Occurrence and Quantification of Antimicrobial Resistance Genes in the Gastrointestinal Microbiome of Two Wild Seabird Species With Contrasting Behaviors
- 1Laboratory of Wildlife Comparative Pathology, Department of Pathology, School of Veterinary Medicine and Animal Sciences, University of São Paulo, São Paulo, Brazil
- 2Group of Epidemiology and Environmental Health, Animal Health Research Centre (INIA-CISA), Madrid, Spain
- 3Facultad de Ciencias de la Vida, Universidad Andres Bello, Santiago, Chile
- 4Associação R3 Animal, Florianópolis, Brazil
Antimicrobial resistance genes (ARGs) are environmental pollutants and anthropization indicators. We evaluated human interference in the marine ecosystem through the ocurrence and quantification (real-time PCRs) of 21 plasmid-mediated ARGs in enema samples of 25 wild seabirds, upon admission into rehabilitation: kelp gull (Larus dominicanus, n = 14) and Magellanic penguin (Spheniscus magellanicus, n = 11). Overall, higher resistance values were observed in kelp gulls (non-migratory coastal synanthropic) in comparison with Magellanic penguins (migratory pelagic non-synanthropic). There were significant differences between species (respectively, kelp gull and Magellanic penguin): ARGs occurrence (blaTEM [p = 0.032]; tetM [p = 0.015]; tetA [p = 0.003]; and sulII [p = 0.007]), mean number of ARGs per sample (p = 0.031), ARGs mean load percentage (aadA [p = 0.045], tetA [p = 0.031], tetM [p = 0.016], blaTEM [p = 0.032], sulII [p = 0.008]), percentage of genes conferring resistance to an antimicrobial class (betalactams [p = 0.036] and sulfonamides [p = 0.033]), mean number of genes conferring resistance to one or more antimicrobial classes (p = 0.024]), percentage of multiresistant microbiomes (p = 0.032), and clustering (p = 0.006). These differences are likely due to these species' contrasting biology and ecology - key factors in the epidemiology of ARGs in seabirds. Additionally, this is the first report of mecA in seabirds in the Americas. Further studies are necessary to clarify the occurrence and diversity of ARGs in seabirds, and their role as potential sources of infection and dispersal within the One Health chain of ARGs.
Introduction
Antimicrobial resistance is an issue of serious public health concern with global economic, social and political implications affecting human and animal populations, as well as the environment (1–3). This worldwide phenomenon is compromising our ability to treat infectious diseases, and undermining or preventing advances in health and medicine (4). Microbial resistance is the result of natural bacteria genetic plasticity and interactions between microbial agents, host organisms and the environment (1, 5), enhanced by the selective pressure exerted by antimicrobial usage and over-prescription in human and veterinary medicine treatments, animal and fish production (i.e., growth promoters and prophylaxis), agriculture and food technologies (1, 5, 6). The consequent remodeling of the existing microbiomes (group of all the genomic elements of a specific microbiota), associated with their dissemination capacity, confer antimicrobial resistance genes (ARGs) the role of environmental pollutants (7, 8) and indicators of environmental anthropization (2, 9, 10).
Seabirds are long-lived, wide-ranging, and upper trophic level marine predators present in all marine ecosystems and oceans of the world, from coastline to pelagic and open seas (11). By acting as predators, scavengers and cross-ecosystem nutrient ancillaries, seabirds play important roles in the processes, function and resilience of island and marine ecosystems (12). Essentially, seabirds respond rapidly to environmental changes, and due to their behavior and population dynamics, are excellent sentinels of the marine ecosystem health, reflecting natural and anthropogenic changes to the environment (13), including pollution by ARGs (14–16). In seabirds, most ARGs studies have focused on synanthropic species, due to their proximity to anthropized areas and feeding habits, and relied on classic microbiological techniques (bacterial culture and sensitivity testing) (9, 17, 18). Nevertheless, recent studies have shown that biological and ecological factors (e.g., migration and feeding niche) are also relevant to the issue of ARGs in wild birds (16, 19, 20). Additionally, only a small fraction of bacteria are cultivable (21, 22). Thus, in order to promote a more comprehensive approach, we employed highly sensitive real time polymerase chain reaction (rtPCR) methods (10, 23) to directly detect and quantify 21 selected plasmid-mediated ARGs in the gastrointestinal microbiome of two wild seabirds species (kelp gulls [Larus dominicanus] and Magellanic penguins [Spheniscus magellanicus]) upon admission to a rehabilitation center. The goals of this study were to (i) assess the presence and load of ARGs in these individuals and (ii) evaluate our findings in light of selected biological and ecological parameters (i.e., dispersal [migratory and non-migratory], feeding niche [coastal and pelagic], and interaction with human-impacted areas [synanthropic and non-synanthropic]). We hypothesized that due to their non-migratory coastal synanthropic behavior (24), kelp gull would present higher occurrence and load of ARGs than the migratory pelagic non-synanthropic Magellanic penguin (25, 26).
