. Mem. Inst. Oswaldo Cruz 107 (6), 713–719. doi: 10.1590/s0074-02762012000600002
Schweighofer, A., Hirt, H., Meskiene, I. (2004). Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 9 (5), 236–243. doi: 10.1016/j.tplants.2004.03.007
Sela, I., Ashkenazy, H., Katoh, K., Pupko, T. (2015). GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43 (W1), W7–W14. doi: 10.1093/nar/gkv318
Sunter, J. D., Moreira-Leite, F., Gull, K. (2018). Dependency relationships between IFT-dependent flagellum elongation and cell morphogenesis in Leishmania. Open Biol. 8 (11), 180124. doi: 10.1098/rsob.180124
Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30 (12), 2725–2729. doi: 10.1093/molbev/mst197
Virshup, D. M., Shenolikar, S. (2009). From promiscuity to precision: protein phosphatases get a makeover. Mol. Cell. 13, 33(5), 537–45. doi: 10.1016/j.molcel.2009.02.015
Wang, B., Zhang, P., Wei, Q. (2008). Recent progress on the structure of Ser/Thr protein phosphatases. Sci. China C. Life Sci. 51 (6), 487–494. doi: 10.1007/s11427-008-0068-y
Keywords: phosphoprotein phosphatase, threonine phosphatases, PP2C, Leishmania mexicana, leishmaniasis
Citation: Escalona-Montaño AR, Zuñiga-Fabián M, Cabrera N, Mondragón-Flores R, Gómez-Sandoval JN, Rojas-Bernabé A, González-Canto A, Gutiérrez-Kobeh L, Pérez-Montfort R, Becker I and Aguirre-García MM (2021) Protein Serine/Threonine Phosphatase Type 2C of Leishmania mexicana. Front. Cell. Infect. Microbiol. 11:641356. doi: 10.3389/fcimb.2021.641356
Received: 14 December 2020; Accepted: 23 March 2021;
Published: 15 April 2021.
Edited by:
Loïc Riviere, Université de Bordeaux, FranceReviewed by:
Chiranjib Pal, West Bengal State University, IndiaShaojun Long, China Agricultural University, China
Copyright © 2021 Escalona-Montaño, Zuñiga-Fabián, Cabrera, Mondragón-Flores, Gómez-Sandoval, Rojas-Bernabé, González-Canto, Gutiérrez-Kobeh, Pérez-Montfort, Becker and Aguirre-García. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Magdalena Aguirre-García, maguirre@unam.mx
 Alma Reyna Escalona-Montaño
Alma Reyna Escalona-Montaño Mariana Zuñiga-Fabián
Mariana Zuñiga-Fabián Nallely Cabrera3
Nallely Cabrera3  Ricardo Mondragón-Flores
Ricardo Mondragón-Flores Jenny Nancy Gómez-Sandoval
Jenny Nancy Gómez-Sandoval Araceli Rojas-Bernabé
Araceli Rojas-Bernabé Augusto González-Canto
Augusto González-Canto Laila Gutiérrez-Kobeh
Laila Gutiérrez-Kobeh Ruy Pérez-Montfort
Ruy Pérez-Montfort Ingeborg Becker
Ingeborg Becker Maria Magdalena Aguirre-García
Maria Magdalena Aguirre-García