Common Dysregulation of Innate Immunity Pathways in Human Primary Astrocytes Infected With Chikungunya, Mayaro, Oropouche, and Zika Viruses
- 1Laboratório de Virologia Molecular, Departamento de Genética, Instituto de Biologia, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil
- 2Laboratório de Biologia Integrativa, Departamento de Genética Ecologia e Evolução, Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Belo Horizonte, Brazil
- 3Laboratório de Bioinformática, Laboratório Nacional de Computação Científica, Ministério de Ciência Tecnologia e Comunicações, Petrópolis, Brazil
- 4Instituto de Microbiologia Paulo de Goes, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil
- 5Instituto de Ciências Biomédicas, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil
- 6Departamento de Biologia Celular e Molecular, Faculdade de Medicina de Ribeirão Preto, Universidade de São Paulo, Ribeirão Preto, Brazil
- 7Hospital Universitário Clementino Fraga Filho, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil
Arboviruses pose a major threat throughout the world and represent a great burden in tropical countries of South America. Although generally associated with moderate febrile illness, in more severe cases they can lead to neurological outcomes, such as encephalitis, Guillain-Barré syndrome, and Congenital Syndromes. In this context astrocytes play a central role in production of inflammatory cytokines, regulation of extracellular matrix, and control of glutamate driven neurotoxicity in the central nervous system. Here, we presented a comprehensive genome-wide transcriptome analysis of human primary astrocytes infected with Chikungunya, Mayaro, Oropouche, or Zika viruses. Analyses of differentially expressed genes (DEGs), pathway enrichment, and interactomes have shown that Alphaviruses up-regulated genes related to elastic fiber formation and N-glycosylation of glycoproteins, with down-regulation of cell cycle and DNA stability and chromosome maintenance genes. In contrast, Oropouche virus up-regulated cell cycle and DNA maintenance and condensation pathways while down-regulated extracellular matrix, collagen metabolism, glutamate and ion transporters pathways. Zika virus infection only up-regulated eukaryotic translation machinery while down-regulated interferon pathways. Reactome and integration analysis revealed a common signature in down-regulation of innate immune response, antiviral response, and inflammatory cytokines associated to interferon pathway for all arboviruses tested. Validation of interferon stimulated genes by reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) corroborated our transcriptome findings. Altogether, our results showed a co-evolution in the mechanisms involved in the escape of arboviruses to antiviral immune response mediated by the interferon (IFN) pathway.
Introduction
Arthropod-borne viruses (i.e. arboviruses) comprehend a large group of viruses belonging to different families that are transmitted mainly by the bite of infected mosquitoes or ticks during the blood meal. As many of them are the etiological agents of relevant human pathologies, they pose a special threat in vulnerable countries around the world. Tropical countries of South America, such as Brazil, are specially in risk for arboviruses outbreaks due to the suitable warm weather, abundance of insect vectors, deforestation, high population concentration near forest areas, social-economical disparities, and overburden of the public healthcare system (Peña-García et al., 2017; Figueiredo, 2019; Medeiros and Vasconcelos, 2019). Many endemic outbreaks along the history, as well the recent introduction of arboviruses in Brazil, have risen the alarm of scientific and medical communities concerning the risk of new outbreaks, co-circulation of those viruses, and increasing of more severe cases (Metsky et al., 2017; Zanotto and Leite, 2018; Lorenz et al., 2019; Gutierrez et al., 2020). Viruses members of Alphavirus, Orthobunyavirus, and Flavivirus genus (Togaviridae, Peribunyaviridae, and Flaviviridae viral families, respectively) present a higher incidence in Brazil and has been associated to a plethora of clinically relevant outcomes, such as mild to debilitating fever, hemorrhagic fever, arthritis, microcephaly, and neurological disorders (Figueiredo, 2015; Santiago et al., 2015; Burt et al., 2017; Azeredo et al., 2018; Vernal et al., 2019; Brito Ferreira et al., 2020; Diagne et al., 2020; Gutierrez et al., 2020).
Alphaviruses as Chikungunya Virus (CHIKV) and Mayaro Virus (MAYV) circulate in Brazil, with many cases reported from Northern to Southeastern regions of the country (Carvalho et al., 2019; Lorenz et al., 2019; Naveca et al., 2019; Vasconcellos et al., 2019). They are enveloped viruses with single positive RNA (+) strand genome, flanked by 5’ and 3’ UTRs (untranslated regions). The genome is ~11.5 kb long and composed by two ORFs (open reading frame): the first one encodes the non-structural proteins nsP1-4 that plays a role in viral replication and host factors interaction and modulation, followed by a second ORF that encodes the structural proteins C (capsid), E3, E2, E1 (envelope glycoproteins) (Acosta-Ampudia et al., 2018; Frolov and Frolova, 2019; Levi and Vignuzzi, 2019). CHIKV are mainly transmitted by mosquito vectors from genera Aedes and MAYV is more restricted to sylvatic species of Sabethes and Haemagogus, reflecting on the epidemic pattern of CHIKV and in local MAYV circulation. Those alphaviruses cause a febrile illness, with common symptoms such as nausea, headache, rash, myalgia, and polyarthralgia. The arthralgia can persist for months after the acute infection, characterizing the main chronic incapacitating symptom of those infections. However, both viruses have been associated with more severe neurological manifestations, such as meningo-encephalitis, Guillain-Barré syndrome, acute disseminated encephalomyelitis (ADEM), and severe myelitis, with increasing reports of neuro-CHIKV in endemic areas such as Brazil (Bandeira et al., 2016; Balavoine et al., 2017; Khatri et al., 2018; Maria et al., 2018; Simon et al., 2018; Hameed and Khan, 2019; Silva et al., 2020).
Oropouche Virus (OROV) is an Orthobunyavirus responsible for many outbreaks, in Brazil and other South America countries (Elliott, 2014; De Regge, 2017). OROV genome is composed of three single strand negative RNAs (−) flanked by 5’ and 3’ UTRs: namely the S (small) RNA, a 961 nt long RNA that encodes the nucleocapsid protein N and the non-structural protein NSs; a 4.5 kb long M (medium) RNA that encodes the viral envelope glycoproteins Gn and Gc, and the non-structural protein NSm; and a 6.9 kb long L (large) RNA that encodes the viral RNA-dependent RNA polymerase (RdRp) (Travassos da Rosa et al., 2017; Sakkas et al., 2018). The midge Culicoides paraensis has been considered the main urban vector of OROV, although it was already isolated from other mosquitoes’ vectors, including species from the genera Aedes and Culex. OROV is the causative agent of Oropouche Fever, a mild febrile illness that shares common symptoms with other arboviral fevers as moderate to high fever, headache, nausea and diarrhea, vomiting, dizziness, and myalgia. Photophobia is also a particular common symptom and, in some cases, OROV has been associated with neurological meningo-encephalitis outcomes (Bastos et al., 2012; Santos et al., 2014; Vernal et al., 2019).
Zika Virus (ZIKV) has raised global attention in the past few years as a potential threat to humans, mainly to pregnant women, due to the infection could promote severe neurological disorders and congenital fetal malformations in Brazil (Duffy et al., 2009). Those clinical outcomes were first described in the last outbreaks, in French Polynesia (2013) and with the arrival and spread of ZIKV throughout the Americas (2014) (Cao-Lormeau et al., 2016; Schuler-Faccini et al., 2016). ZIKV is a flavivirus with a genome composed of a single positive strand RNA of ~11 kb, flanked by 5’ and 3’ UTR regions coding a precursor polyprotein that is processed by viral and cellular protease to generate three structural proteins C (capsid), prM, and E (envelope) and seven non-structural proteins NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5. The epidemics in South America between 2014 and 2016 brought light to the severe neurological manifestations of ZIKV infection, named Congenital Zika Syndrome (CZS), that affects the nervous system and multiple organs of the developing fetus of infected pregnant women. The CZS comprehend a myriad of clinical neurological manifestations including microcephaly, hydrocephaly and lissencephaly as well as ventricular calcifications and arthrogryposis (congenital joints contracture) (Liang et al., 2019). In adults, ZIKV infection can also be associated with encephalitis, ADEM, and Guillain-Barré Syndrome (Bautista, 2019).
Despite all the description of neurological associated manifestations of arboviruses, the neurotropism of viruses such as MAYV and OROV is still elusive. CHIKV and ZIKV can infect central nervous system (CNS) cells such as: neurons, astrocytes, and microglia (Klein et al., 2019). Astrocytes play a pivotal role in the CNS and are highly associated with encephalitis caused by those viruses (Soung and Klein, 2018). Astrocytes are responsible for the maintenance of the Blood-Brain Barrier (BBB) interacting with the brain microvascular endothelial cells (BMECs) (Cardoso et al., 2010), secretion of proinflammatory cytokines (Voet et al., 2019), recruitment of T cells upon brain injury or infection (Zhang et al., 2008), regulation of synapses, and regulation of glutamate uptake (Olsen et al., 2015). Viral infections can induce apoptosis in neuronal cells and dysregulate astrocyte functions increasing the BBB permeability and neuroinvasion by up-regulating metalloproteinases expression (Wang et al., 2008; Ju et al., 2009; Yang et al., 2012), promoting injury derived from long-term astrocytic inflammatory response (Garber et al., 2018), and neurotoxicity by accumulation of extracellular glutamate (Cisneros and Ghorpade, 2012), which all together contributes to the encephalitis process.
Here, we present a comparative transcriptome study of human primary astrocytes cells infected with CHIKV, MAYV, OROV, or ZIKV describing the specific genes and pathways modulated by each virus and the integration of common pathways by system biology strategy. Human primary astrocytes were highly permissive to all viruses with consequent down-modulation of IFN and interferon-stimulated genes (ISGs) pathways as a viral escape strategy to the innate antiviral immune response.
Materials and Methods
Primary Human Astrocyte Isolation
Adult primary human astrocytes were isolated from surgically resected anterior temporal lobe tissue, from patients selected for surgical treatment of temporal lobe epilepsy associated with hippocampal sclerosis. The selected patients were evaluated by video-electroencephalography monitoring with a 132-channel Nihon-Kohden® apparatus, and the ictal onset zone was concordant with neuroimaging and semiology data. The pathological tissue targeted in surgery for these cases is the gliotic hippocampus, and the anterior temporal lobe resection is used merely as a surgical pathway to the diseased area. All patients gave written consent to the study, and the procedures were in agreement with the Brazilian Ministry of Health Ethics Committee (Certification of Presentation for Ethical Approval, CAAE, submission number 69409617.9.0000.5258, decision number 2.792.114). As previously described (Sebollela et al., 2012), only healthy cortical tissue was used to produce astrocyte cultures. Experimental protocols were performed as described previously (Diniz et al., 2012; Dezonne et al., 2017). Briefly, tissues were washed in Dulbecco’s Modified Eagle’s Mediu (DMEM), mechanically dissociated, chopped into small (<2 mm3) pieces with a sterile scalpel, and incubated in 10 ml of 0.25% trypsin solution at 37°C for 10 min. After centrifugation for 10 min, the cell pellet was resuspended in DMEM/F-12 growth medium supplemented with 10% Fetal Calf Serum (FCS) and plated onto tissue culture plates in a humidified 5% CO2, 95% air atmosphere at 37°C for 2 h in order to achieve adherence of microglial cells. The non-adherent astrocytes were transferred to other culture plates, previously coated with poly-L-lysine. Adherent astrocytes were allowed to grow by replacing the medium once a week. New passages of cells were generated by harvesting confluent astrocyte cultures using trypsin-EDTA (Ethylenediamine tetraacetic acid) solution (0.25% trypsin with EDTA; Thermo Fisher Scientific). Human astrocytes from up to the seventh passage and from two different donors were used in the study.
Cell Lines, Viruses, and Infections
Vero cells (ATCC, CCL-81) were maintained in DMEM (Gibco) and primary astrocytes were maintained in DMEM/F12 (Gibco), both supplemented with 10% v/v Fetal Bovine Serum (FBS) (Gibco) and 1% v/v of penicillin-streptomycin (10.000 U/ml-10.000 µg/ml) (Gibco). DMEM/F12 was also supplemented with 1% v/v of glutamine (200 mM, Gibco), 0.6% g/v of glucose (Sigma-Aldrich), and 0.13% g/v of sodium bicarbonate (Sigma-Aldrich). All cells were incubated at 37°C and 5% CO2. CHIKV isolate BHI3745/H804709 (GenBank accession #KP164570.1), MAYV strain 4675 (ATCC 66, GenBank accession #MK070492.1), OROV strain BeAn19991 (GenBank accession #KP052852.1, #KP052851.1, #KP052850.1), and ZIKV strain PE/243 (GenBank accession #KX197192.1) were used to perform infection. Viral stocks used here are expressed as Plaque Forming Units/ml (CHIKV, 1.6 × 108 PFU/ml; MAYV, 7 × 107 PFU/ml; OROV, 2 × 106 PFU/ml; and ZIKV, 2.6 × 106 PFU/ml) and were propagated by serial passages in Vero cells by routine methods using DMEM. Astrocyte infections were performed at multiplicity of infection (MOI) 1 (2 × 105 cells/well) for all four viruses in 12-well plate during 1 h at 37°C and 5% CO2 in medium without FBS, under biosafety level 3 conditions at a Biosafety Level-3 laboratory at Federal University of Rio de Janeiro. For each infection, uninfected cells from the same donor were used in a pairwise experiment. Virus titration was performed by plaque assay in Vero cells plated at 3 × 105 cells/well in 12-well plates 1 day prior to infection. After 1 h incubation with the virus, cells medium was replenished by DMEM supplemented with 1% v/v FBS, 1% v/v antibiotics, and 1% v/v carboxymethyl cellulose (CMC) (Sigma-Aldrich), and incubated at 37°C and 5% CO2 during 2 days for CHKV and MAYV, and 5 days for OROV and ZIKV. Cells were fixed with 4% formaldehyde for 20 min at room temperature, washed in Phosphate Buffered Saline (PBS) (Gibco), and stained with 20% v/v ethanol-violet crystal solution for 15 min for visual counting. Astrocytes viability assay upon virus infection was evaluated by CellTiter-Blue (Promega) according to manufacturer’s instructions. The fluorescence was measured at SpectraMax Paradigm Multi-Mode Detection Platform (Molecular Devices).
