Editorial: Autophagy-Mediated Cell Survival and Death in Disease Progression and Treatment
- 1CancerCare Manitoba Research Institute, CancerCare Manitoba, University of Manitoba, Winnipeg, MB, Canada
- 2Department of Hepatobiliary and Pancreatic Surgery, Zhejiang Provincial Key Laboratory of Pancreatic Disease, The First Affiliated Hospital, Institute of Translational Medicine, Cancer Center, Zhejiang University School of Medicine, Zhejiang University, Hangzhou, China
- 3Department of Cellular and Integrative Physiology, University of Nebraska Medical Center, Omaha, NE, United States
Editorial on the Research Topic
Autophagy-Mediated Cell Survival and Death in Disease Progression and Treatment
Introduction
Rationale for This Research Topic
Autophagy (macro-autophagy) is an evolutionarily conserved, intracellular “self-digestion” catabolic process characterized by the formation of double-membrane autophagosomes that sequester and enclose cytoplasmic materials (cargos) and fusion of autophagosome with lysosome for degradation (Figure 1). Autophagy is a stress-responsive mechanism that supports eukaryotic cells to survive harsh environments benefiting their growth. However, deregulated autophagy triggers cell death. A wide array of research studies support the crucial role of autophagy in a variety of diseases. Cell death instigates the pathogenesis of several diseases such as liver and heart disease while it prevents and treats cancer. Autophagy has dual roles in both preventing and inducing cell death by scavenging damaged and aged cell organelles and excessively degrading essential cellular organelles such as mitochondria, respectively. Understanding the roles and mechanisms of autophagy in cell death and survival during disease progression could provide a novel and effective strategy to prevent and treat disease. Autophagy is a dynamic process, which alters in a temporal fashion and depends on various factors including stress type, stress duration, and cell type. Thus, the level of autophagy does not remain constant, and it varies in a time and context-dependent manner. Therefore, unraveling the specific roles of autophagy in different types of diseases and different pathological conditions is warranted. The published studies on this topic have elucidated novel regulatory roles of autophagy in hepatocellular carcinoma, pancreatic cancer, glioblastoma, triple negative breast cancer, intervertebral disc degeneration, and liver disease. These studies will further our understanding of the crucial roles of autophagy in cell death and survival and trigger future related studies in other diseases.
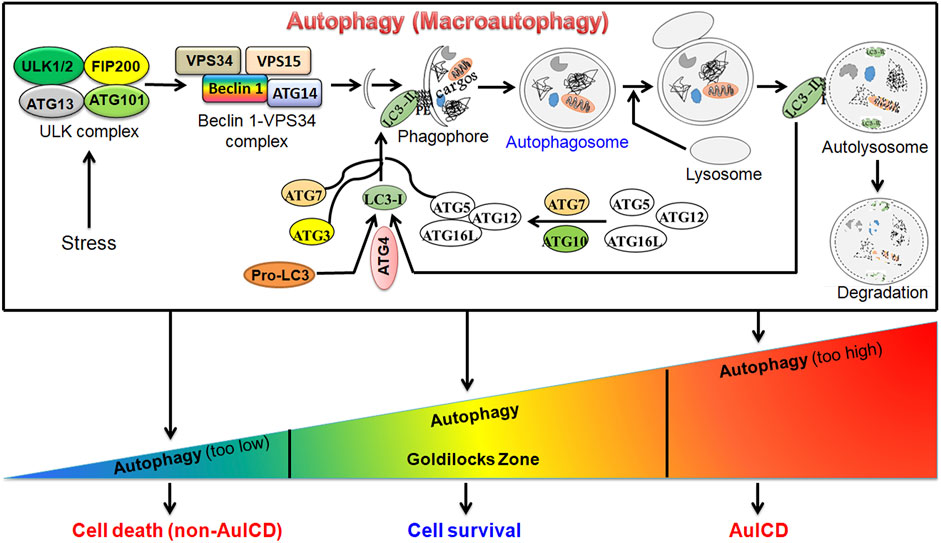
FIGURE 1. A graphic model for the canonical autophagy pathway and roles of autophagy in cell survival and death. Under stress (e.g., starvation), autophagy is induced. This is followed by the formation of autophagosome, a double-membrane vesicle, which fuses with the lysosome to form autolysosome to degrade cargo(s) by lysosomal enzymes. Autophagy can be at different levels. In the homeostatic “Goldilocks Zone” condition, autophagy promotes cell survival and prevents cell death. When it is deregulated, cell death can be induced via two mechanisms. First, when autophagy is highly downregulated, the accumulation of damaged cellular organelles, such as dysfunctional mitochondria, or other components triggers cell death. Second, when autophagy is highly upregulated, it degrades essential cellular compoents or organelles such as mitochondria leading to autophagy-induced cell death (AuICD).
