Blockchain Biology
- Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program, New York, NY, United States
Introduction
Taking the world by storm in recent years, blockchain technology revolutionizes the way we transact assets, manage data, and enforce agreements. Originally developed by Satoshi Nakamoto for the cryptocurrency Bitcoin, blockchain has been adapted for diverse data management applications such as streamlining remittances, enhancing food traceability, securing electronic health records, ensuring genomic data privacy, training artificial intelligence, bolstering cybersecurity, tackling climate change, and supporting clinical trials (Chapron, 2017; Grishin et al., 2019; Howson, 2019; Wong et al., 2019; Krittanawong et al., 2020; Reina, 2020).
Blockchains are decentralized, append-only ledgers. Instead of a centralized entity, for example a bank, controlling an entire ledger, multiple parties (nodes) form a network to maintain a synchronized, distributed, and identical record. Decentralization safeguards the integrity of the ledger when individual nodes are lost. The ledger is comprised of blocks that store data, such as the details of a financial transaction, and are linked chronologically to create a metaphorical chain of blocks. The append-only design of blockchain guarantees a complete, traceable, and virtually tamper-proof ledger.
Despite its implementation in many industries, blockchain has never been harnessed to directly study biological mechanisms. Current uses of blockchain technology in biology and medicine has been limited to peripheral applications such as storing sequencing data or preventing tampering of clinical trial data. Although longstanding problems in computational biology mirror those addressed by blockchain, the technology has never been exploited to answer fundamental biological questions. Proposed here is a conceptual framework for employing blockchain technology to probe biological mechanisms. How principles of decentralization, synchronicity, immutability, and contracts can be utilized for cancer evolution and synthetic biology are explored.
Decentralized Ledgers and Modeling Cancer Evolution
The rigorous recordkeeping capabilities of blockchain can be harnessed to probe cancer evolution and lineage tracing. Clonal evolution in cancer exhibits strikingly similar features as blockchain (Figure 1A). Accruing genetic and epigenetic alterations in a stepwise, sequential manner, cancer cell clones are subject to Darwinian natural selection throughout their growth. Clonal architectures involve a founder mutation, for example ETV6-RUNX1 fusion in acute lymphoblastic leukemia, that drives clonal expansion and subsequent diversification (Greaves and Maley, 2012). Defining the ledger as the complete history of a cancer, this critical origin can be represented by the genesis block of a blockchain. The dataset in each block harbors a snapshot of the cancer state in time, ideally the entire single-cell omics signature. Accordingly, appending a new block to the ledger corresponds to adding an updated snapshot of the cancer state to the cancer history. Appending new blocks is critical because cancer cells are constantly subjected to dynamic evolutionary pressures, including resource competition, microenvironmental constraint, and therapeutic intervention (Ferrando and López-Otín, 2017). Decentralization can be achieved by treating every cell as an individual node and connections as intercellular relationships. Reconstructing the ledger necessitates integration of the intrinsic omics of a single cell and all its intercellular relationships. In this model, despite heterogeneity across cells, they are synchronous in their ability to contribute to the reconstruction of a cancer history ledger. Establishing nodal connections are realistic given the significant computational advances in characterizing cell-cell communication (Efremova et al., 2020).
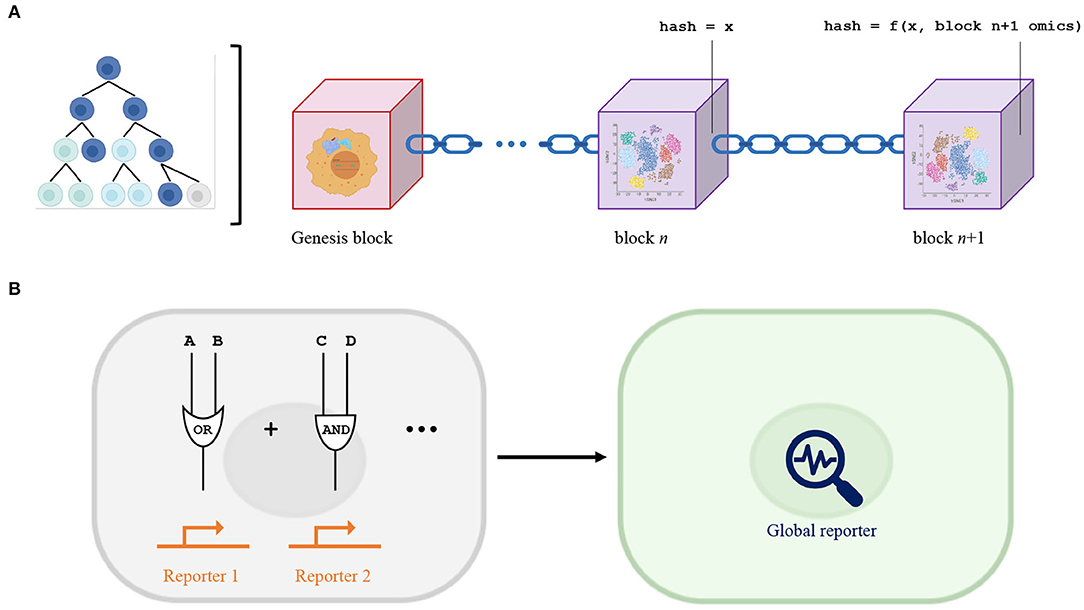