Flow Cytometry, Immunofluorescence, and Antibodies
Virus infectivity was evaluated using specific antibodies against viral proteins as follows. Briefly, astrocytes (2 × 105 cells/sample) were fixed with 4% paraformaldehyde for 20 min and permeabilized in 1% v/v Triton X-100 PBS solution. Blocking was performed in 5% v/v Donkey Serum (Sigma-Aldrich) PBS solution for 1h at 37°C. Primary antibodies for each virus were diluted as follows: for CHIKV was used 1:50 of mouse monoclonal J31F (Thermo Fisher Scientific), for MAYV was used 1:300 of mouse monoclonal anti-alphavirus E1 antibody (MAB8754, Sigma-Aldrich), for OROV was used 1:300 of mouse polyclonal anti-OROV antibody, and for ZIKV was used 1:300 of mouse monoclonal anti-NS1 antibody (Abcam, ab218546), all diluted in blocking solution at 37°C for 30 min. Cells were then washed thrice in PBS and incubated with 2 µg/ml Donkey anti-mouse AlexaFluor 488 secondary antibody (Thermo Fisher Scientific) at 37°C for 30 min. After incubation with the secondary antibody, cells were washed and resuspended in PBS. Flow cytometry was performed using Accuri C6 cytometer (BD Biosciences). At least 10,000 gated events were counted per experimental replica at Fluorescein isothiocyanate (FITC) channel. Astrocytes specific markers were confirmed by immunofluorescence. For that, cells were fixed and permeabilized as described above, followed by blocking in PBS 3% Bovine Serum Albumin (BSA) (Sigma-Aldrich) and 5% Normal Goat Serum (NGS) (Thermo Fisher Scientific) for 1 h and followed by overnight incubation at 4°C with the primary antibodies. Primary antibodies were diluted in block solution as follows: 1:500 of rabbit anti-GFAP (Glial Fibrillary Acidic Protein) (Dako, Glostrup, DK), 1:200 of mouse anti-GS (Glutamine Synthetase) (Merck-Milipore), 1:200 of rabbit anti-GLAST(Glutamate Aspartate Transporter 1) (Abcam), and 1:200 of rabbit anti-GLT1 (Glutamate Transporter 1) (Abcam). Cells were washed with PBS and incubated for 2 h with 1:400 dilution of goat anti-rabbit or anti-mouse IgG AlexaFluor 488, or 1:1,000 dilution of goat anti-rabbit IgG AlexaFluor 546 secondary antibodies (Thermo Fisher Scientific). Nuclei was stained with 4’,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI) (Sigma-Aldrich). Cells were mounted with mounting medium (DakoCytomation) and imaged under a confocal microscope (Leica TCS SPE).
RNA Isolation, Quality Assessment, and RNA-Seq
Astrocytes were plated at 2 × 105 cells/well and infected at MOI 1 with either CHIKV, MAYV, OROV, or ZIKV. Eighteen h post-infection (hpi) for CHIKV, MAYV, and OROV or 48 hpi for ZIKV, cells were washed with PBS, trypsinized, centrifuged, and the pellet were used for RNA extraction. Six replicates of non-infected astrocytes and three replicates of infected astrocytes were used for each virus. Total cellular RNA was isolated using RNeasy Mini kit (Qiagen) according to manufacturer’s instructions. RNA quantification was performed using Qubit RNA HS Assay kit and Qubit 3 Fluorometer (Thermo Fisher Scientific). Genomic DNA contamination was avoided by DNase treatment (TURBO DNA-free Kit, Thermo Fisher Scientific) before RNA-Seq and RT-qPCR validation. Only samples with RNA integrity number (RIN) ≥ 9 were used, as verified by RNA 6000 Pico Kit and 2100 Bioanalyzer (Agilent Technologies). Ribossomal RNA was depleted using Ribo-Zero Gold (Illumina) and 200 ng of total RNA for each sample were used for library preparation with TruSeq Stranded Total RNA Library Prep (Illumina) according to manufacturer’s instructions. RNA-seq were performed in 2 × 9 samples using NextSeq 500/550 High Output v2 Kit (150 cycles) (Illumina) in a NextSeq 550 platform (Illumina).
RNA-Seq Statistical Analysis
The RNA sequence files (fastq) were applied to the software BBDuk2 (https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-user-guide/bbduk-guide) to trim low quality-related sequences (bellow Q30) and remove Illumina adapter-contaminant tags. The FASTQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc) reports the overall quality of the sequencing. The aligner HiSat2 (Kim et al., 2015) mapped the filtered reads to the human genome assembly GRCh38.p12 presented at the Ensembl database (https://www.ensembl.org/Homo_sapiens). The R/Bioconductor package Rsubread using the function of feature counts (Liao et al., 2019) performed the counting table of the mapped reads for the following statistical analysis. The R/Bioconductor package DESeq2 (Love et al., 2014) conducts the differential gene expression (DGE) test. It is also applied to the R/Bioconductor package apeglm (Zhu et al., 2019) to shrink log-fold change. The criteria of the adjusted p-value (corrected by false discovery rate—FDR) below 0.05 and log2 fold change (FC) above the absolute value of 1.0 was followed to classify genes as differentially expressed. The GOstats (Falcon and Gentleman, 2007) performed the Gene Ontology (GO) enrichment analysis. The package Pathview (Luo and Brouwer, 2013) visualizes the KEGG pathways and shows the associated genes. The ReactomePA package (Yu and He, 2016) leads the Reactome database analysis applying enrichment and allowing to create graphics. All the results and additional information about the RNAseq analysis were stored in the relational database PostgreSQL 12. The data can be easily accessed at the site: http://biotools.labinfo.lncc.br/astrocitovirus_data. The dataset used in our study is publicly available in SRA-NCBI (www.ncbi.nlm.nih.gov/sra), SRA accession PRJNA662366.
RT-qPCR Validation
Cells were seeded (2 × 105 cells/sample) and infected at MOI 1 as described above. The RNA was extracted and treated as described above at the indicated times points post infection and cDNA was produced using High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) and 500 ng of RNA. Quantitative PCR was performed in three biological replicates per condition using 20 ng of RNA input and SYBR Green PCR Master Mix (Applied Biosystems) on 7500 Fast Real-Time PCR System (Thermo Fisher Scientific) according to manufacturer’s instructions. Hypoxanthine Phosphoribosyltransferase 1 (HPRT1) gene was used as normalization and pre-designed PrimeTime primers (Integrated DNA Technologies) were used for selected target genes according to manufacturer’s instruction (for primers sequences see Table S1). Normalized gene expression was compared between experimental and mock non-infected conditions using Mann-Whitney test. Fold change and p-value for each comparison are presented using volcano plots.
Results
Human Primary Astrocytes Are Permissive to Different Arboviruses Infection
In order to investigate the neuropathogenesis of arboviruses, we infected human cortical primary astrocytes (hAST) with CHIKV, MAYV, OROV, or ZIKV due to those cells multiple roles in maintaining the homeostasis of CNS and respond to infections. We confirmed the expression of classical astrocytes markers such as: glial protein (GFAP), glutamine synthetase (GS), and glutamate transporters (GLT1 and GLAST); which corroborates the same previously demonstrated astrocytic phenotype derived from induced pluripotent stem cells (iPSC) (Figure 1A) (Sebollela et al., 2012; Diniz et al., 2014; Garcia et al., 2014; Dezonne et al., 2017; Eugenio et al., 2018; da Silva et al., 2019). No cytopathic effects were observed in response to CHIKV, MAYV, and OROV until 48 h post infection (hpi) and until 72 h post ZIKV infection (Figure 1B). We also assured that at least 80% of cells were infected in that time span at MOI 1, as demonstrated by flow cytometry using antibodies against each virus protein (Figure 1C). Flaviviruses as ZIKV presents a slow replication rate compared to alphaviruses such as CHIKV and MAYV, taking more time to reach the same levels of replication. Even in the earliest time point (i.e., 6 hpi for CHIKV, MAYV, and OROV, and 24 hpi for ZIKV) 80% of cells were infected with all viruses tested, reinforcing that hAST is highly susceptible to different arboviruses infections, including MAYV and OROV, which have never been described before. To assure that infections were productive in astrocytes cells, we quantified the viral titer derived from the supernatant of infected cells. All arboviruses were able to produce infectious particles as evaluated by plaque assay (Figure 1D). Alphaviruses infection were more productive in those cells, followed by Orthobunyavirus and Flavivirus, respectively. CHIKV produced the highest titer: 1.52 × 106 PFU/ml as soon as 6 hpi, peaking 1.24 × 107 PFU/ml at 18 hpi; followed by MAYV and OROV, respectively. Again, ZIKV took additional 24 h to reach the same levels of virus production compared to the other viruses, confirming its lower replication rate in hAST compared with CHIKV, MAYV, and OROV (Figure 1D). With exception of ZIKV (Hamel et al., 2017; Kozak et al., 2017; Meertens et al., 2017; Chen et al., 2018; Stefanik et al., 2018; Sher et al., 2019), this is the first study to demonstrate the capacity of CHIKV, MAYV, and OROV to infect hAST, corroborating their potential as agents of neuro-associated outcomes.
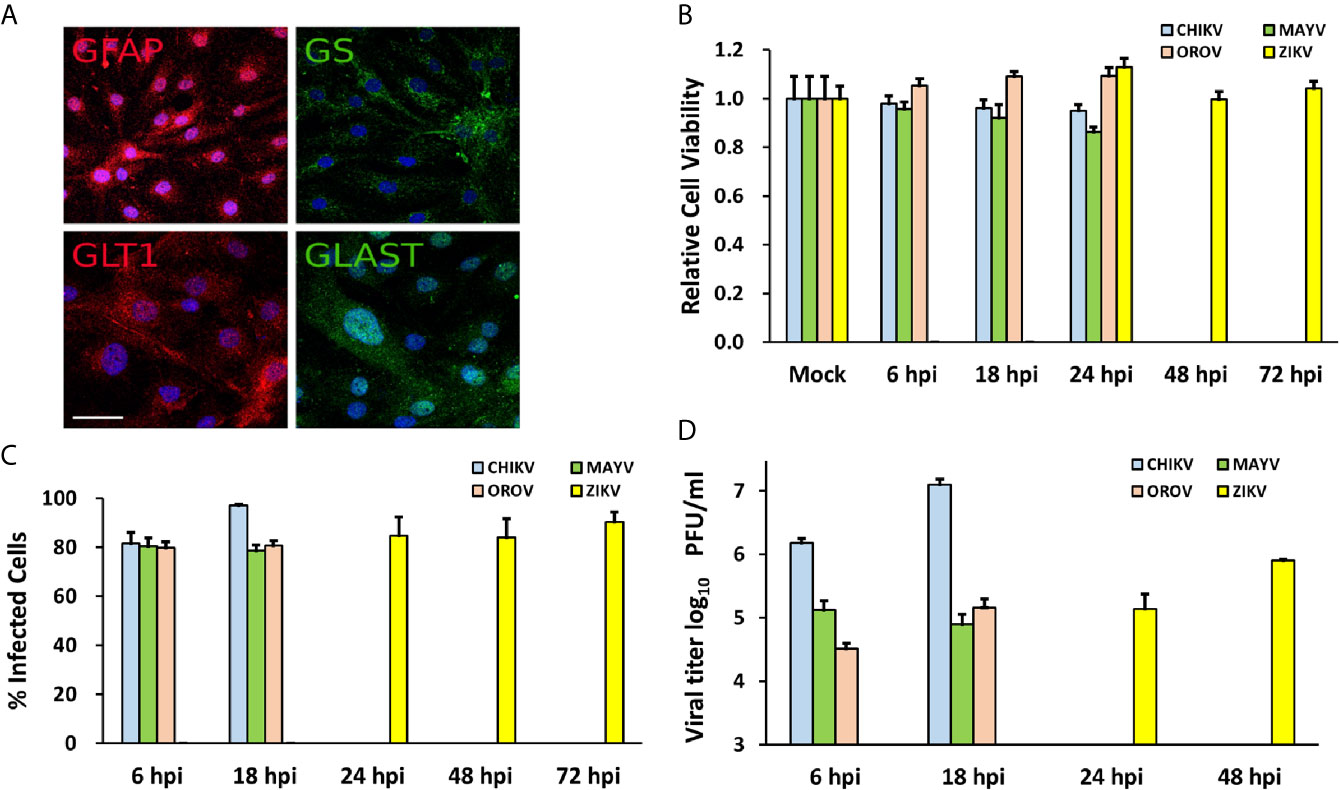
Figure 1 Human primary astrocytes are permissive to CHIKV, MAYV, OROV, and ZIKV infection. Human primary astrocytes were isolated from brain surgery and infected with CHIKV, MAYV, OROV, or ZIKV at MOI 1 in different time points. (A) Immunofluorescence microscopy showing astrocytes specific cell markers: GFAP, Glial Fibrillary Acidic Protein, and GLT1, Glutamate Transporter 1 (both in red); GS, Glutamine Synthetase, and GLAST, Glutamate Aspartate Transporter 1 (both in green). Nuclei are stained with DAPI (cyan). Scale bar = 50 µm. (B) Cell viability post infection was measured at indicated time points. All infected cells were normalized by mock (uninfected) cells. (C) Virus infectivity was measured by flow cytometry using primary antibodies for virus proteins: CHIKV (J31F, anti-E1 antibody); MAYV (anti-alphavirus E1 antibody); OROV (anti-OROV serum), and ZIKV (anti-NS1 antibody) and plotted as percentage of infected cells in gated population. 10.000 events were recorded. (D) Viral production measured by plaque assay from the supernatants of infected cells. Viral titers were plotted as log10 Plaque Forming Units/ml. All the experiments were performed on three replicates from two independent experiments presented here as media and standard deviation (SD).