New Findings Reported in This Research Topic
It should be noted that a comprehensive inclusion of the current new findings on this research topic could not be possible due to the authors’ diverse choices in journals. We have collected five original research articles and one review article on this research topic, which provides a glimpse of the ongoing research in the area of autophagy regulation in cell death and survival in diseases.
More than 30 autophagy related (ATG) genes and their proteins are involved in the canonical autophagy pathway. In recent years, there is an emerging interest in investigating the roles of individual ATG proteins in disease progression and treatment. In this research topic, two studies have reported the roles of ATG4 proteins in cancer cell death and proliferation. In mammals, four isoforms of ATG4 (ATG4A, ATG4B, ATG4C, and ATG4D) exist. Chen et al. (Chen et al.) reported that the long non-coding RNA (lncRNA) colorectal neoplasia differentially expressed (CRNDE) promotes autophagy by stabilizing ATG4B mRNA to upregulate ATG4B protein. This is involved in promoting sorafenib-induced drug resistance in hepatocellular carcinoma (HCC) cells and tumors. In contrast, Nie et al. (Nie et al.) demonstrated that ATG4D-dependent mitophagy (mitochondria-selective autophagy) promotes PPARγ antagonist (drug T0070907)-induced apoptotic cell death and suppresses tumor growth of pancreatic cancer cells. Although the roles of ATG4B and ATG4D in drug resistance and tumor growth of the same type of cancer cells need to be investigated in future studies, the results from these two studies support that autophagy induction driven by different ATG proteins could have opposite effects on cancer drug resistance and tumor progression.
Similar to the pro-cell survival mechanism of ATG4B-dependent autophagy in HCC (Chen et al.), Eicosapentaenoic acid (EPA)-induced autophagy inhibits apoptosis, suppresses endoplasmic reticulum stress and the degradation of extracellular matrix in a rat model. Altogether, they alleviate intervertebral disc degeneration (IDD), which is a major cause of low back pain (Lin et al.). Similar to the report on the cell death-promoting effect of ATG4D-dependent autophagy (Nie et al.), Chen and Gibson (Chen and Gibson) demonstrated that knockout of the novel tumor suppressor TSSC4 (tumor suppressing subtransferable candidate 4) increases autophagy-induced cell death (AuICD) instigated by temozolomide (TMZ) in glioblastoma (GBM) cells.
Studies in recent years have suggested that ATG proteins can function independently of autophagy. Here, Rajak et al. (Rajak et al.) have comprehensively reviewed the current understanding of autophagy-dependent and -independent functions of hepatic ULK1 signaling in liver disease.
Finally, based on the DNA homologous recombination repair deficiency and bioinformatics analysis, Liao et al. (Liao et al.) published a model to predict anticancer drugs suitable for treating triple-negative breast cancer. As autophagy plays a critical role in DNA homologous recombination repair, understanding the roles of autophagy on anticancer activities of these predicted drugs may lead to new strategies to improve cancer treatment.
Future Perspectives
The majority of this research topic included new original studies revealing pro-cell survival and pro-cell death roles of autophagy in disease progression and treatment, which supports the notion that the functions of autophagy are context-dependent. Based on the diseases studied in this research topic, novel therapeutic strategies can be utilized to target the pro-cell survival or pro-cell death roles of autophagy in various diseases. For example, the pro-cell survival mechanism of autophagy can be targeted to treat cancer cells. The levels of autophagy must be maintained in a “Goldilocks Zone” to maintain cellular homeostasis as suppressing or exacerbating autophagy could lead to cell death via autophagy-independent mechanisms or autophagy-induced cell death (AuICD). Driving autophagy out of the Goldilocks Zone in cancer cells has the potential to overcome drug resistance for cancer treatment (Figure 1).