Figure 1. (A) Blockchain model of cancer evolution. Founder mutation (represented by lavender and turquoise fusion protein) initiates genesis block. Contents of every block encompass the complete single-cell omics of the cancer at a certain time point. Each block is marked by a unique hash that is a function of the hash of the previous block and its own contents. Proof-of-work determines the timeframe elapsed between each block. The entire ledger is decentralized across every cancer cell. (B) Model for logic-based smart contracts integrated with biological Boolean logic gates. Complex synthetic biological circuits can involve many unique signal inputs into multiple Boolean logic gates of different types, each requiring an independent reporter (orange). Implementing logic-based smart contracts eliminates the need for individual reporters to validate an individual logic gate, potentially allowing for a general global, blockchain-based reporter (blue) that can model dynamic and multiplexed circuits.
What guarantees that a newly appended block is an accurate updated snapshot of the cancer state? The cryptographic hash and proof-of-work mechanisms of a blockchain can guarantee that the evolutionary trajectory is faithfully documented (temporal, lineage, and omic accuracy). Cryptographic hash functions are one-way functions (inputs can only be determined by trial-and-error, not rationally, from outputs) that map an arbitrary dataset to a fixed value such as a string of binary digits. Each block contains the hash of the previous block and its own unique hash that is a function of both its intrinsic data and the previous hash, enabling an append-only chain. A cryptographic hash function can map a single-cell omics signature to a dimension-reduced fingerprint of the cancer. Such processing is realistic given the substantial progress made in computational methods for multimodal integration of single-cell omics data (Efremova and Teichmann, 2020). The linear organization of blocks ensures that changes during the inter-block timeframe in any arbitrary feature of the cancer, say flux through a signaling pathway in a specific cell, can be determined by comparing the contents of block “n+1” and block “n.” Proof-of-work dictates that hashes need to meet certain conditions, thus requiring brute force computations as a prerequisite for adding new blocks due to the one-way nature of cryptographic hash functions. Because adjusting the hash conditions modulates the difficulty of adding new blocks, proof-of-work establishes the inter-block timeframe and tunes the temporal resolution of the cancer history.
By establishing a high fidelity cancer history, a blockchain model of cancer evolution may be a powerful model for retrospective lineage tracing. Retrospectively reconstructing cell lineage information is valuable for understanding human diseases because experimental manipulation is impossible (Baron and van Oudenaarden, 2019). Naturally occurring mutations, such as copy number variations, single-nucleotide variants, LINE-1 transpositions, microsatellite mutations, and mtDNA mutations, can moonlight as endogenous lineage barcodes (Woodworth et al., 2017), which can serve as a starting point for reconstructing the cancer history blockchain. Integrating a blockchain model with current genetic methods that probe biological memory, such as MemorySeq (Shaffer et al., 2020), may expand the comprehensiveness and utility of retrospective lineage tracing.
Smart Contracts and Biological Boolean Logic Gates
Smart contracts make blockchain an attractive platform to encode Boolean logic gates for biological systems. Originally conceptualized by Nick Szabo and eventually integrated with the Ethereum blockchain by Vitalik Buterin, smart contracts are protocols that automatically execute upon fulfillment of certain conditions and enjoy all the cardinal features of blockchain such as decentralization, immutability, and validity. For example, instead of hiring a real estate broker, smart contracts on a blockchain can automatically process the sale of property via an agreement that cannot be lost or fraudulently altered.
Both smart contracts and Boolean logic gates share core principles of conditionality. Boolean logic applies logic operators, such as conjunction (AND), disjunction (OR), negation (NOT), and exclusivity (XOR), to binary values (true and false or 1 and 0). Smart contracts are classically programmed using the procedural language Solidity. Procedural languages outline step-by-step how a process is performed, whereas declarative languages define what goal must be met. Considerable efforts have been made to shift toward declarative programming to create logic-based smart contracts that are less error-prone and ambiguous than traditional smart contracts (Idelberger et al., 2016; Hu and Zhong, 2018).