Alphaviruses (CHIKV and MAYV) Replicate Better in Human Astrocytes Cells and Modulate a Higher Number of Cellular Genes
To analyze host cellular genes and pathways modulated upon infection, we performed a transcriptome-wide sequencing of uninfected (n = 6) and infected hAST with each virus (n = 3). Transcriptome analyses were performed at 18 hpi for CHIKV, MAYV, and OROV and 48 hpi for ZIKV, following the infection peak and absence of cytopathic effect for each virus (Figures 1B, C). We found an increasing number of RNA-seq reads that mapped on CHIKV and MAYV genomes compared with OROV and ZIKV suggesting that alphaviruses present higher replication rates in hAST (Figure 2A). As a reflection of higher titers of viral replication for alphaviruses, we found an increasing number of cellular genes dysregulated by CHIKV and MAYV compared with OROV and ZIKV (Figure 2B and Table S2).
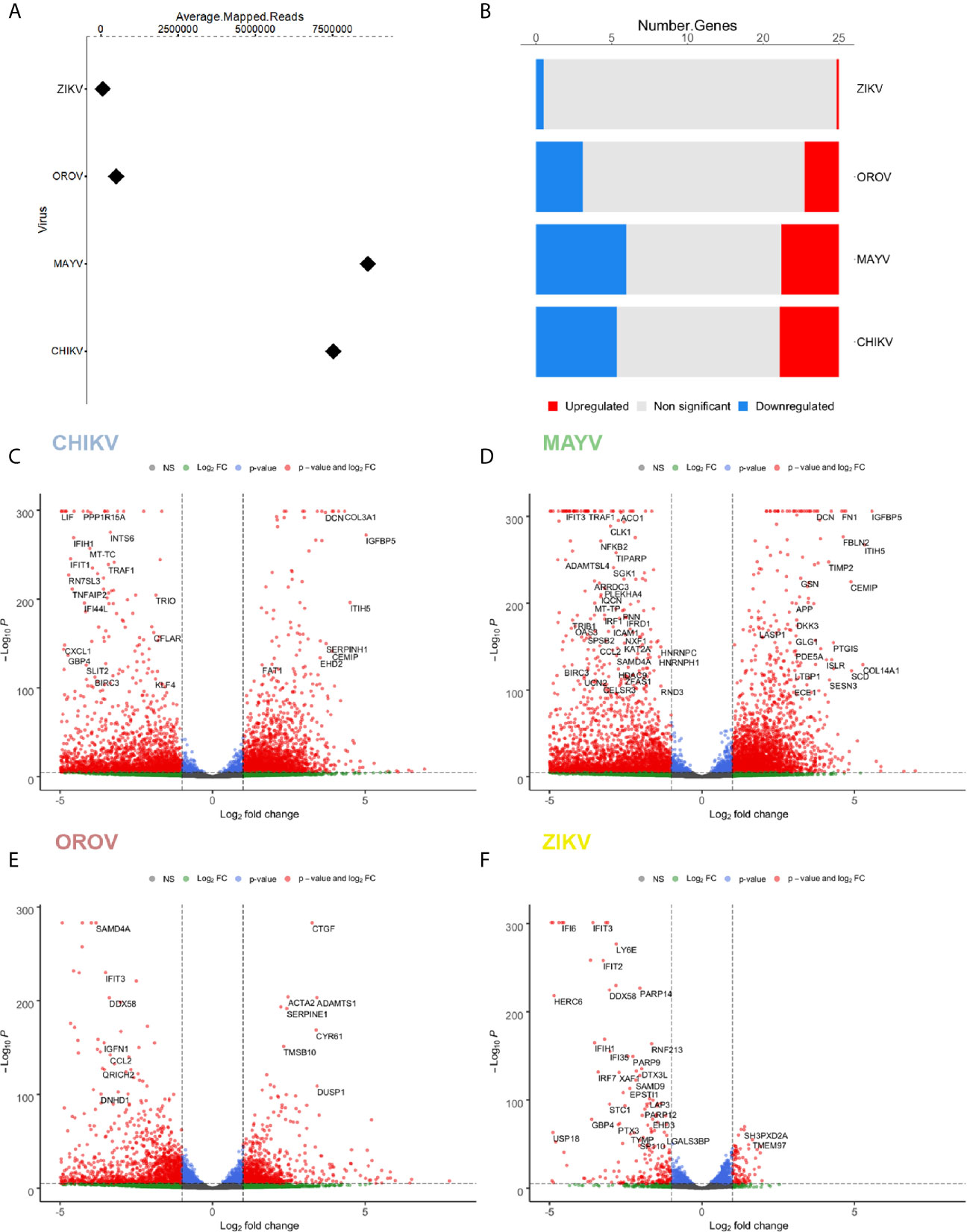
Figure 2 Differentially expressed genes (DEGs) signature for CHIKV, MAYV, OROV, and ZIKV in human primary astrocytes. Signature of DEGs detected in RNA-Seq of infected (n = 3) over uninfected (n=6) cells. (A) Relative abundance (average of mapped reads) of viral RNA in hAST infected with CHIKV, MAYV, OROV, and ZIKV. (B) Number of up- and down-regulated genes for CHIKV, MAYV, OROV, and ZIKV with statistical significance (p ≤ 0.05 and at least 1 log2FC dysregulation over uninfected cells). Volcano plots for the differentially expressed genes (DEGs) for CHIKV (C), MAYV (D), OROV (E), and ZIKV (F). RNA-seq from triplicates were performed from CHIKV, MAYV, and OROV infected cells 18 hpi and ZIKV infected cells 48 hpi. X-axis represents the log2 Fold Change of DEG relative to uninfected cells and Y-axis depicts significance. Hpi, hours post infection.
Briefly, we found 3578, 4104, 2056, and 133 up-regulated genes for CHIKV, MAYV, OROV, and ZIKV infected cells, respectively, in contrast with 4883, 6388, 2802, and 415 down-regulated genes (p ≤ 0.05 and at least 1 log2FC dysregulation over uninfected cells) (Figures 2C–F). Those results reflect an increasing impact in the modulation of differentially expressed genes (DEGs) that corroborates with the crescent genus-dependent viral replication as follows: flavivirus, orthobunyavirus, and alphaviruses. Altogether, the four viruses interfere with astrocyte gene expression down-regulating a higher number of genes compared with up-regulated genes (Figures 2C–F and Table S2).
CHIKV Interferes With Extracellular Matrix, N-Glycosylation, Translation, Cell Cycle, and Interferon Pathways
Our reactome analysis showed that CHIKV infected astrocytes presented an enrichment of up-regulated DEGs in pathways related to protein translation, post-translational modification of glycoproteins (N-linked glycosylation and metabolism of aminoacid), remodeling of extracellular matrix, protein translation, and targeting to endoplasmic reticulum (ER) membrane (Figure 3A and Table S3). The most significant genes associated with those pathways were: extracellular matrix/elastic fiber formation, as fibronectin 1 (FN1), fibulin (EGF Containing Fibulin Extracellular Matrix Protein 1, EFEMP1; Fibulin 1, FBLN), and elastin (Elastin Microfibril Interfacer 2, EMILIN2); amino acid activation, including aminoacyl-tRNA synthetases (Seryl-tRNA Synthetase, SARS; Bifunctional Aminoacyll-tRNA Synthetase, EPRS; Glycyl-tRNA Synthetase 1, GARS; Alanyl-tRNA Synthetase 1, AARS; Tyrosyl-Trna Synthetase 1, YARS); ER associated translation (Translocation Associated Membrane Protein 1, TRAM1; Protein Disulfide Isomerase Family A Member 3, PDIA3; Calreticulin, CALR; KDEL Endoplasmic Reticulum Protein Retention Receptor 1, KDELR1); and glycosylation (LEM Domain Containing 3, MAN1; Beta-1,4-Mannosyl-Glycoprotein 4-Beta-N-Acetylglucosaminyltransferase, MGAT3; Dolichyl-Diphosphooligosaccharide–Protein Glycosyltransferase Non-Catalytic Subunit, DDOST; Alpha-1,3-Mannosyl-Glycoprotein 2-Beta-N-Acetylglucosaminyltransferase, MGAT1). Translation factors are usually modulated by RNA viruses to promote viral protein production (Au and Jan, 2014). Indeed, the glycosylation of virus envelope proteins are crucial to increase CHIKV virulence and the up-regulation of genes related to asparagine N-linked glycosylation reinforces this hypothesis (Acharya et al., 2015; Lancaster et al., 2016). Cytoskeleton tubulin isoforms predominantly found in differentiated neurologic cells (Tubulin Alpha 1a, TUBA1A) and transmembrane vesicular protein involved in traffic (Transmembrane P24 Trafficking Protein 10, TMED10) were highly induced by CHIKV (Figure 3A), showing the importance of glycosylation pathways not only for envelope proteins, but also in virus assembly and budding.
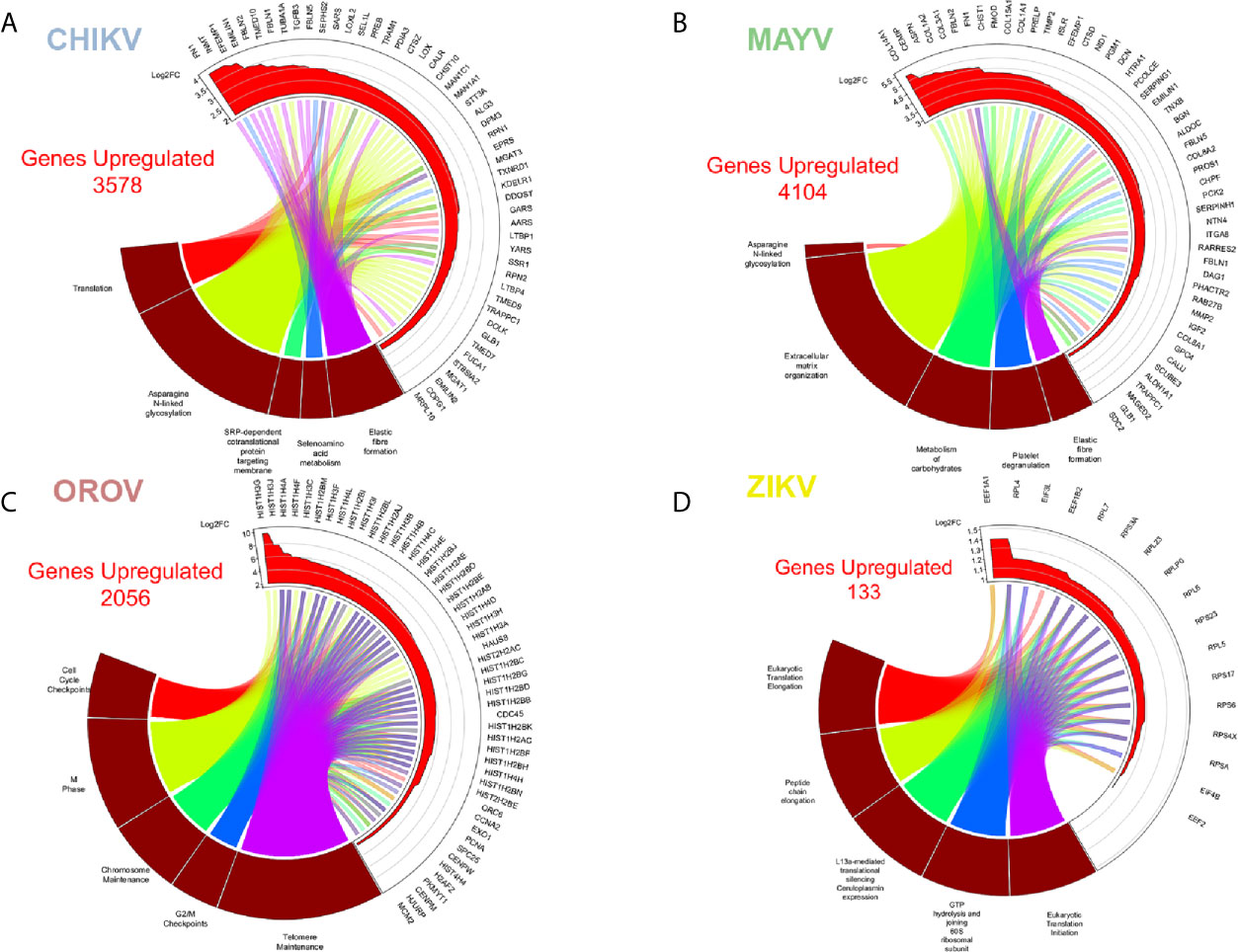
Figure 3 Enriched up-regulated pathways in CHIKV, MAYV, OROV, and ZIKV infected astrocytes. Cell pathways with up-regulated genes in (A) CHIKV (18 hpi), (B) MAYV (18 hpi), (C) OROV (18 hpi), and (D) ZIKV (48 hpi) infected cells. Circus plot of Gene Set Enrichment Analysis (GSEA) of the five up-regulated most significantly enriched Reactome pathways (dark red sectors at the bottom of the circus plot) with the 50 associated DEGs. The 20 most significant up-regulated pathways are depicted. In the upper sector of the circus plot the genes were ordered by modulus of log2 Fold Change relative to uninfected cells.
CHIKV down-regulated pathways in astrocytes included: chromosome stability, DNA repair and maintenance; and cell cycle (in special, nucleosome assembly and prometaphase directed steps) (Figure 4A and Table S4). It is known that viruses always interfere with cell cycle to increase their own replication, promoting virus translation and avoiding apoptosis (Bressy et al., 2019; Caller et al., 2019). In fact, our reactome analysis of down-regulated pathways showed a consistent relationship within cell cycle controlling genes, indicating a possible G2 arrest phenotype (Figure 4A). Among the down-regulated genes were: genes from the cell division processes, as centrosome, centromere, and tubulin associated genes (Centrosomal Protein, CEPs; Centromere Protein, CENPs; and Tubulin, TUBs); cell cycle (Cell Division Cycle, CDCs); and chromosome stability, such as histones (HISTs) and DNA polymerase subunits (POLs) (Figure 4A). This goes in agreement with previous reports of CHIKV interfering with cell cycle in other cell models (Saxena et al., 2013; Thio et al., 2013). The Aurora Kinase B (AURKB), a chromosome segregation and cytokinesis promoting gene, was the most down-regulated gene in CHIKV infected cells (Figure 4A). Although the role of AURKB depletion has been controversially described for flaviviruses (Madejón et al., 2015; Pérez-Olais et al., 2019), its impact remains elusive for alphaviruses; nonetheless, it could be speculated as a reinforcer of G2 arrest.