Methods
Sample Collection
Fresh fecal samples were immediately obtained by enema (16) in 25 physically restrained birds (14 kelp gulls and 11 Magellanic penguins) upon admission at the wildlife rehabilitation center (Associação R3 Animal, Florianópolis, Santa Catarina state, southern Brazil), and stored at −20°C until analyses. All birds included in the study came directly from their rescue sites (beach), and did not receive previous veterinary care prior to their arrival at the center. Total DNA extraction was carried out by a pressure filtration technique (QuickGene DNA tissue kit S, Fujifilm, Tokyo, Japan), according with the manufacturer's instructions. The 16S rRNA gene was amplified by real time PCR (rtPCR) in 10-fold dilutions of each extracted sample [(27, 28), Supplementary Materials] to verify adequate concentration of bacterial DNA. A sample was considered validated when its 10-fold dilution showed a cycle threshold (Ct) <25 (29). To normalize the study, ct was obtained based on the fluorescence variation value [(ΔF/ΔC) = 0.02] (30). Once validated, samples were analyzed by rtPCR for 21 selected ARGs encoding resistance to eight antimicrobial classes: tetracyclines (tet(A), tet(B), tet(Y), tet(K), tet(M), tet(Q), tet(S), and tet(W) (28), aminoglycosides [aadA (31) and str (32)], sulfonamides (sulI, sulII), chloramphenicols [catI and catII (28)], macrolides [erm(B), erm(F) (33)], quinolones [qnrB (34) and qnrS (35)]; betalactams [blaTEM (31) and mecA (36)], and polymyxins [mcr-1 (30)] (Supplementary Materials). The estimation of the percentage of bacteria harboring ARGs (mean load percentage of each ARG), was based on the formula % gene X = 10[2+0.33(ct16S−ctgeneX)], with ct as the cycle threshold (16S rRNA regarding bacterial determination and X for each evaluated gene), and 0.33 as the mean slope for all the evaluated genes. Results were expressed in log10 scale of the hypothetical percentage of bacteria presenting each gene, ranging from −8 (sample considered negative) to +2 (when 100% of the bacteria in the sample presented the ARG) (30). The same thermal cycle was used for all rtPCR reactions [6′ 95°C, 40x (10″ 95°C, 30″ 60°C)], with alignment and extension in the same step, at constant 60°C. A melting curve step was performed at the end of the rtPCR reaction (30). As per (37), we applied the term “multiresistant microbiome” when a fecal sample presented at least three ARGs encoding resistance to different classes of antimicrobials (10, 29, 30). All samples used in this study were collected as part of the Santos Basin Beach Monitoring Project (Projeto de monitoramento de Praias da Bacia de Santos - PMP-BS), licensed by the Brazilian Institute of the Environment and Renewable Natural Resources (IBAMA) of the Brazilian Ministry of Environment (ABIO N° 640/2015), and in full compliance with the Biodiversity Information and Authorization System (SISBIO 59150-4). All procedures were performed according to the Ethical Committee in Animal Research of the School of Veterinary Medicine and Animal Sciences, University of São Paulo (process number 1753110716).