As knowledge of molecular mechanisms and signaling pathways rapidly grows, Boolean logic gates offer a powerful approach to model complex networks and extract relevant biological relationships (Morris et al., 2010). Beyond modeling and analysis, boolean logic gates are integral for synthetic biological systems and networks with wide-ranging applications such as biosensing, pharmaceuticals, and biofuels (Khalil and Collins, 2010). Boolean logic gates are experimentally encoded by various synthetic DNA, RNA, protein, and photosensitive molecules (Miyamoto et al., 2013; Erbas-Cakmak et al., 2018). Importantly, Boolean logic gating facilitates the development of highly specific and selective therapeutics, particularly monoclonal antibodies and chimeric antigen receptor (CAR) T cells. Conditionally functional AND-gated antibodies based on binary toggling between phosphorylated and non-phosphorylated states have been synthesized (Gunnoo et al., 2014). Multi-antigen targeting CAR-T cells can be engineered to exhibit AND, OR, and NOT logic gating with the goal of restricting antigen escape and toxicity (Han et al., 2019).
Because Boolean logic gates are central to synthetic biology, logic-based smart contracts present a novel computational approach to modeling biochemical circuits. A central component of synthetic biological circuits is assaying output and performance, and this is often achieved by detecting fluorescent reporters (Brophy and Voigt, 2014). However, fluorescent reporters have limitations such as a requirement for artificial overexpression and susceptibility to protein degradation. Furthermore, encoding more advanced outputs such as oscillation, which requires co-expression of repressors (Gilad and Shapiro, 2017), and permitting multiplexing can be challenging. Because smart contracts serve to eliminate third-party confirmation, logic-based smart contracts can eliminate the need for individual reporters directly downstream of individual biological Boolean logic gates and shift the burden of verification to a global, blockchain-based reporter instead (Figure 1B). Confidence that biological Boolean logic gates function correctly can be attributed to trust in a blockchain, which can be designed to be a ledger of the state of a particular cell for example. This simplification is valuable for complex networks and may facilitate efforts to engineer dynamic and multiplexed circuits. As evidenced by recent advances in adapting machine learning algorithms to design gene circuits (Hiscock, 2019), computational methods like blockchain should be utilized in tandem with experimental techniques to maximize synthetic biology capabilities.
Discussion
Blockchain technology remains under-tapped. Outlined here are two applications of “blockchain biology,” the application of blockchain principles to directly study and model biological mechanisms. Specifically, blockchain-based retrospective lineage tracing and monitoring multiplexed biochemical circuits are proposed. Considerable development is needed to advance blockchain technology to a functional computational biology paradigm. For example, what data should go on-chain vs. off-chain? How will available experimental methods inform blockchain models in biology? In addition to expanding the range of biological contexts amenable to interrogation by blockchain principles, significant methods development is crucial. From proof-of-work vs. proof-of-stake to lightning network addressing scalability, the numerous possibilities for blockchain infrastructure is clearly evidenced by the diverse forms of cryptocurrency. Biology remains an uncharted territory for the immense potential of blockchain, a future ripe to begin building block by block.
Author Contributions
ACC wrote the manuscript.
Funding
ACC was supported by NIH Medical Scientist Training Program Training Grant T32GM007739.
Conflict of Interest
ACC trades cryptocurrencies, primarily Bitcoin and Ethereum.