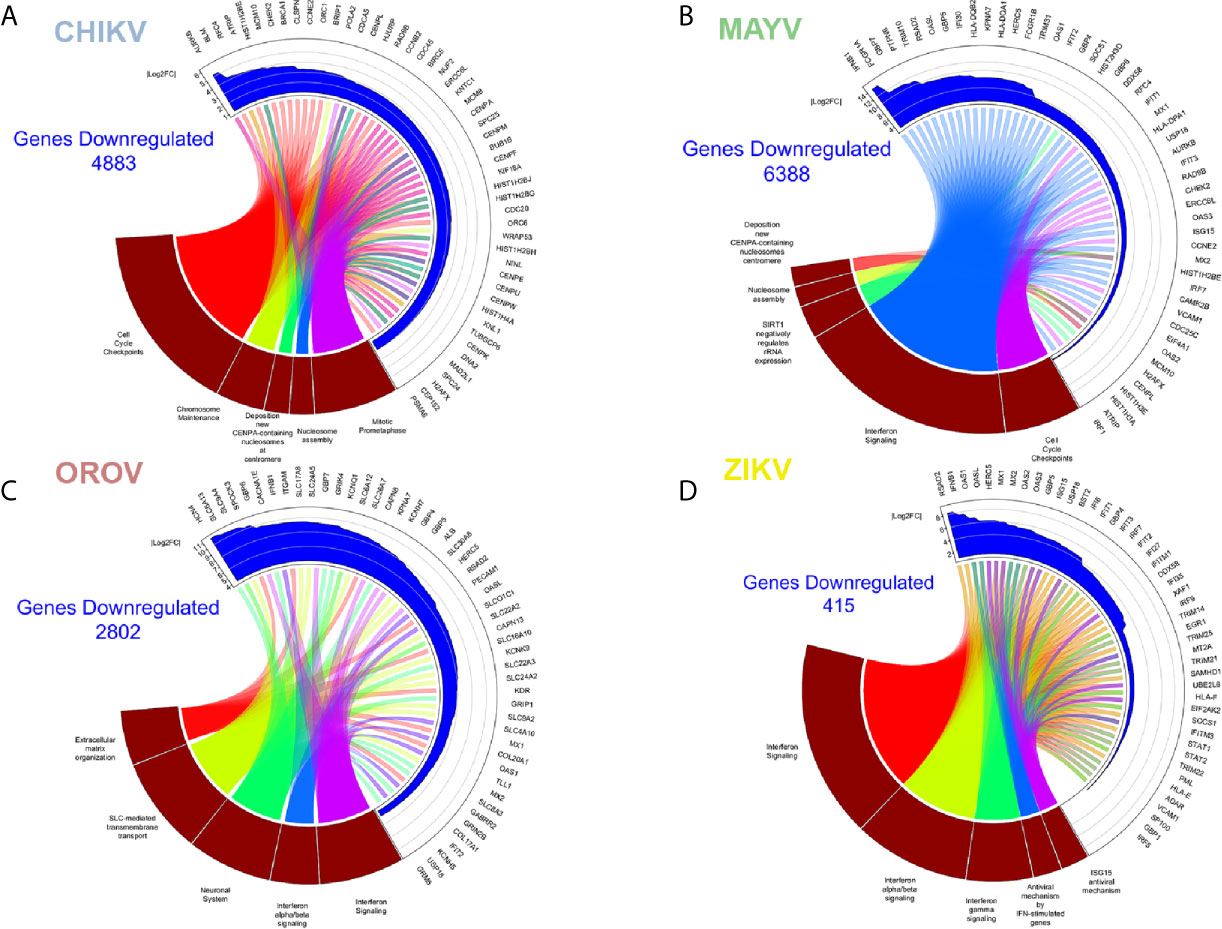
Figure 4 Enriched down-regulated pathways in CHIKV, MAYV, OROV, and ZIKV infected astrocytes. Cell pathways with down-regulated genes in (A) CHIKV (18 hpi), (B) MAYV (18 hpi), (C) OROV (18 hpi), and (D) ZIKV (48 hpi) infected cells. Circus plot of Gene Set Enrichment Analysis (GSEA) of the five down-regulated most significantly enriched Reactome pathways (dark red sectors at the bottom of the circus plot) with the 50 associated DEGs. The 20 most significant down-regulated pathways are depicted. In the upper sector of the circus plot the genes were ordered by modulus of log2 Fold Change relative to uninfected cells.
MAYV Dysregulates the Same Profile of CHIKV Associated Pathways, With Prominent Down-Modulation of Interferon Response Genes
MAYV infection led to an enrichment and up-regulation of elastic fiber formation and N-glycosylation pathways, as in CHIKV infection; carbohydrates metabolism, platelet degranulation, and extracellular matrix organization (Figure 3B). The gene interactome modulated by MAYV is highly heterogeneous compared with CHIKV. Nonetheless, many genes related to elastic fiber/extracellular matrix pathways as integrins (ITGA8), fibronectin (FN1, also highly up-regulated in CHIKV infection), collagens (COLs), and metalloproteinases (MMP2) were up-regulated by MAYV (Figure 3B). Metalloproteinases are involved in extracellular matrix degradation and are associated with neurological outcomes in viral infections (Savarin et al., 2011; Shukla et al., 2016; Xing et al., 2017; Barkhash et al., 2018; Song et al., 2018; Chopra et al., 2019).
MAYV shares common down-modulated pathways with CHIKV, such as as Type-I interferon signaling (Table S2), nucleossome assembly and formation, and cell cycle (Figure 4B and Table S4). Reactome analysis showed two main clusters: (i) one related to interferon signaling and innate immunity, and (ii) another one related to pathways associated to cell cycle arrest, and repression of DNA replication and cell division (Figure 4B). MAYV and CHIKV, both alphaviruses, down-regulated common genes related to cell cycle and chromosome stability, such as CENPs, HISTs, and CDCs (Figures 4A, B). Indeed, interferon (IFNB1), interferon stimulated genes—ISGs (Interferon Induced Protein With Tetratricopeptide Repeats, IFITs; Radical S-Adenosyl Methionine Domain Containing 2, RSAD2; MX Dynamin Like GTPase 1, MX1; 2’-5’-Oligoadenylate Synthetase, OASs; Tripartite Motif Containing, TRIMs) and agonists of interferon response (DExD/H-Box Helicase 58, DDX58; Interferon Regulatory Factor 7, IRF7; Interferon Regulatory Factor 1, IRF1) were represented among the most down-regulated genes by MAYV, with higher fold change values, up to 12 (IFNB1) (Figure 4B).
OROV Infection Interferes With Ion Transport, Neuronal Synapses Regulation and Type-I Interferon Pathways, Contributing to Neuropathogenesis
OROV infection in human astrocytes, contrasting with alphaviruses infection, up-regulated pathways related to cell cycle, cell division, DNA maintenance and replication, and extracellular matrix remodeling (Figure 3C and Table S3). The opposite direction of pathways regulated by Alphavirus and Orthobunyavirus demonstrates the complexity and diversity in host factors modulation and may represent different strategies of virus replication. The pathway reactome corroborates the overall tendency in OROV infection, up-regulating pathways related to DNA condensation (mainly through the core H2, H3, and H4 histones), cell cycle checkpoints and G2 arrest, and telomere maintenance (Figure 3C and Table S3). Some proteins from related orthobunyaviruses were already demonstrated to have nuclear localization, interfering with cell cycle and hampering global cellular transcription through histone modification (Copeland et al., 2013; Gouzil et al., 2017). The genes in the reactome corroborated this hypothesis, as many histones, CENPs, and other G2/M step related proteins were up-regulated. The G2/M arrest could increase the availability of capped-mRNAs, which serves as primer donors for bunyaviruses translation: a process named cap-snatching. Corroborating our findings, and in contrast with bunyaviruses, alphaviruses can cap their own RNAs using nsP activities (Hopkins and Cherry, 2013) (Figure 3C).
Overall, the pathways found down-regulated by OROV in astrocytes corroborate the dysregulation of interferon response; highlighting genes with antiviral activities (RSAD2, also known as viperin; OASL; OAS1; MX1/2; IFIT2). However, unlike alphaviruses, OROV infection also down-regulated extracellular matrix remodeling genes [such as SPARC (Osteonectin), Cwcv And Kazal Like Domains Proteoglycan 3, SPOCK3, and Integrin Subunit Alpha M, ITGAM] and collagen metabolism pathways (Collagen Type XVII Alpha 1 Chain, COL17A1, and Collagen Type XX Alpha 1 Chain, COL20A1) (Figure 4C and Table S4). Moreover, OROV infection also down-modulates ion channels, transmembrane solute carriers, and synapses regulation pathways that play crucial roles in astrocytes functions. Genes related to potassium channels (Potassium Voltage-Gated Channel, KCNs, and Hyperpolarization Activated Cyclic Nucleotide Gated Potassium Channel 4, HCN4) and solute transporters from the family of Solute Carrier (SLCs) were the most representative clusters (Figure 4C) down-modulated by OROV. Recently, it was demonstrated that K+ channels are up-regulated and therefore required for endosomal trafficking and high infectivity of bunyaviruses, corroborating our findings (Hover et al., 2016; Hover et al., 2018). SLC family of transporters includes Excitatory Amino Acid Transporter 1 (EAAT1, also known asSLC1A3/GLAST) and Excitatory Amino Acid Transporter 2 (EAAT2, also known as SLC1A2/GLT1), both glutamate transporters whose dysfunction have been implicated in increasing neurotoxicity driven by accumulation of extracellular glutamate (Legay et al., 2003; Wang et al., 2003; Moidunny et al., 2016). Those pathways, together with down-regulation of Type-I interferon signaling, could increase the neuropathological manifestations of OROV infection.
ZIKV Up-Regulated Eukaryotic and Viral Translation Pathways, and Down-Regulated Type-I Interferon Signaling Proteins
ZIKV infection modulated the least number of genes (133 up- and 415 down-regulated, respectively) compared with the other viruses. This observation is in agreement with the lower replication rates expected for flaviviruses compared with alphaviruses or orthobunyaviruses (Figures 1B–D). However, it is important to note that Asian derived strains (as the Brazilian one used here) have usually lower virulence and replication kinetics compared to African strains (Simonin et al., 2016; Hamel et al., 2017). Nonetheless, the most enriched up-regulated pathways upon ZIKV infection were viral mRNA translation, translation elongation, aminoacid synthesis, ribosomal assembly, and cap-dependent translation (Figure 3D and Table S3); all related to host translation machinery. All up-regulated genes were either eukaryotic initiation (EIFs) or elongation factors (EEFs), and ribosomal proteins (RPSsand RPLs) (Figure 3D). Most of the pathways down-regulated by ZIKV in astrocytes were related to interferon-induced antiviral innate immune response (Figure 4D and Table S4). The clusterization of the pathways classified them into type I and II interferon signaling, and antiviral interferon-responsive genes (Figure 4D). Transcriptional factors related to interferon pathways were also down-modulated (IRFs and Signal Transducer And Activator of Transcription, STATs), reinforcing the relevance of ISGs in controlling ZIKV infection. As demonstrated by other cellular models, most genes down-regulated by ZIKV were ISGs: RSAD2, IFITs, Guanylate Binding Protein (GBPs), MXs, and OASs (Figure 4D). Cytoplasmatic RNA sensor (DDX58, also known as Retinoic Acid Inducible Gene I, RIG-I) and classical interferon induced antiviral genes that direct block virus replication at several steps (SAM And HD Domain Containing Deoxynucleoside Triphosphate Triphosphohydrolase 1, SAMHD1: RNA virus replication; TRIMs: virus uncoating; and Bone Marrow Stromal Cell Antigen 2, BST2: virus budding, respectively) were also down-modulated by ZIKV infection. All this scenario suggests that ZIKV establishes stable infection in astrocytes counteracting and down-modulating interferon pathway at several steps.
Viral Infection Leads to an Overall Down-Regulation of Innate Immunity Genes in Astrocytes
We performed RNA-seq integration to identify common pathways modulated by the four viruses. Only 9 up-regulated and 171 down-regulated DEGs were co-modulated by CHIKV, OROV, MAYV, and ZIKA (Figures 5A, B, respectively and Table S5). As expected, CHIKV and MAYV shared a higher number of up- (2,450) and down- (2,528) regulated genes among themselves compared with other viruses analyzed, suggesting a signature for alphaviruses (Figures 5A, B). Gene ontology analysis of the 171 common down-regulated genes indicated that most of those genes belongs to pathways related to virus RNA sensing (RIG-1), interferon signaling (including ISGs), cytokines and chemokine signaling, JAK-STAT (Janus Kinase-STAT), and Toll receptors signaling, suggesting a global modulation of innate immune response and inflammatory response in primary astrocytes infected by RNA viruses (Figure 5C and Table S5). The alphaviruses CHIKV and MAYV also cluster together when analyzing only the common dysregulated genes for all viruses, as expected (Figure 5D). Deeper analysis of the virus RNA sensing and interferon signaling pathways down-modulated by the four viruses identified antiviral genes triggered by interferon stimulation (ISGs), such as: ISG15, MX1-2, DDX58, CT And RLD Domain Containing E3 Ubiquitin Protein Ligase 5 (HERC5), OAS, Interferon Induced Protein With Tetratricopeptide Repeats (IFIT), Adenosine Deaminase RNA Specific (ADAR), SAMHD1, Interferon Induced Transmembrane Protein 1 (IFITM1), and TRIMs; all those already described for other RNA and DNA viruses infections.