Statistical Analysis
The k-means clustering method was used to investigate the resistance patterns (GENESIS software v. 1.7.7, Graz University of Technology, Graz, Austria), by assigning each sample to one cluster (Figure 1). Two clusters were selected, corresponding to low (value = 0) and high (value = 1) levels of ARGs. The Mann-Whitney U non-parametric test was used to establish the differences between species regarding: ARGs occurrence, mean number of ARGs per sample, mean load percentage of each ARG, the mean number of genes conferring resistance to one or more antimicrobial classes in each sample, percentage of multiresistant microbiomes and resistance patterns. Such statistical analyses were performed in R software (R Development Core Team 3.0.1., 2013), with a significance level of p < 0.05.
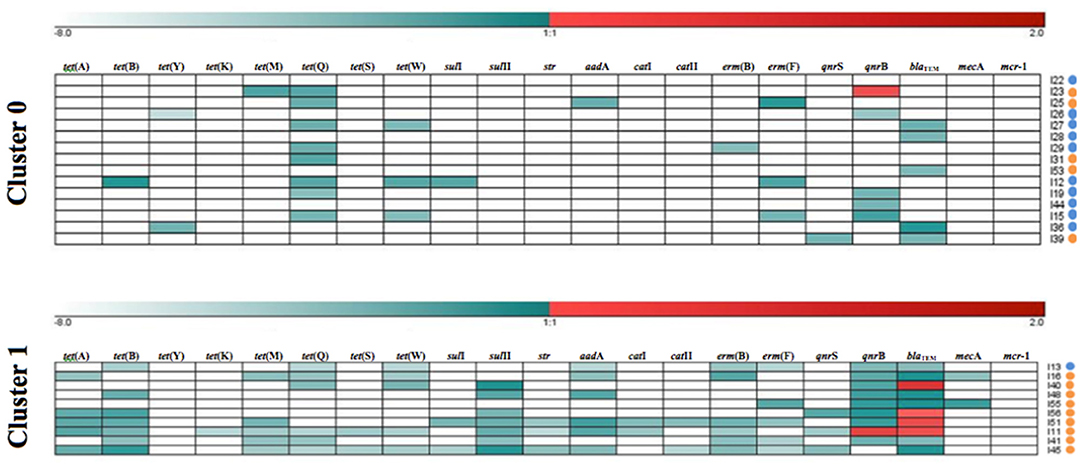
Figure 1. Resistance patterns of kelp gull (Larus dominicanus) and Magellanic penguin (Spheniscus magellanicus) samples obtained by k-means clustering of each antimicrobial resistance gene (ARG). Cluster 1 shows samples with high relative load percentage and Cluster 0 shows samples with low relative load percentage. Relative load percentage is expressed in a color scale (white for negative [−8] and dark red for the maximum value [+2]). The species are indicated on the right side (kelp gull [orange dots] and Magellanic penguin [blue dots]).
Results
All the tested samples validated for the 16S rRNA gene. All animals, with the exception of one individual (96%, 24/25), were positive to at least one ARG (Table 1). ARGs results according with the species are described below.
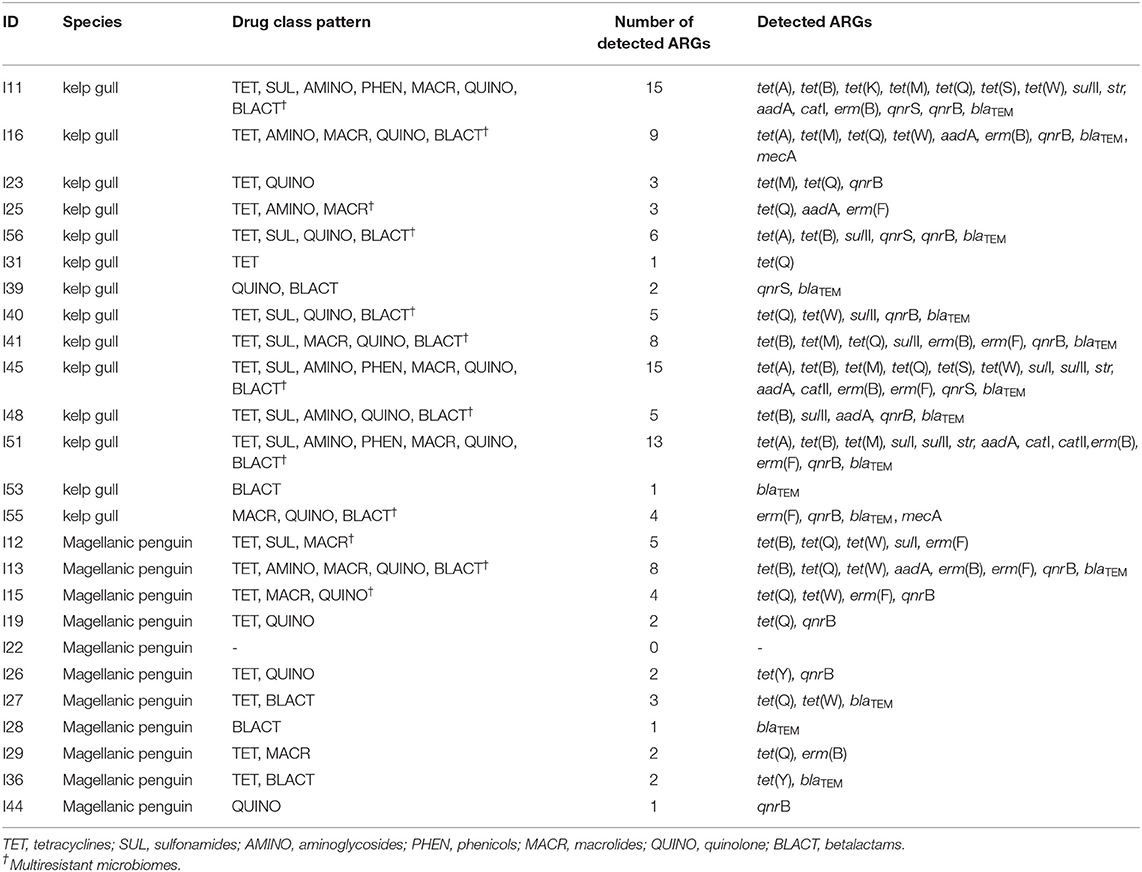
Table 1. Microbiome patterns, number of detected genes per sample, and detected genes according with the animal ID and species (kelp gull Larus dominicanus and Magellanic penguin Spheniscus magellanicus).
Kelp Gull