References
Baron, C. S., and van Oudenaarden, A. (2019). Unravelling cellular relationships during development and regeneration using genetic lineage tracing. Nat. Rev. Mol. Cell Biol. 20, 753–765. doi: 10.1038/s41580-019-0186-3
Brophy, J. A., and Voigt, C. A. (2014). Principles of genetic circuit design. Nat. Methods 11, 508–520. doi: 10.1038/nmeth.2926
Chapron, G. (2017). The environment needs cryptogovernance. Nature 545, 403–405. doi: 10.1038/545403a
Efremova, M., and Teichmann, S. A. (2020). Computational methods for single-cell omics across modalities. Nat. Methods 17, 14–17. doi: 10.1038/s41592-019-0692-4
Efremova, M., Vento-Tormo, M., Teichmann, S. A., and Vento-Tormo, R. (2020). CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484–1506. doi: 10.1038/s41596-020-0292-x
Erbas-Cakmak, S., Kolemen, S., Sedgwick, A. C., Gunnlaugsson, T., James, T. D., Yoon, J., et al. (2018). Molecular logic gates: the past, present and future. Chem. Soc. Rev. 47, 2228–2248. doi: 10.1039/C7CS00491E
Ferrando, A. A., and López-Otín, C. (2017). Clonal evolution in leukemia. Nat. Med. 23, 1135–1145. doi: 10.1038/nm.4410
Gilad, A. A., and Shapiro, M. G. (2017). molecular imaging in synthetic biology, and synthetic biology in molecular imaging. Mol. Imaging Biol. 19, 373–378. doi: 10.1007/s11307-017-1062-1
Greaves, M., and Maley, C. C. (2012). Clonal evolution in cancer. Nature 481, 306–313. doi: 10.1038/nature10762
Grishin, D., Obbad, K., and Church, G. M. (2019). Data privacy in the age of personal genomics. Nat. Biotechnol. 37, 1115–1117. doi: 10.1038/s41587-019-0271-3
Gunnoo, S. B., Finney, H. M., Baker, T. S., Lawson, A. D., Anthony, D. C., and Davis, B. G. (2014). Creation of a gated antibody as a conditionally functional synthetic protein. Nat. Commun. 5:4388. doi: 10.1038/ncomms5388
Han, X., Wang, Y., Wei, J., and Han, W. (2019). Multi-antigen-targeted chimeric antigen receptor T cells for cancer therapy. J. Hematol. Oncol. 12:128. doi: 10.1186/s13045-019-0813-7
Hiscock, T. W. (2019). Adapting machine-learning algorithms to design gene circuits. BMC Bioinformatics 20:214. doi: 10.1186/s12859-019-2788-3
Howson, P. (2019). Tackling climate change with blockchain. Nat. Clim. Chang. 9, 644–645. doi: 10.1038/s41558-019-0567-9
Hu, J., and Zhong, Y. (2018). “A method of logic-based smart contracts for blockchain system,” in Proceedings of the 4th International Conference on Data Processing and Applications (Guangdong: ACM), 58–61. doi: 10.1145/3224207.3224218
Idelberger, F., Governatori, G., Riveret, R., and Sartor, G. (2016). “Evaluation of logic-based smart contracts for blockchain systems,” in Rule technologies. Research, Tools, and Applications. RuleML 2016. Lecture Notes in Computer Science, Vol. 9718, eds J. Alferes, L. Bertossi, G. Governatori, P. Fodor, and D. Roman (Stony Brook, NY: Springer), 167–183.
Khalil, A. S., and Collins, J. J. (2010). Synthetic biology: applications come of age. Nat. Rev. Genet. 11, 367–379. doi: 10.1038/nrg2775
Krittanawong, C., Rogers, A. J., Aydar, M., Choi, E., Johnson, K. W., Wang, Z., et al. (2020). Integrating blockchain technology with artificial intelligence for cardiovascular medicine. Nat. Rev. Cardiol. 17, 1–3. doi: 10.1038/s41569-019-0294-y
Miyamoto, T., Razavi, S., DeRose, R., and Inoue, T. (2013). Synthesizing biomolecule-based Boolean logic gates. ACS Synth. Biol. 2, 72–82. doi: 10.1021/sb3001112
Morris, M. K., Saez-Rodriguez, J., Sorger, P. K., and Lauffenburger, D. A. (2010). Logic-based models for the analysis of cell signaling networks. Biochemistry 49, 3216–3224. doi: 10.1021/bi902202q
Reina, A. (2020). Robot teams stay safe with blockchains. Nat. Mach. Intell. 2, 240–241. doi: 10.1038/s42256-020-0178-1
Shaffer, S. M., Emert, B. L., Reyes Hueros, R. A., Cote, C., Harmange, G., Schaff, D. L., et al. (2020). Memory sequencing reveals heritable single-cell gene expression programs associated with distinct cellular behaviors. Cell 182, 947–959.e17. doi: 10.1016/j.cell.2020.07.003
Wong, D. R., Bhattacharya, S., and Butte, A. J. (2019). Prototype of running clinical trials in an untrustworthy environment using blockchain. Nat. Commun. 10:917. doi: 10.1038/s41467-019-08874-y
Keywords: blockchain, genetics, cancer, systems biology, network biology
Citation: Chin AC (2020) Blockchain Biology. Front. Blockchain 3:606413. doi: 10.3389/fbloc.2020.606413
Received: 14 September 2020; Accepted: 17 November 2020;
Published: 01 December 2020.
Edited by:
Immaculate Dadiso Motsi-Omoijiade, University of Birmingham, United KingdomReviewed by:
Natalie Pankova, Metadvice Ltd, United KingdomCopyright © 2020 Chin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alfred C. Chin, acc4002@med.cornell.edu
 Alfred C. Chin
Alfred C. Chin