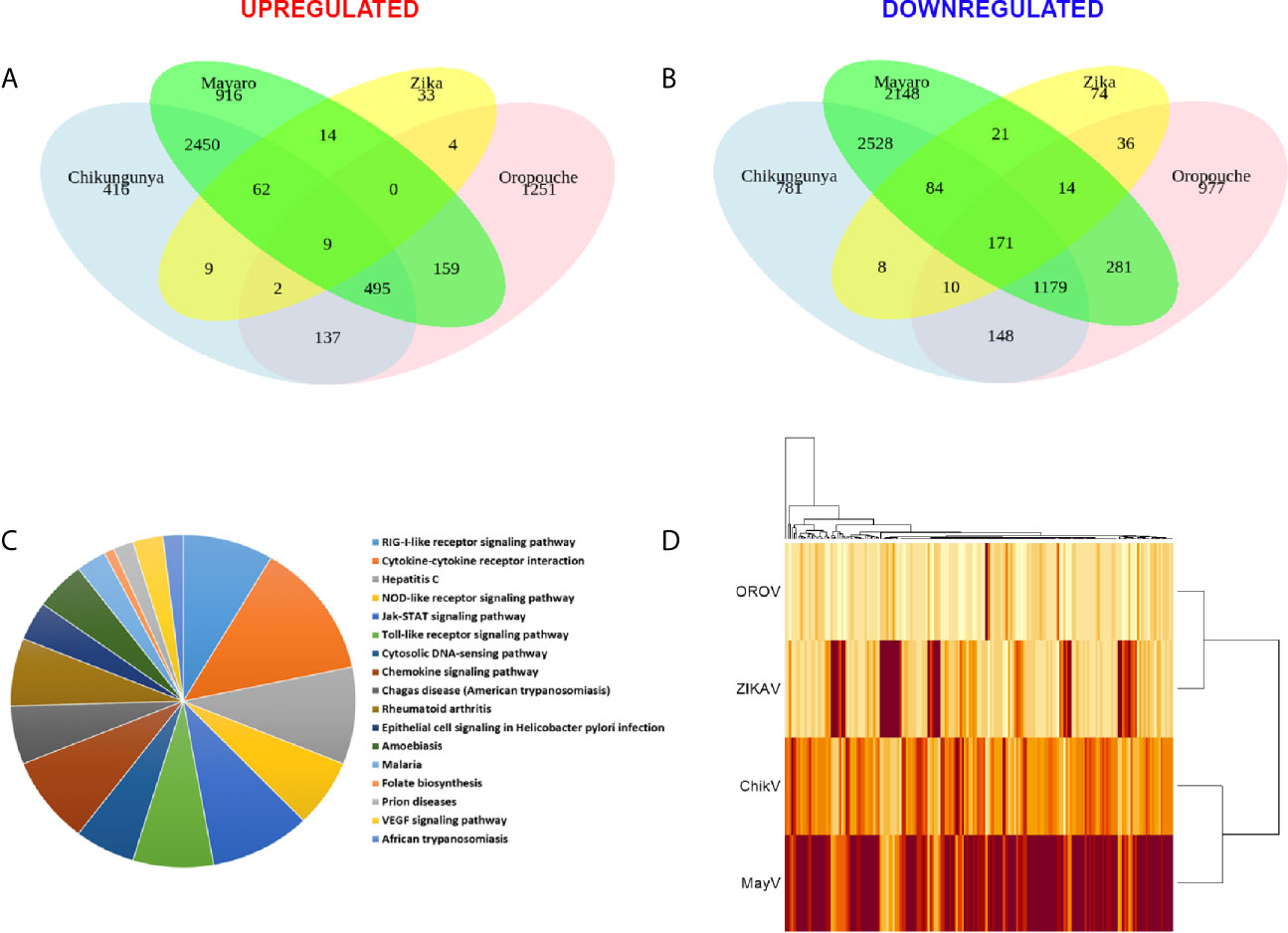
Figure 5 Common differentially expressed genes (DEGs) signature for CHIKV, MAYV, OROV, and ZIKV in human primary astrocytes. Signature of DEGs detected in RNA-Seq of infected (n = 3) over mock uninfected (n = 6) hAST cells. Venn diagram showing the quantity of common and exclusive up-regulated (A) and down-regulated (B) genes found in the RNA-Seq dataset. Only DEGs with at least 1 log2 Fold Change and an adjusted P value ≤ 0.05 are shown. (C) Gene ontology by biological processes of 171 common down-regulated genes by all viruses (CHIKV, MAYV, OROV, and ZIKV). (D) Heatmap of the common up-regulated (9) and down-regulated (171) genes among all viruses here included. Dendrogram of Euclidian hierarchical clusterization among the four viruses are shown. Color scale represents baseMean (estimative of abundance) of up-regulated and down-regulated genes (light yellow represents lower and brown represents higher abundance).
We performed RT-qPCR of 16 common genes dysregulated for all viruses in order to validate our RNA-seq data (Figure 6). We selected genes belonging to cytoskeleton and motility (Actin Alpha 2, ACTA2) (Rockey et al., 2019), autophagy (Decidual Protein Induced By Progesterone, DEPP1), L-serine synthesis (Phosphoserine Aminotransferase 1, PSAT1) (Jiang et al., 2020), inflammatory response (Interleukin 7 Receptor, IL7R, and Major Histocompatibility Complex, Class II, DO Beta, HLA-DOB) (Inchley et al., 2013; Alsadeq et al., 2018), axonal guidance and neuropathology (Semaphorin 3A, SEMA3A) (Niclou et al., 2003; Yang et al., 2019), interferon induction (DDX58, Interferon Induced With Helicase C Domain 1, IFIH1, TRIM25, and IRF7), and interferon responsive/agonist genes with antiviral activity (IFIT1, IFIT2, MX1, MX2, ISG15, and RSAD2) (García-Sastre, 2017; Schoggins, 2019).
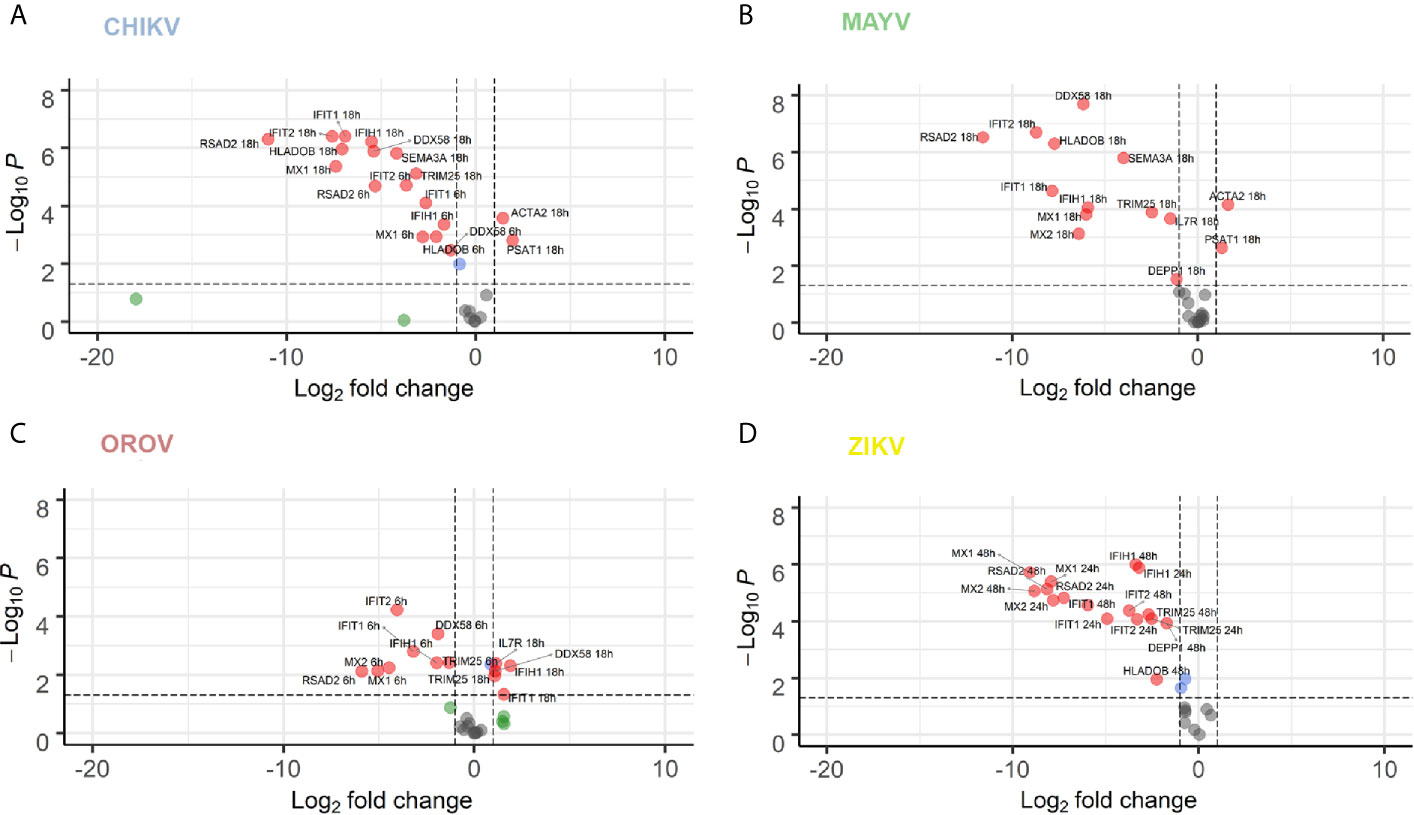
Figure 6 RT-qPCR validation of 16 selected DEGs co-modulated by CHIKV, MAYV, OROV, and ZIKV. hAST cells were infected with MOI 1 of CHIKV (A), MAYV (B), OROV (C) at 6 and 18 hpi and ZIKV (D) at 24 and 48 hpi and RT-qPCR was performed at indicated time points and compared to uninfected cells (n = 3 for each time point and condition). Data denotes p-values (y-axis) and mean fold change (x-axis) of infected cells relative to uninfected cells for 16 DEGs selected from commonly modulated genes in RNA-Seq dataset. Gene expression was normalized by endogenous HPRT1 levels. Red dots represent DEGs with at least 1 log2 Fold Change and P value ≤ 0.05. Blue dots represent DEGs with only P value ≤ 0.05. Green dots represent DEGs with at least 1 log2 Fold Change. Grey dots represent non-significant DEGs.
Overall, the majority of genes tested by RT-qPCR confirmed the dysregulation in expression found in our RNA-Seq data. To validate our findings, we also perform a kinetics of DEGs during virus infection at increasing time points. Most of the tested genes presented an increasing time-dependent down-regulation in later time points for each virus infection (6 and 18 hpi for CHIKV, MAYV, and OROV; 24 and 48 hpi for ZIKV) (Figures 6A–D, respectively). The only exceptions found were DDX58, IFIT, and IFIH1, that were down-modulated at early times of OROV infection and lose significance at later points. PSAT1 and ACTA2 were up-regulated in response to all viral infections, with higher induction in cells infected with CHIKV and MAYV at 18 hpi. Those genes are related to cell cytoskeleton and invasion, suggesting the virus dependence of intracellular trafficking to sustain replication. ISGs and immune response genes such as IFIT1, IFIT2, IFIH1, MX1, MX2, and RSAD2 were highly down-regulated in response to all viruses at later time points of infection, confirming our transcriptome data. The intracellular virus RNA sensor DDX58 (RIG-I) was down-regulated by all viruses, regardless the virus family, showing the central importance of this pathway in recognize viruses in infected cells and stimulate IFN and ISGs. Taken together, those results corroborate our transcriptome analysis showing that viruses from different families, such as CHIKV, MAYV, OROV, and ZIKV, interfere with antiviral and interferon mediated immune response to stablish an efficient and persistent infection in human primary astrocytic cells.
Discussion
Arboviruses from Togaviridae, Peribunyaviridae, and Flaviviridae families account for the most epidemics and public health system burden in tropical countries, mainly in South and Central Americas. Although the most common manifestations of acute infection involve self-limiting febrile illness with mild to moderate symptoms, in some cases those viruses can invade CNS and generate severe neurological disorders, some of them with mimicking phenotypes of inflammatory autoimmune nervous system diseases. Even so, the mechanisms of neuropathogenesis and host interaction remain poorly understood for most of them. Here, we presented a comprehensive genome-wide transcriptome analysis of human primary astrocytes infected with CHIKV, MAYV, OROV, or ZIKV.