The blaTEM gene presented the highest occurrence (79%, 11/14), followed by qnrB (64%, 9/14), tet(Q) (57%, 8/14), sulII (50%, 7/14), tet(B), tet(M), and aadA (43%, 6/14), tet(A), erm(B) and erm(F) (36%, 5/14), tet(W), and qnrS (29%, 4/29), str (21%, 3/21), tet(S), sulI, catI, catII, and mecA (14%, 2/14), and tet(K) (7%, 1/14). The tet(Y) and mcr-1 genes were not detected in this group. The mean number of ARGs per sample was 6.4 (with min = 1 and max = 15). The blaTEM gene presented the highest mean load percentage (−2.2) (considering ≥-3 as the median value, with −8 [min] and +2 [max]).
When clustered by antimicrobial class, kelp gulls were positive to one or more genes encoding resistance to tetracycline, quinolone and betalactams (79%, 11/14), sulfonamides and macrolides (50%, 7/14), aminoglycosides (43%, 6/14), and phenicols (21%, 3/14). No gulls presented ARGs encoding polymyxin resistance (mcr-1). The mean number of genes conferring resistance to one or more antimicrobial classes presented in each gull sample was four. Additionally, 71% (10/14) of the gulls presented multiresistant microbiomes (Table 1), of these, five presented two similar patterns: a tetracycline, sulfonamide, quinolone, betalactam, aminoglycoside, phenicol and macrolide combination (30%; 3/10), and a tetracycline, sulfonamide, quinolone and betalactam combination (20%; 2/10).
Magellanic Penguin
The tet(Q) gene presented the highest occurrence (55%, 6/11), followed by qnrB (45%, 5/11), blaTEM and tet(W) (36%, 4/11), erm(F) (27%, 3/11), tet(B), tet(Y), and erm(B) (18%, 2/11), sulI and aadA (9%, 1/11). Genes tet(A), tet(K), tet(M), tet(S), sulII, str, catI, catII, qnrS, mecA, and mcr-1 were not detected. The mean number of ARGs per sample was 2.7 (with a maximum of eight genes per individual). Only one penguin did not present any of the tested ARGs. None of the genes presented mean load percentage ≥−3.