As reported in this research topic, an emerging focus in the field of autophagy is to study the roles of individual ATG proteins in human diseases, where autophagy-independent functions of these proteins may play a role. ULK1, an ATG protein, has autophagy-independent roles in the liver, which needs further investigation in liver disease and potentially other metabolic diseases.
Author Contributions
YC wrote the first draft of the manuscript. YZ and PM reviewed and approved the manuscript.
Funding
YZ is supported by the National Natural Science Foundation of China (92053117 and 81972591). PKM is supported by the National Institutes of Health (NIH) grant P20GM104320, Nebraska Center for the Prevention of Obesity Diseases, and Nebraska Research Initiative Collaborative Seed Grant.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank all the authors, reviewers, and editors of this research topic for their participation and contributions. We would also like to thank the editorial staff of the Frontiers in Cell and Developmental Biology for their support in the successful completion of this research topic.
References
Chen, L., Sun, L., Dai, X., Li, T., Yan, X., Zhang, Y., et al. (2021). LncRNA CRNDE Promotes ATG4B-Mediated Autophagy and Alleviates the Sensitivity of Sorafenib in Hepatocellular Carcinoma Cells. Front. Cell. Dev. Biol. 9, 687524. doi:10.3389/fcell.2021.687524
Chen, Y., and Gibson, S. B. (2022). Tumor Suppressing Subtransferable Candidate 4 Expression Prevents Autophagy-Induced Cell Death Following Temozolomide Treatment in Glioblastoma Cells. Front. Cell. Dev. Biol. 10, 823251. doi:10.3389/fcell.2022.823251
Liao, G., Yang, Y., Xie, A., Jiang, Z., Liao, J., Yan, M., et al. (2022). Applicability of Anticancer Drugs for the Triple-Negative Breast Cancer Based on Homologous Recombination Repair Deficiency. Front. Cell. Dev. Biol. 10, 845950. doi:10.3389/fcell.2022.845950
Lin, Z., Ni, L., Teng, C., Zhang, Z., Wu, L., Jin, Y., et al. (2021). Eicosapentaenoic Acid-Induced Autophagy Attenuates Intervertebral Disc Degeneration by Suppressing Endoplasmic Reticulum Stress, Extracellular Matrix Degradation, and Apoptosis. Front. Cell. Dev. Biol. 9, 745621. doi:10.3389/fcell.2021.745621
Nie, S., Shi, Z., Shi, M., Li, H., Qian, X., Peng, C., et al. (2022). PPARγ/SOD2 Protects against Mitochondrial ROS-dependent Apoptosis via Inhibiting ATG4D-Mediated Mitophagy to Promote Pancreatic Cancer Proliferation. Front. Cell. Dev. Biol. 9, 745554. doi:10.3389/fcell.2021.745554
Keywords: autophagy, ATG4B, ATG4D, cell death, eicosapentaenoic acid (EPA), TSSC4 (tumor suppressing subtransferable candidate 4), ULK1, breast cancer
Citation: Chen Y, Zhao Y and Mishra PK (2022) Editorial: Autophagy-Mediated Cell Survival and Death in Disease Progression and Treatment. Front. Cell Dev. Biol. 10:916347. doi: 10.3389/fcell.2022.916347
Received: 09 April 2022; Accepted: 15 June 2022;
Published: 04 July 2022.
Edited by:
Craig Michael Walsh, University of California, United StatesReviewed by:
Alejandro Ropolo, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaCopyright © 2022 Chen, Zhao and Mishra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongqiang Chen, yongqiangchen2007@gmail.com
 Yongqiang Chen
Yongqiang Chen Yongchao Zhao
Yongchao Zhao Paras Kumar Mishra
Paras Kumar Mishra