The capacity to infect and sustain productive infection in astrocytes was previously demonstrated for ZIKV (Hamel et al., 2017; Kozak et al., 2017; Meertens et al., 2017; Chen et al., 2018; Stefanik et al., 2018; Sher et al., 2019). However, to date there was no report of the same capacity for CHIKV, MAYV, and OROV in human cells. Those viruses were not only capable of infecting but demonstrated a more prominent viral production than ZIKV (Figures 1C, D, and 2A). Those findings demonstrated that arthritogenic alphaviruses and OROV have also neurotropic potential like already described for encephalic alphaviruses (Ronca et al., 2016) and encephalic orthobunyaviruses (Evans and Peterson, 2019). Our RNA-Seq data showed an greater number of DEGs and different cellular pathways modulated by viruses presenting higher viremia since the beginning of infection (CHIK, MAYV e OROV), compared with lower replication rates of ZIKV in human primary astrocytes. Indeed, viruses from the same family, such as CHIKV and MAYV (Togaviridae), dysregulate a higher number of common genes and cellular pathways compared to OROV and ZIKV that belong to Peribunyaviridae and Flaviviridae families, respectively. Those results suggest that the virus genome RNA polarity (positive or negative) and replication strategy could differentially interfere with cellular pathways. However, the integration analysis showed a common signature of DEGs by those different viruses, reflecting the down-regulation of antiviral innate immune response and interferon pathway. This observation suggests that to stablish an efficient and productive infection in human primary astrocytes, viruses from different families need to counteract the cellular and immune response against the virus replication (Figure 5). DDHX58 (RIG-I), a cytosolic dsRNA-sensing protein, was consistently down-regulated among all viruses. Besides, IFN-β was the second most commonly down-regulated gene to all viral infections (Table S5). Transcription factors that control interferon induction such as IRFs, mainly IRF-7, was also consistently down-regulated. Since the upstream RIG-I along with IRFs were down-regulated, it is reasonable to estimate the suppression in IFN-β and ISGs expression (Figure 5 and Table S5). However, other RNA-Seq studies in mice (Wilson et al., 2017) and in human cohort (Soares-Schanoski et al., 2019) showed that CHIKV infection led to a global up-regulation in innate immunity and pro-inflammatory genes. The same was observed in neuroprogenitors cells infected with ZIKV (Lima et al., 2019). In case of ZIKV, though induced in neuroprogenitor cells, interferon, innate immunity, and inflammation were repressed in microglia (Tiwari et al., 2017) and dendritic cells (Sun et al., 2017), indicating cell type-specific dynamic in regulation of antiviral response. Aside of cell or tissue specific responses, another hypothesis that could explain contradictory innate immunity regulation among the datasets is the time-dependent effect on gene expression. In general, previous studies showed an induction of IFN pathways triggered by RIG-I recognition of virus double strand RNA structures at early points of RNA viruses infection, followed by antiviral and ISGs stimulation. Our results showed a common down-regulation of this pathway in later steps of CHIKV, MAYV, OROV, and ZIKV infection, possibly as a viral escape response to innate immune antiviral genes, allowing their replication in human astrocytic cells. A recent study using Vesicular Stomatitis Virus (VSV) as model proposed that a social evolution dictates the innate immunity evasion (Domingo-Calap et al., 2019). According to this model, IFN suppression is a costly mechanism, but it is evolutionary necessary to establish an efficient and productive infection. Due to high infectivity (about 80% of cell population) in astrocytes in a relative short time span (Figure 1B), it is likely that innate immunity suppression is occurring in early moments after infection. Among the various ISGs found, IFITs and MXs were also consistently down-regulated (Figure 6). From those, RSAD2, also known as Viperin, is a well characterized antiviral protein that already was demonstrated to restrict ZIKV (Panayiotou et al., 2018) and CHIKV (Teng et al., 2012), but its antiviral activity against MAYV and OROV was never described. Interestingly, in opposition to our results, in another study was demonstrated that RSAD2 as wells as IFN response is induced upon ZIKV infection in mice astrocytes (Lindqvist et al., 2016); however, RSAD2 regulation and its capacity to restrict viral replication in human astrocytes remain to be investigated.
Eukaryotic translation factors and ribosomal proteins were up-regulated upon ZIKV (Figure 3D), which reflects the viral recruitment of host translation machinery for viral protein synthesis (Walsh et al., 2013; Li, 2019). CHIKV and MAYV both up-regulated N-glycosylation machinery (Figures 3A, B), which, coupled with the up-regulation of translation machinery, suggests a multiple-step retargeting of host cell machinery towards viral protein synthesis and maturation (Acharya et al., 2015; Lancaster et al., 2016). Moreover, metalloproteinases, collagen and integrin genes were found up-regulated by CHIKV and MAYV infection but down-regulated in response to OROV infection (Figure 4C), suggesting opposite regulation of extracellular matrix and adhesion proteins between the two viral families. Regulation of CNS extracellular matrix by astrocytes is fundamental to neurogenesis, neuronal migration, and formation of new synapses (Cope and Gould, 2019); therefore, any unbalancing in the processes could lead to neuropathological effects.
Another interesting divergent regulation was observed between alphaviruses and OROV. Cyclin Dependent Kinase 1 (CDK1) and cyclins (CCNs) A, B, and E were down-regulated in CHIKV and MAYV infection, while they were up-regulated in OROV infection (Table S2). Those results suggest a G1 cell cycle arrest induced by alphaviruses, and a G2/M arrest for OROV. G1 arrest was already described to be beneficial to viral production (Lundberg et al., 2018; Wang et al., 2018), which corroborates the overall recruitment of host translation and glycoprotein processing machineries in order to favor alphavirus production. Rift Valley Fever Virus (RVFV), a bunyavirus from Phleboviridae family, induces G2 cell cycle arrest for optimal cap-snatching and viral replication (Hopkins et al., 2013). It is possible that OROV induces the same cell cycle arrest in G2, what should be further investigated, since cap-snatching was never proved for OROV, but it is common for other bunyaviruses. OROV infection also down-regulated many membrane transporters and ion channels (Figure 4C). From those, SLC1A2/EAAT2 (see Table S2) is implicated in glutamate uptake, suggesting that its down-regulation lead to accumulation of extracellular glutamate, which is often associated to neurotoxicity (Legay et al., 2003; Wang et al., 2003; Moidunny et al., 2016). We previously demonstrated the down-regulation of this gene in OROV infected HuH-7 cells and its possible association with upregulation of miR-217 (Geddes et al., 2018). Taken together, that suggests that EAAT2 might be a regulated target in OROV infected cells that could explain the neuropathogeny associated to the virus.
In this study, we provided an integrated genome-wide transcriptome analysis of human primary astrocytes, crucial cells responsible for a variety of functions in the CNS, infected with the emergent and clinically alarming human pathogens CHIKV, MAYV, OROV, and ZIKV. To our knowledge, this is the first study to present a transcriptome for MAYV and OROV, as well as first to present CHIKV transcriptome in cells from CNS. This work shed a light upon the mechanisms and viral-host interactions in the neuropathologies associated to those widespread human arboviruses regarding nervous system outcomes.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
Conceptualization, VG and RA. Methodology, VG, OB, LR, JS, AV, and RA. Validation, VG, OB, and LC. Formal analysis, OB, FM, and RS. Investigation, VG, LC, FC, AG, AL, CF, and LD. Resources, CF, EN, AT, IA-M, SA-L, LR, JS, AV, and RA. Data curation, OB. Writing—original draft, VG, OB, and RA. Writing—review and editing, VG, OB, LC, FC, RS, IA-M, SA-L, LR, JS, AV, and RA. Visualization, VG, OB, AV, and RA. Supervision, AV and RA. Project Administration, AV and RA. Funding Acquisition, AV and RA. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by FINEP (grant no. 01.16.0078.00) and the European Union’s Horizon 2020 research and innovation program through the ZIKAlliance project (grant agreement no. 734548). VG is supported by CAPES under the grant number: 1545985. RA is supported by CNPQ 312688/2017-2, 439119/2018-9; FAPERJ 202.922/2018. AV is supported by CNPq (303170/2017-4) and FAPERJ (26/202.903/20). LC is supported by RABICO/CAPES Project 88887.333817/2019-00. IA-M is supported by CNPq (436933/2018-7) and FAPERJ (203.225/2017).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.641261/full#supplementary-material
Supplementary Table 1 | Oligonucleotides used in this study.
Supplementary Table 2 | All DEGs found in transcriptome for each virus.
Supplementary Table 3 | 50 most up-regulated genes for each virus used in Circoplot.
Supplementary Table 4 | 50 most down-regulated genes for each virus used in Circoplot.
Supplementary Table 5 | Common DEG among CHIKV, MAYV, OROV, and ZIKV.
References
Acharya D., Paul A. M., Anderson J. F., Huang F., Bai F. (2015). Loss of Glycosaminoglycan Receptor Binding after Mosquito Cell Passage Reduces Chikungunya Virus Infectivity. PLoS Negl. Trop. Dis. 9, e0004139. doi: 10.1371/journal.pntd.0004139
Acosta-Ampudia Y., Monsalve D. M., Rodríguez Y., Pacheco Y., Anaya J.-M., Ramírez-Santana C. (2018). Mayaro: an emerging viral threat? Emerg. Microbes Infect. 7, 163. doi: 10.1038/s41426-018-0163-5
Alsadeq A., Lenk L., Vadakumchery A., Cousins A., Vokuhl C., Khadour A., et al. (2018). IL7R is associated with CNS infiltration and relapse in pediatric B-cell precursor acute lymphoblastic leukemia. Blood 132, 1614–1617. doi: 10.1182/blood-2018-04-844209
Au H. H. T., Jan E. (2014). Novel viral translation strategies. Wiley Interdiscip. Rev. RNA 5, 779–801. doi: 10.1002/wrna.1246
Azeredo E. L., Dos Santos F. B., Barbosa L. S., Souza T. M. A., Badolato-Corrêa J., Sánchez-Arcila J. C., et al. (2018). Clinical and Laboratory Profile of Zika and Dengue Infected Patients: Lessons Learned From the Co-circulation of Dengue, Zika and Chikungunya in Brazil. PLoS Curr. 10. ecurrents.outbreaks.0bf6aeb4d30824de63c4d5d745b217f5. doi: 10.1371/currents.outbreaks.0bf6aeb4d30824de63c4d5d745b217f5
Balavoine S., Pircher M., Hoen B., Herrmann-Storck C., Najioullah F., Madeux B., et al. (2017). Guillain-Barré Syndrome and Chikungunya: Description of All Cases Diagnosed during the 2014 Outbreak in the French West Indies. Am. J. Trop. Med. Hyg. 97, 356–360. doi: 10.4269/ajtmh.15-0753
Bandeira A. C., Campos G. S., Sardi S. I., Rocha V. F. D., Rocha G. C. M. (2016). Neonatal encephalitis due to Chikungunya vertical transmission: First report in Brazil. IDCases 5, 57–59. doi: 10.1016/j.idcr.2016.07.008
Barkhash A. V., Yurchenko A. A., Yudin N. S., Ignatieva E. V., Kozlova I. V., Borishchuk I. A., et al. (2018). A matrix metalloproteinase 9 (MMP9) gene single nucleotide polymorphism is associated with predisposition to tick-borne encephalitis virus-induced severe central nervous system disease. Ticks Tick. Borne. Dis. 9, 763–767. doi: 10.1016/j.ttbdis.2018.02.010
Bastos M., de S., Figueiredo L. T. M., Naveca F. G., Monte R. L., Lessa N., et al. (2012). Identification of Oropouche Orthobunyavirus in the cerebrospinal fluid of three patients in the Amazonas, Brazil. Am. J. Trop. Med. Hyg. 86, 732–735. doi: 10.4269/ajtmh.2012.11-0485
Bautista L. E. (2019). Zika virus infection and risk of Guillain-Barré syndrome: A meta-analysis. J. Neurol. Sci. 403, 99–105. doi: 10.1016/j.jns.2019.06.019
Bressy C., Droby G. N., Maldonado B. D., Steuerwald N., Grdzelishvili V. Z. (2019). Cell Cycle Arrest in G(2)/M Phase Enhances Replication of Interferon-Sensitive Cytoplasmic RNA Viruses via Inhibition of Antiviral Gene Expression. J. Virol. 93 (4), e01885–18. doi: 10.1128/JVI.01885-18
Brito Ferreira M. L., Militão de Albuquerque M., de F. P., de Brito C. A. A., de Oliveira França R. F., Porto Moreira Á. J., et al. (2020). Neurological disease in adults with Zika and chikungunya virus infection in Northeast Brazil: a prospective observational study. Lancet Neurol. 19, 826–839. doi: 10.1016/S1474-4422(20)30232-5