When clustered by antimicrobial class, individuals were positive to one or more genes encoding resistance to tetracyclines (73%, 8/11), quinolone (45%, 5/11), macrolides and betalactams (36%, 4/11), and sulfonamides and aminoglycosides (9%, 1/11). None of the individuals presented ARGs encoding chloramphenicol or polimyxin resistance. The mean number of genes conferring resistance to one or more antimicrobial classes presented in each sample was 2.1. Mutiresistant microbiomes were found in 27% (3/11) of the penguins (Table 1). Although no common patterns were observed, genes conferring resistance to tetracycline and macrolides were detected in the microbiomes of the three individuals presenting multiresistant profiles.
Qualitative Analysis
There were significant differences between species (respectively, kelp gull and Magellanic penguin) in regards to: ARG occurrence (blaTEM [79 and 36%. p = 0.032]; tet(M) [43 and 0%. p = 0.015]; tet(A) [36 and 0%. p = 0.003]; and sulII [50 and 0%. p = 0.007]), mean number of ARGs per sample (6.4 and 2.7. p = 0.031), ARG mean load percentage (aadA [−5.4 and −7.7. p = 0.045], tet(A) [−5.8 and −8. p = 0.031]; tet(M) [−5.8 and −8. p = 0.016]; blaTEM [−2.2 and −5.8. p = 0.032]; sulII [−4.8 and −8. p = 0.008]), percentage of genes potentially conferring resistance to an antimicrobial class (betalactams [79 and 36%. p = 0.036] and sulfonamides [50 and 9%. p = 0.033]), mean number of genes conferring resistance to one or more antimicrobial classes (4 and 2.1. p = 0.024]), percentage of multiresistant microbiomes (71 and 27%. p = 0.032]), and clustering (0.6 and 0.1. p = 0.006]). Statistically significant differences are summarized in Table 2.
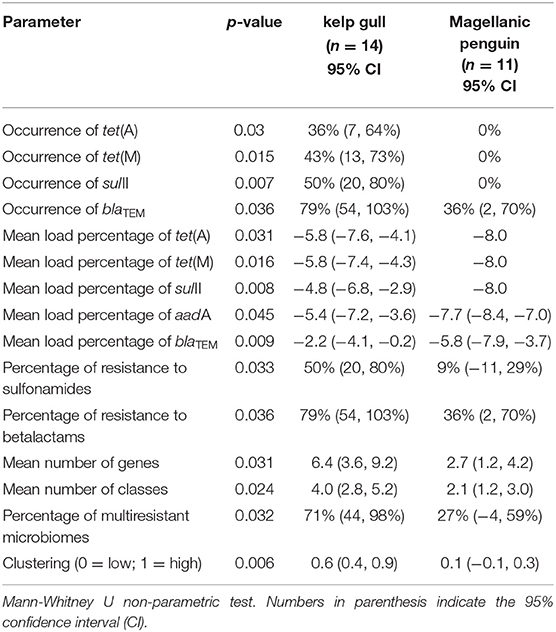
Table 2. Statistically significant differences between kelp gull (Larus dominicanus) and Magellanic penguin (Spheniscus magellanicus): ARG occurrence, mean number of ARGs per sample, mean load percentage of each ARG, the mean number of antimicrobial classes presented in each sample, percentage of multiresistant microbiomes, and resistance patterns.