Burt F. J., Chen W., Miner J. J., Lenschow D. J., Merits A., Schnettler E., et al. (2017). Chikungunya virus: an update on the biology and pathogenesis of this emerging pathogen. Lancet Infect. Dis. 17, e107–e117. doi: 10.1016/S1473-3099(16)30385-1
Caller L. G., Davies C. T. R., Antrobus R., Lehner P. J., Weekes M. P., Crump C. M. (2019). Temporal Proteomic Analysis of BK Polyomavirus Infection Reveals Virus-Induced G(2) Arrest and Highly Effective Evasion of Innate Immune Sensing. J. Virol. 93 (16), e00595–19. doi: 10.1128/JVI.00595-19
Cao-Lormeau V.-M., Blake A., Mons S., Lastère S., Roche C., Vanhomwegen J., et al. (2016). Guillain-Barré Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet (London England) 387, 1531–1539. doi: 10.1016/S0140-6736(16)00562-6
Cardoso F. L., Brites D., Brito M. A. (2010). Looking at the blood-brain barrier: molecular anatomy and possible investigation approaches. Brain Res. Rev. 64, 328–363. doi: 10.1016/j.brainresrev.2010.05.003
Carvalho F. R., Medeiros T., Vianna R. A., de O., Douglass-Jaimes G., Nunes P. C. G., et al. (2019). Simultaneous circulation of arboviruses and other congenital infections in pregnant women in Rio de Janeiro, Brazil. Acta Trop. 192, 49–54. doi: 10.1016/j.actatropica.2019.01.020
Chen J., Yang Y.-F., Yang Y., Zou P., Chen J., He Y., et al. (2018). AXL promotes Zika virus infection in astrocytes by antagonizing type I interferon signalling. Nat. Microbiol. 3, 302–309. doi: 10.1038/s41564-017-0092-4
Chopra S., Overall C. M., Dufour A. (2019). Matrix metalloproteinases in the CNS: interferons get nervous. Cell. Mol. Life Sci. 76, 3083–3095. doi: 10.1007/s00018-019-03171-9
Cisneros I. E., Ghorpade A. (2012). HIV-1, methamphetamine and astrocyte glutamate regulation: combined excitotoxic implications for neuro-AIDS. Curr. HIV Res. 10, 392–406. doi: 10.2174/157016212802138832
Cope E. C., Gould E. (2019). Adult Neurogenesis, Glia, and the Extracellular Matrix. Cell Stem Cell 24, 690–705. doi: 10.1016/j.stem.2019.03.023
Copeland A. M., Altamura L. A., Van Deusen N. M., Schmaljohn C. S. (2013). Nuclear relocalization of polyadenylate binding protein during rift valley fever virus infection involves expression of the NSs gene. J. Virol. 87, 11659–11669. doi: 10.1128/JVI.01434-13
da Silva V. D., de Faria B. M., Colombo E., Ascari L., Freitas G. P. A., Flores L. S., et al. (2019). Design, synthesis, structural characterization and in vitro evaluation of new 1,4-disubstituted-1,2,3-triazole derivatives against glioblastoma cells. Bioorg. Chem. 83, 87–97. doi: 10.1016/j.bioorg.2018.10.003
De Regge N. (2017). Akabane, Aino and Schmallenberg virus-where do we stand and what do we know about the role of domestic ruminant hosts and Culicoides vectors in virus transmission and overwintering? Curr. Opin. Virol. 27, 15–30. doi: 10.1016/j.coviro.2017.10.004
Dezonne R. S., Sartore R. C., Nascimento J. M., Saia-Cereda V. M., Romão L. F., Alves-Leon S. V., et al. (2017). Derivation of Functional Human Astrocytes from Cerebral Organoids. Sci. Rep. 7, 45091. doi: 10.1038/srep45091
Diagne C. T., Bengue M., Choumet V., Hamel R., Pompon J., Missé D. (2020). Mayaro Virus Pathogenesis and Transmission Mechanisms. Pathog. (Basel Switzerland) 9, 738. doi: 10.3390/pathogens9090738
Diniz L. P., Almeida J. C., Tortelli V., Vargas Lopes C., Setti-Perdigão P., Stipursky J., et al. (2012). Astrocyte-induced synaptogenesis is mediated by transforming growth factor β signaling through modulation of D-serine levels in cerebral cortex neurons. J. Biol. Chem. 287, 41432–41445. doi: 10.1074/jbc.M112.380824
Diniz L. P., Tortelli V., Garcia M. N., Araújo A. P. B., Melo H. M., Silva G. S. S., et al. (2014). Astrocyte transforming growth factor beta 1 promotes inhibitory synapse formation via CaM kinase II signaling. Glia 62, 1917–1931. doi: 10.1002/glia.22713
Domingo-Calap P., Segredo-Otero E., Durán-Moreno M., Sanjuán R. (2019). Social evolution of innate immunity evasion in a virus. Nat. Microbiol. 4, 1006–1013. doi: 10.1038/s41564-019-0379-8
Duffy M. R., Chen T.-H., Hancock W. T., Powers A. M., Kool J. L., Lanciotti R. S., et al. (2009). Zika virus outbreak on Yap Island, Federated States of Micronesia. N. Engl. J. Med. 360, 2536–2543. doi: 10.1056/NEJMoa0805715
Elliott R. M. (2014). Orthobunyaviruses: recent genetic and structural insights. Nat. Rev. Microbiol. 12, 673–685. doi: 10.1038/nrmicro3332
Eugenio M., Campanati L., Müller N., Romão L. F., de Souza J., Alves-Leon S., et al. (2018). Silver/silver chloride nanoparticles inhibit the proliferation of human glioblastoma cells. Cytotechnology 70, 1607–1618. doi: 10.1007/s10616-018-0253-1
Evans A. B., Peterson K. E. (2019). Throw out the Map: Neuropathogenesis of the Globally Expanding California Serogroup of Orthobunyaviruses. Viruses 11 (9), 794. doi: 10.3390/v11090794
Falcon S., Gentleman R. (2007). Using GOstats to test gene lists for GO term association. Bioinformatics 23, 257–258. doi: 10.1093/bioinformatics/btl567
Figueiredo L. T. M. (2015). The recent arbovirus disease epidemic in Brazil. Rev. Soc Bras. Med. Trop. 48, 233–234. doi: 10.1590/0037-8682-0179-2015
Figueiredo L. T. M. (2019). Human Urban Arboviruses Can Infect Wild Animals and Jump to Sylvatic Maintenance Cycles in South America. Front. Cell. Infect. Microbiol. 9, 259. doi: 10.3389/fcimb.2019.00259
Frolov I., Frolova E. I. (2019). Molecular Virology of Chikungunya Virus. Curr. Top. Microbiol. Immunol. 1–31. doi: 10.1007/82_2018_146
Garber C., Vasek M. J., Vollmer L. L., Sun T., Jiang X., Klein R. S. (2018). Astrocytes decrease adult neurogenesis during virus-induced memory dysfunction via IL-1. Nat. Immunol. 19, 151–161. doi: 10.1038/s41590-017-0021-y
Garcia C., Dubois L. G., Xavier A. L., Geraldo L. H., da Fonseca A. C. C., Correia A. H., et al. (2014). The orthotopic xenotransplant of human glioblastoma successfully recapitulates glioblastoma-microenvironment interactions in a non-immunosuppressed mouse model. BMC Cancer 14, 923. doi: 10.1186/1471-2407-14-923
García-Sastre A. (2017). Ten Strategies of Interferon Evasion by Viruses. Cell Host Microbe 22, 176–184. doi: 10.1016/j.chom.2017.07.012
Geddes V. E. V., de Oliveira A. S., Tanuri A., Arruda E., Ribeiro-Alves M., Aguiar R. S. (2018). MicroRNA and cellular targets profiling reveal miR-217 and miR-576-3p as proviral factors during Oropouche infection. PLoS Negl. Trop. Dis. 12, e0006508. doi: 10.1371/journal.pntd.0006508
Gouzil J., Fablet A., Lara E., Caignard G., Cochet M., Kundlacz C., et al. (2017). Nonstructural Protein NSs of Schmallenberg Virus Is Targeted to the Nucleolus and Induces Nucleolar Disorganization. J. Virol. 91 (1), e01263–16. doi: 10.1128/JVI.01263-16
Gutierrez B., Wise E. L., Pullan S. T., Logue C. H., Bowden T. A., Escalera-Zamudio M., et al. (2020). Evolutionary Dynamics of Oropouche Virus in South America. J. Virol. 94, e01127–e01119. doi: 10.1128/JVI.01127-19
Hameed S., Khan S. (2019). Rare variant of Guillain-Barré syndrome after chikungunya viral fever. BMJ Case Rep. 12 (4), e228845. doi: 10.1136/bcr-2018-228845
Hamel R., Ferraris P., Wichit S., Diop F., Talignani L., Pompon J., et al. (2017). African and Asian Zika virus strains differentially induce early antiviral responses in primary human astrocytes. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 49, 134–137. doi: 10.1016/j.meegid.2017.01.015
Hopkins K., Cherry S. (2013). Bunyaviral cap-snatching vs. decapping: recycling cell cycle mRNAs. Cell Cycle 12, 3711–3712. doi: 10.4161/cc.26878
Hopkins K. C., McLane L. M., Maqbool T., Panda D., Gordesky-Gold B., Cherry S. (2013). A genome-wide RNAi screen reveals that mRNA decapping restricts bunyaviral replication by limiting the pools of dcp2-accessible targets for cap-snatching. Genes Dev. 27, 1511–1525. doi: 10.1101/gad.215384.113
Hover S., King B., Hall B., Loundras E.-A., Taqi H., Daly J., et al. (2016). Modulation of Potassium Channels Inhibits Bunyavirus Infection. J. Biol. Chem. 291, 3411–3422. doi: 10.1074/jbc.M115.692673
Hover S., Foster B., Fontana J., Kohl A., Goldstein S. A. N., Barr J. N., et al. (2018). Bunyavirus requirement for endosomal K+ reveals new roles of cellular ion channels during infection. PLoS Pathog. 14, e1006845. doi: 10.1371/journal.ppat.1006845
Inchley C. S., Osterholt H. C. D., Sonerud T., Fjærli H. O., Nakstad B. (2013). Downregulation of IL7R, CCR7, and TLR4 in the cord blood of children with respiratory syncytial virus disease. J. Infect. Dis. 208, 1431–1435. doi: 10.1093/infdis/jit336
Jiang J., Zhang L., Chen H., Lei Y., Zhang T., Wang Y., et al. (2020). Regorafenib induces lethal autophagy arrest by stabilizing PSAT1 in glioblastoma. Autophagy 16, 106–122. doi: 10.1080/15548627.2019.1598752
Ju S. M., Song H. Y., Lee J. A., Lee S. J., Choi S. Y., Park J. (2009). Extracellular HIV-1 Tat up-regulates expression of matrix metalloproteinase-9 via a MAPK-NF-kappaB dependent pathway in human astrocytes. Exp. Mol. Med. 41, 86–93. doi: 10.3858/emm.2009.41.2.011
Khatri H., Shah H., Roy D., Tripathi K. M. (2018). A Case Report on Chikungunya Virus-Associated Encephalomyelitis. Case Rep. Infect. Dis. 2018, 8904753. doi: 10.1155/2018/8904753
Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Klein R. S., Garber C., Funk K. E., Salimi H., Soung A., Kanmogne M., et al. (2019). Neuroinflammation During RNA Viral Infections. Annu. Rev. Immunol. 37, 73–95. doi: 10.1146/annurev-immunol-042718-041417
Kozak R. A., Majer A., Biondi M. J., Medina S. J., Goneau L. W., Sajesh B. V., et al. (2017). MicroRNA and mRNA Dysregulation in Astrocytes Infected with Zika Virus. Viruses 9 (10), 297. doi: 10.3390/v9100297
Lancaster C., Pristatsky P., Hoang V. M., Casimiro D. R., Schwartz R. M., Rustandi R., et al. (2016). Characterization of N-glycosylation profiles from mammalian and insect cell derived chikungunya VLP. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 1032, 218–223. doi: 10.1016/j.jchromb.2016.04.025
Legay V., Deleage C., Beaulieux F., Giraudon P., Aymard M., Lina B. (2003). Impaired glutamate uptake and EAAT2 downregulation in an enterovirus chronically infected human glial cell line. Eur. J. Neurosci. 17, 1820–1828. doi: 10.1046/j.1460-9568.2003.02621.x
Levi L. I., Vignuzzi M. (2019). Arthritogenic Alphaviruses: A Worldwide Emerging Threat? Microorganisms 7 (5), 133. doi: 10.3390/microorganisms7050133
Li S. (2019). Regulation of Ribosomal Proteins on Viral Infection. Cells 8 (5), 508. doi: 10.3390/cells8050508
Liang B., Guida J. P., Costa Do Nascimento M. L., Mysorekar I. U. (2019). Host and viral mechanisms of congenital Zika syndrome. Virulence 10, 768–775. doi: 10.1080/21505594.2019.1656503
Liao Y., Smyth G. K., Shi W. (2019). The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47, e47. doi: 10.1093/nar/gkz114
Lima M. C., de Mendonça L. R., Rezende A. M., Carrera R. M., Aníbal-Silva C. E., Demers M., et al. (2019). The Transcriptional and Protein Profile From Human Infected Neuroprogenitor Cells Is Strongly Correlated to Zika Virus Microcephaly Cytokines Phenotype Evidencing a Persistent Inflammation in the CNS. Front. Immunol. 10, 1928. doi: 10.3389/fimmu.2019.01928
Lindqvist R., Mundt F., Gilthorpe J. D., Wölfel S., Gekara N. O., Kröger A., et al. (2016). Fast type I interferon response protects astrocytes from flavivirus infection and virus-induced cytopathic effects. J. Neuroinflamm. 13, 277. doi: 10.1186/s12974-016-0748-7
Lorenz C., Freitas Ribeiro A., Chiaravalloti-Neto F. (2019). Mayaro virus distribution in South America. Acta Trop. 198, 105093. doi: 10.1016/j.actatropica.2019.105093
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Lundberg L., Fontenot J., Lin S.-C., Pinkham C., Carey B. D., Campbell C. E., et al. (2018). Venezuelan Equine Encephalitis Virus Capsid Implicated in Infection-Induced Cell Cycle Delay in vitro. Front. Microbiol. 9, 3126. doi: 10.3389/fmicb.2018.03126
Luo W., Brouwer C. (2013). Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29, 1830–1831. doi: 10.1093/bioinformatics/btt285
Madejón A., Sheldon J., Francisco-Recuero I., Perales C., Domínguez-Beato M., Lasa M., et al. (2015). Hepatitis C virus-mediated Aurora B kinase inhibition modulates inflammatory pathway and viral infectivity. J. Hepatol. 63, 312–319. doi: 10.1016/j.jhep.2015.02.036
Maria A., Vallamkonda N., Shukla A., Bhatt A., Sachdev N. (2018). Encephalitic presentation of Neonatal Chikungunya: A Case Series. Indian Pediatr. 55, 671–674. doi: 10.1007/s13312-018-1356-7
Medeiros D. B. A., Vasconcelos P. F. C. (2019). Is the brazilian diverse environment is a crib for the emergence and maintenance of exotic arboviruses? An. Acad. Bras. Cienc. 91, e20190407. doi: 10.1590/0001-3765201920190407