Discussion
In accordance with our hypothesis, kelp gulls presented higher occurrence and load of ARGs than Magellanic penguins, findings that may potentially be influenced by the contrasting behaviors of these two seabird species in regard to feeding niches, interaction with human-impacted areas and dispersal. The kelp gull is the most widespread and abundant gull species in the Southern Hemisphere (38–40). Like other gull species, kelp gulls are extremely opportunistic and generalist feeders, very adapted to exploiting a wide variety of human-impacted and highly populated areas, and food subsidies (e.g., fishing discards and refuse disposals) (40–42). Such behaviors have been associated with the presence of ARGs in kelp gulls in Argentina (43), as well as in other gull species worldwide (17, 44, 45). Conversely, the Magellanic penguin is a migratory upper trophic level predator and the most abundant penguin in temperate areas, widely distributed along the southern coast of South America (24). Magellanic penguins remain in their colonies during breeding and molting periods, adopting a pelagic behavior while migrating along the continental shelf off the coast of northern Argentina, Uruguay, and southern Brazil (25, 26). Although scarce, studies on the presence of ARGs in penguins have associated ARGs occurrence with anthropization in remote locations (20, 46).
The mecA gene was detected in 14% (2/14) of kelp gulls, but not in penguins. This gene was reported in other wild bird groups in Brazil [passerines (47)] and Europe [corvids (48, 49), storks (50), and vultures (49)]. Nevertheless, to the best of our knowledge, this is the first report of mecA in seabirds in the Americas, only previously reported in European herring gulls (Larus argentatus) in Lithuania through metagenomics (51). The mecA gene is widely disseminated among Staphylococcus aureus and other staphylococcal species (52), encoding resistance to methicillin and cross-resistance to other β-lactam antimicrobials (52–54). Methicillin-resistant staphylococci are disseminated worldwide, frequently causing health care- and community-associated infections (52, 55), being considered one of the leading causes of nosocomial infection in Latin America (56), where it was also reported in animals, food products and the environment (57–59).
The blaTEM gene was detected in kelp gulls and Magellanic penguins, being the most prevalent gene in the former species (79%; 11/14). BlaTEM also presented the highest mean load percentage in this study (−2.2, in kelp gull), indicating an increased dissemination potential in comparison with the other ARGs detected here. Furthermore, the Furthermore, the blaTEM gene presented significant differences in kelp gull in comparison with Magellanic penguin in regards to occurrence (79 and 36%. p = 0.032) and mean load percentage (−2.2 and −5.8. p = 0.032). This gene has been previously described in seabirds in Brazil (16), the United States (14, 60), and Europe Europe (51, 61–64). The TEM betalactamases confer resistance to cephalosporins and penicillins (65), one of the oldest and most widely used antimicrobial classes in humans and veterinary medicine (66, 67), partialy explaining their dissemination in the tested seabirds. Recently, a similar study in Brazil, that evaluated the microbiome of six species of wild seabirds (overall, 304 individuals), found that the blaTEM occurrence and percentage loads ranged from 0 to 25% and −8 to −0.6, respectively, and that the blaTEM prevalence was significantly higher in migratory in comparison with non-migratory species (16). Interestingly, despite the considerable differences regarding species and sampling size, herein we found higher blaTEM occurrence and mean load percentages in kelp gull and Magellanic penguin, and higher blaTEM occurrence in the non-migratory synanthropic species (kelp gull). Epidemiologically, our findings are very concerning, because while the migratory species evaluated by Ewbank et al. (16) were using a pristine habitat (Rocas Atoll), kelp gulls and Magellanic penguins are using anthropized environments. Kelp gulls especially, are using heavily anthropized areas, which likely influence not only the acquisition and potential transmission of ARGs, but also their development and maintenance, once these individuals are continuously more exposed to ARGs sources (e.g., landfills, wastewater), and consequently, to reinfection.
The genes encoding tetracycline resistance (tet) were the most prevalent in this study (79%; 11/14 in kelp gull and 73%; 8/11 in Magellanic penguin): tet(A), tet(M) and tet(W) in kelp gull, and tet(Q) in Magellanic penguin. Additionally, tet(Q) was the most prevalent gene in the penguin group (55%, 6/11). Interestingly, Ewbank et al. (16) found that tetracycline-encoding genes were also the most prevalent antimicrobial class (ranging from 64.5 to 87.9%), significantly greater than the rest of the other ARGs classes (16). Moreover, we observed significant differences between kelp gull and Magellanic penguin in terms of tet(M) and tet(A) occurrence (43 and 0%. p = 0.015, and 36 and 0%. p = 0.003, respectively), and mean load percentage (−5.8 and −8. p = 0.016, and −5.8 and −8. p = 0.031, respectively). The high tet occurrence found herein was not surprising, once it had been previously detected in other seabirds in Brazil (16), and its extensive use in human and veterinary medicine, and in agriculture (68, 69). Tet genes have been reported in gulls in the Americas (16, 70, 71) and Europe (9, 51, 61–64, 72), and in wild penguins in Antarctica (46, 73) and Brazil (74).
Genes sulI and sulII were detected in kelp gull (sulII: 50% [7/14]) and in a Magellanic penguin (sulI: 9% [1/11]). SulI and sulII encode resistance to sulfonamides and have been previously reported in wild seabirds in Brazil (16), with the former also reported in gulls in Europe (61–63, 72). SulII presented significant differences in kelp gulls in comparison with Magellanic penguin regarding its occurrence (50% and 0%. p =0.007) and mean load percentage (−4.8 and −8. p = 0.008). Additionally, resistance to sulfonamides was significantly different in kelp gull in comparison with Magellanic penguin (50 and 9%. p = 0.033). Interestingly, the prevalences of sulfonamide and sulII gene were statistically significant higher in seabirds from an anthropized in comparison with a pristine environment (16). Sulfonamides are among the oldest synthesized antimicrobials, used in several medical therapies (75). This antimicrobial class is known to persist in the environment (76), and to resist biodegradation in wastewater-treatment processes and in media with elevated microbial activity, such as byproduct sludge (77, 78). Thus, the fact that such antimicrobial class presented more significant findings in the synanthropic coastal species (kelp gull), likely indicates higher ARGs pollution of coastal environments due to anthropogenic impact and environmental contamination (e.g., WWTP effluents and wastewater discharge) (10, 16).
Finally, we also observed significant differences in the aadA mean load percentage between kelp gull and Magellanic penguin (respectively, −5.4 and −7.7. p = 0.045). The aadA gene encodes resistance to two aminoglycosides: streptomycin and spectinomycin (79). Aminoglycosides are used against several aerobic Gram-negative bacilli, many staphylococci, some streptococci, and mycobacteria. Of note, streptomycin is used in multidrug treatments against multidrug-resistant M. tuberculosis infections (80). AadA has been previously reported in gull species (61, 63, 72), and in little penguins (Eudyptula minor) (81).
Our findings, especially the detection of the public health relevant mecA and blaTEM genes, are very concerning. The present study evaluated samples collected upon the individuals' admission into a rehabilitation center. Thus, the ARGs detected here were acquired in the wild, most likely in the environment (either in anthropized (e.g., landfills, sewage) or natural (e.g., aquatic, continental shelf) epidemiological settings), but potentially from other sources as well, such as infected food items (82) and through intra and/or interspecific interactions (e.g., kleptoparasitism). Wildlife is not naturally exposed to antimicrobial therapy in the wild, but once under treatment in rehabilitation centers, the presence of ARGs in their microbiome may interfere, and even prevent, successful therapy. Similarly to nosocomial settings, due to the intense use of antimicrobials, rehabilitation centers may be highly contaminated by these drugs and their metabolites, as well as by ARGs, and exert intense selective pressure over the local resistome (83, 84). As a consequence, rehabilitation centers may be hot spots for ARGs acquisition, interaction, and development, facilitating resistance exchanges among wildlife, humans (e.g., staff) and the environment, both while in-care and upon release (84). Thus, rehabilitation centers are very important and informative settings for the study of ARGs within the One Health interface.