Meertens L., Labeau A., Dejarnac O., Cipriani S., Sinigaglia L., Bonnet-Madin L., et al. (2017). Axl Mediates ZIKA Virus Entry in Human Glial Cells and Modulates Innate Immune Responses. Cell Rep. 18, 324–333. doi: 10.1016/j.celrep.2016.12.045
Metsky H. C., Matranga C. B., Wohl S., Schaffner S. F., Freije C. A., Winnicki S. M., et al. (2017). Zika virus evolution and spread in the Americas. Nature 546, 411–415. doi: 10.1038/nature22402
Moidunny S., Matos M., Wesseling E., Banerjee S., Volsky D. J., Cunha R. A., et al. (2016). Oncostatin M promotes excitotoxicity by inhibiting glutamate uptake in astrocytes: implications in HIV-associated neurotoxicity. J. Neuroinflamm. 13, 144. doi: 10.1186/s12974-016-0613-8
Naveca F. G., Claro I., Giovanetti M., de Jesus J. G., Xavier J., Iani F. C., et al. (2019). Genomic, epidemiological and digital surveillance of Chikungunya virus in the Brazilian Amazon. PLoS Negl. Trop. Dis. 13, e0007065–e0007065. doi: 10.1371/journal.pntd.0007065
Niclou S. P., Franssen E. H. P., Ehlert E. M. E., Taniguchi M., Verhaagen J. (2003). Meningeal cell-derived semaphorin 3A inhibits neurite outgrowth. Mol. Cell. Neurosci. 24, 902–912. doi: 10.1016/S1044-7431(03)00243-4
Olsen M. L., Khakh B. S., Skatchkov S. N., Zhou M., Lee C. J., Rouach N. (2015). New Insights on Astrocyte Ion Channels: Critical for Homeostasis and Neuron-Glia Signaling. J. Neurosci. 35, 13827–13835. doi: 10.1523/JNEUROSCI.2603-15.2015
Panayiotou C., Lindqvist R., Kurhade C., Vonderstein K., Pasto J., Edlund K., et al. (2018). Viperin Restricts Zika Virus and Tick-Borne Encephalitis Virus Replication by Targeting NS3 for Proteasomal Degradation. J. Virol. 92 (7), e02054–17. doi: 10.1128/JVI.02054-17
Peña-García V. H., McCracken M. K., Christofferson R. C. (2017). Examining the potential for South American arboviruses to spread beyond the New World. Curr. Clin. Microbiol. Rep. 4, 208–217. doi: 10.1007/s40588-017-0076-4
Pérez-Olais J. H., Ruiz-Jiménez F., Calderón-Garcia E. J., De Jesús-González L. A., Hernández-Rivas R., Del Angel R. M. (2019). The activity of Aurora kinase B is required for dengue virus release. Virus Res. 274, 197777. doi: 10.1016/j.virusres.2019.197777
Rockey D. C., Du Q., Weymouth N. D., Shi Z. (2019). Smooth Muscle α-Actin Deficiency Leads to Decreased Liver Fibrosis via Impaired Cytoskeletal Signaling in Hepatic Stellate Cells. Am. J. Pathol. 189, 2209–2220. doi: 10.1016/j.ajpath.2019.07.019
Ronca S. E., Dineley K. T., Paessler S. (2016). Neurological Sequelae Resulting from Encephalitic Alphavirus Infection. Front. Microbiol. 7, 959. doi: 10.3389/fmicb.2016.00959
Sakkas H., Bozidis P., Franks A., Papadopoulou C. (2018). Oropouche Fever: A Review. Viruses 10 (4), 175. doi: 10.3390/v10040175
Santiago F. W., Halsey E. S., Siles C., Vilcarromero S., Guevara C., Silvas J. A., et al. (2015). Long-Term Arthralgia after Mayaro Virus Infection Correlates with Sustained Pro-inflammatory Cytokine Response. PLoS Negl. Trop. Dis. 9, e0004104–e0004104. doi: 10.1371/journal.pntd.0004104
Santos R. I., Bueno-Júnior L. S., Ruggiero R. N., Almeida M. F., Silva M. L., Paula F. E., et al. (2014). Spread of Oropouche virus into the central nervous system in mouse. Viruses 6, 3827–3836. doi: 10.3390/v6103827
Savarin C., Stohlman S. A., Rietsch A. M., Butchi N., Ransohoff R. M., Bergmann C. C. (2011). MMP9 deficiency does not decrease blood-brain barrier disruption, but increases astrocyte MMP3 expression during viral encephalomyelitis. Glia 59, 1770–1781. doi: 10.1002/glia.21222
Saxena T., Tandon B., Sharma S., Chameettachal S., Ray P., Ray A. R., et al. (2013). Combined miRNA and mRNA signature identifies key molecular players and pathways involved in chikungunya virus infection in human cells. PLoS One 8, e79886. doi: 10.1371/journal.pone.0079886
Schoggins J. W. (2019). Interferon-Stimulated Genes: What Do They All Do? Annu. Rev. Virol 6, 567–584. doi: 10.1146/annurev-virology-092818-015756
Schuler-Faccini L., Ribeiro E. M., Feitosa I. M. L., Horovitz D. D. G., Cavalcanti D. P., Pessoa A., et al. (2016). Possible Association Between Zika Virus Infection and Microcephaly - Brazi. MMWR Morb. Mortal. Wkly. Rep. 65, 59–62. doi: 10.15585/mmwr.mm6503e2
Sebollela A., Freitas-Correa L., Oliveira F. F., Paula-Lima A. C., Saraiva L. M., Martins S. M., et al. (2012). Amyloid-β oligomers induce differential gene expression in adult human brain slices. J. Biol. Chem. 287, 7436–7445. doi: 10.1074/jbc.M111.298471
Sher A. A., Glover K. K. M., Coombs K. M. (2019). Zika Virus Infection Disrupts Astrocytic Proteins Involved in Synapse Control and Axon Guidance. Front. Microbiol. 10, 596. doi: 10.3389/fmicb.2019.00596
Shukla V., Shakya A. K., Shukla M., Kumari N., Krishnani N., Dhole T. N., et al. (2016). Circulating levels of matrix metalloproteinases and tissue inhibitors of matrix metalloproteinases during Japanese encephalitis virus infection. Virusdisease 27, 63–76. doi: 10.1007/s13337-015-0301-9
Silva V. P., Costa D. S., Carvalho V.C.C.V.L., Garcês T. C. C. S., Barros E. L. T., Oliveira J. S., et al. (2020). Peripheral polyneuropathy associated with Chikungunya virus infection. J. Neurovirol. 26, 122–126. doi: 10.1007/s13365-019-00782-7
Simon F., Barnay J.-L., Lannuzel A. (2018). The wide spectrum of neurological consequences of chikungunya disease. Rev. Med. Virol. 28, e1999. doi: 10.1002/rmv.1999
Simonin Y., Loustalot F., Desmetz C., Foulongne V., Constant O., Fournier-Wirth C., et al. (2016). Zika Virus Strains Potentially Display Different Infectious Profiles in Human Neural Cells. EBioMedicine 12, 161–169. doi: 10.1016/j.ebiom.2016.09.020
Soares-Schanoski A., Baptista Cruz N., de Castro-Jorge L. A., de Carvalho R. V. H., Santos C. A., Dos, Rós N., et al. (2019). Systems analysis of subjects acutely infected with the Chikungunya virus. PLoS Pathog. 15, e1007880. doi: 10.1371/journal.ppat.1007880
Song J., Hu Y., Li H., Huang X., Zheng H., Hu Y., et al. (2018). miR-1303 regulates BBB permeability and promotes CNS lesions following CA16 infections by directly targeting MMP9. Emerg. Microbes Infect. 7, 155. doi: 10.1038/s41426-018-0157-3
Soung A., Klein R. S. (2018). Viral Encephalitis and Neurologic Diseases: Focus on Astrocytes. Trends Mol. Med. 24, 950–962. doi: 10.1016/j.molmed.2018.09.001
Stefanik M., Formanova P., Bily T., Vancova M., Eyer L., Palus M., et al. (2018). Characterisation of Zika virus infection in primary human astrocytes. BMC Neurosci. 19, 5. doi: 10.1186/s12868-018-0407-2
Sun X., Hua S., Chen H.-R., Ouyang Z., Einkauf K., Tse S., et al. (2017). Transcriptional Changes during Naturally Acquired Zika Virus Infection Render Dendritic Cells Highly Conducive to Viral Replication. Cell Rep. 21, 3471–3482. doi: 10.1016/j.celrep.2017.11.087
Teng T.-S., Foo S.-S., Simamarta D., Lum F.-M., Teo T.-H., Lulla A., et al. (2012). Viperin restricts chikungunya virus replication and pathology. J. Clin. Invest. 122, 4447–4460. doi: 10.1172/JCI63120
Thio C. L.-P., Yusof R., Abdul-Rahman P. S. A., Karsani S. A. (2013). Differential proteome analysis of chikungunya virus infection on host cells. PLoS One 8, e61444. doi: 10.1371/journal.pone.0061444
Tiwari S. K., Dang J., Qin Y., Lichinchi G., Bansal V., Rana T. M. (2017). Zika virus infection reprograms global transcription of host cells to allow sustained infection. Emerg. Microbes Infect. 6, e24. doi: 10.1038/emi.2017.9
Travassos da Rosa J. F., de Souza W. M., Pinheiro F., de P., Figueiredo M. L., Cardoso J. F., et al. (2017). Oropouche Virus: Clinical, Epidemiological, and Molecular Aspects of a Neglected Orthobunyavirus. Am. J. Trop. Med. Hyg. 96, 1019–1030. doi: 10.4269/ajtmh.16-0672
Vasconcellos A. F., Silva J. M. F., de Oliveira A. S., Prado P. S., Nagata T., Resende R. O. (2019). Genome sequences of chikungunya virus isolates circulating in midwestern Brazil. Arch. Virol. 164, 1205–1208. doi: 10.1007/s00705-019-04174-4
Vernal S., Martini C. C. R., da Fonseca B. A. L. (2019). Oropouche Virus-Associated Aseptic Meningoencephalitis, Southeastern Brazil. Emerg. Infect. Dis. 25, 380–382. doi: 10.3201/eid2502.181189
Voet S., Srinivasan S., Lamkanfi M., van Loo G. (2019). Inflammasomes in neuroinflammatory and neurodegenerative diseases. EMBO Mol. Med. 11 (6), e10248. doi: 10.15252/emmm.201810248
Walsh D., Mathews M. B., Mohr I. (2013). Tinkering with translation: protein synthesis in virus-infected cells. Cold Spring Harb. Perspect. Biol. 5, a012351. doi: 10.1101/cshperspect.a012351
Wang Z., Pekarskaya O., Bencheikh M., Chao W., Gelbard H. A., Ghorpade A., et al. (2003). Reduced expression of glutamate transporter EAAT2 and impaired glutamate transport in human primary astrocytes exposed to HIV-1 or gp120. Virology 312, 60–73. doi: 10.1016/S0042-6822(03)00181-8
Wang P., Dai J., Bai F., Kong K.-F., Wong S. J., Montgomery R. R., et al. (2008). Matrix metalloproteinase 9 facilitates West Nile virus entry into the brain. J. Virol. 82, 8978–8985. doi: 10.1128/JVI.00314-08
Wang Z., Wang Y., Wang S., Meng X., Song F., Huo W., et al. (2018). Coxsackievirus A6 Induces Cell Cycle Arrest in G0/G1 Phase for Viral Production. Front. Cell. Infect. Microbiol. 8, 279. doi: 10.3389/fcimb.2018.00279
Wilson J. A. C., Prow N. A., Schroder W. A., Ellis J. J., Cumming H. E., Gearing L. J., et al. (2017). RNA-Seq analysis of chikungunya virus infection and identification of granzyme A as a major promoter of arthritic inflammation. PLoS Pathog. 13, e1006155. doi: 10.1371/journal.ppat.1006155
Xing Y., Shepherd N., Lan J., Li W., Rane S., Gupta S. K., et al. (2017). MMPs/TIMPs imbalances in the peripheral blood and cerebrospinal fluid are associated with the pathogenesis of HIV-1-associated neurocognitive disorders. Brain Behav. Immun. 65, 161–172. doi: 10.1016/j.bbi.2017.04.024
Yang C.-M., Lin C.-C., Lee I.-T., Lin Y.-H., Yang C. M., Chen W.-J., et al. (2012). Japanese encephalitis virus induces matrix metalloproteinase-9 expression via a ROS/c-Src/PDGFR/PI3K/Akt/MAPKs-dependent AP-1 pathway in rat brain astrocytes. J. Neuroinflamm. 9, 12. doi: 10.1186/1742-2094-9-12
Yang M., Wang X., Fan Y., Chen Y., Sun D., Xu X., et al. (2019). Semaphorin 3A Contributes to Secondary Blood-Brain Barrier Damage After Traumatic Brain Injury. Front. Cell. Neurosci. 13, 117. doi: 10.3389/fncel.2019.00117
Yu G., He Q.-Y. (2016). ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 12, 477–479. doi: 10.1039/C5MB00663E
Zanotto P. M. A., Leite L. C. C. (2018). The Challenges Imposed by Dengue, Zika, and Chikungunya to Brazil. Front. Immunol. 9, 1964. doi: 10.3389/fimmu.2018.01964
Zhang B., Chan Y. K., Lu B., Diamond M. S., Klein R. S. (2008). CXCR3 mediates region-specific antiviral T cell trafficking within the central nervous system during West Nile virus encephalitis. J. Immunol. 180, 2641–2649. doi: 10.4049/jimmunol.180.4.2641
Keywords: Chikungunya, Mayaro, Oropouche, Zika, innate immunity, astrocytes
Citation: Geddes VEV, Brustolini OJB, Cavalcante LTdF, Moreira FRR, de Castro FL, Guimarães APdC, Gerber AL, Figueiredo CM, Diniz LP, Neto EdA, Tanuri A, Souza RP, Assunção-Miranda I, Alves-Leon SV, Romão LF, de Souza JPBM, de Vasconcelos ATR and de Aguiar RS (2021) Common Dysregulation of Innate Immunity Pathways in Human Primary Astrocytes Infected With Chikungunya, Mayaro, Oropouche, and Zika Viruses. Front. Cell. Infect. Microbiol. 11:641261. doi: 10.3389/fcimb.2021.641261
Received: 13 December 2020; Accepted: 28 January 2021;
Published: 15 March 2021.
Edited by:
Tatiana Barichello, University of Texas Health Science Center at Houston, United StatesReviewed by:
Jaqueline Generoso, Universidade do Extremo Sul Catarinense, BrazilCharles T. Spencer, The University of Texas at El Paso, United States
Tejaswini Doifode, Carilion Clinic, United States
Copyright © 2021 Geddes, Brustolini, Cavalcante, Moreira, de Castro, Guimarães, Gerber, Figueiredo, Diniz, Neto, Tanuri, Souza, Assunção-Miranda, Alves-Leon, Romão, de Souza, de Vasconcelos and de Aguiar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana Tereza Ribeiro de Vasconcelos, atrv@lncc.br; Renato Santana de Aguiar, santanarnt@ufmg.br
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share last authorship
 Victor Emmanuel Viana Geddes1,2†
Victor Emmanuel Viana Geddes1,2†  Otávio José Bernardes Brustolini
Otávio José Bernardes Brustolini Ana Paula de Campos Guimarães
Ana Paula de Campos Guimarães Alexandra Lehmkuhl Gerber
Alexandra Lehmkuhl Gerber Luan Pereira Diniz
Luan Pereira Diniz Eurico de Arruda Neto
Eurico de Arruda Neto Amilcar Tanuri
Amilcar Tanuri Renan Pedra Souza
Renan Pedra Souza Iranaia Assunção-Miranda
Iranaia Assunção-Miranda Soniza Vieira Alves-Leon
Soniza Vieira Alves-Leon Ana Tereza Ribeiro de Vasconcelos
Ana Tereza Ribeiro de Vasconcelos Renato Santana de Aguiar
Renato Santana de Aguiar