Magellanic penguins are a migratory species. Bird migrations may cover great distances, through natural bio-barriers such as oceans, thus considered as holders of a potential central epidemiological role in the dissemination of ARGs, even to remote locations (3, 16, 44). Because migratory birds are capable of acquiring ARGs from humans, domestic animals and the environment (15, 17, 20, 44, 45, 85–89), this group has been largely suggested as reservoirs and dispersers of antimicrobial resistance (45, 88, 90). Despite a recent experimental study in captive ring-billed gulls (Larus delawarensis) in which the individuals were able to shed and contaminate the artificial environment and infect cospecifics in a controlled setting (91), further studies under natural conditions are necessary to confirm such hypothesis. Herein, migration may have not been a key factor from an epidemiological perspective of ARGs dispersal affecting humans, because despite our significant findings in Magellanic penguin [e.g., detection of ARGs in 10 out of the 11 individuals and of a gene of great public health importance (blaTEM)], this is a highly pelagic species that spends a great part of its life cycle in the oceans (26), sustaining limited direct contact with humans. By contrast, kelp gulls are not migratory, only capable of small geographical dislocations (24). Such species presents synanthropic behavior and adaptability to highly anthropized areas, in closer contact with humans and food-producing animals, consequently playing a more relevant role than Magellanic penguins in the epidemiological chain of ARGs within the human-animal-environmental interface. These findings show that all geographical dislocations – from great migrations to small geographical movements, must be considered in the study of ARGs dispersal and epidemiology.
Herein, we showed that the biological and ecological parameters evaluated in this study (i.e., dispersal [migratory and non-migratory], feeding niche [coastal and pelagic], and interaction with human-impacted areas [synanthropic and non-synanthropic]) are key factors in the complex epidemiology of ARGs in wild seabirds. Additionally, we reported the first detection of the mecA gene in seabirds in the Americas. Our findings greatly contribute to the current knowledge on ARGs in wild birds both nationally and worldwide, emphasize the importance of ARGs studies in wildlife rehabilitation settings, and reinforce the utility of culture-free highly sensitive molecular diagnostics to assess ARGs in the microbiome of wild birds. Nevertheless, it is important to consider the limitations of our study: (1) our techniques characterize the resistance genotype, not the phenotype, (2) microbiomes were evaluated at the exact point in time of each sample collection, and host-bacteria could eventually lose ARGs-containing plasmids prior to transmission and/or dispersal, and (3) our small sampling size. Admission and pre-release sampling and analysis would allow future assessment of rehabilitation centers as epidemiological settings. Further studies on ARGs in the microbiome of a greater number of seabirds, considering biological and ecological parameters, and the species' natural history (e.g., feeding strategy, habitat, territory), are necessary to broaden our understanding regarding the occurrence and diversity of ARGs in seabirds, and their role as potential sources of infection and dispersal within the One Health chain of ARGs acquisition, interaction, and dissemination.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by the Ethical Committee in Animal Research of the School of Veterinary Medicine and Animal Sciences, University of São Paulo (process number 1753110716).
Author Contributions
AE, FE, CS, and JC-D: conceptualization. AE, CS, IS, and EN: formal analysis. AE, CS, IS, SC-S, MA, JR, and CK: original draft preparation. All authors: review and editing.
Funding
This study was financed by Coordination and Improvement of Higher Level or Education Personnel (CAPES), Brazilian National Council for Scientific and Technological Development (CNPq) ([grant numbers 304999-18 and 165364/2018-1]), São Paulo Research Foundation (FAPESP) (grant numbers [2016/20956-0 and 2018/25069-7]). The samples used in this study were collected as part of the Santos Basin Beach Monitoring Project (Projeto de Monitoramento de Praias da Bacia de Santos, PMP-BS), under ABIO N° 640/2015, required by the Brazilian Institute of the Environment and Renewable Natural Resources (IBAMA) of the Brazilian Ministry of Environment for the environmental licensing process of the oil production and transport by Petrobras at the pre-salt province (25°05′S 42°35′W to 25°55′S 43° 34′W), between 2,100 and 2,300 m isobaths.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the staff of Associação R3 Animal and INIA-CISA.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.651781/full#supplementary-material
References
1. Da Costa PM, Loureiro L, Matos AJ. Transfer of multidrug-resistant bacteria between intermingled ecological niches: the interface between humans, animals and the environment. Int J Environ Res Public Health. (2013) 10:278–94. doi: 10.3390/ijerph10010278
2. Jobbins SE, Alexander KA. From whence they came - antibiotic-resistant Escherichia coli in African wildlife. J Wildl Dis. (2015) 51:811–20. doi: 10.7589/2014-11-257
3. Smith S, Wang J, Fanning S, McMahon BJ. Antimicrobial resistant bacteria in wild mammals and birds: a coincidence or cause for concern? Ir Vet J. (2014) 67:8. doi: 10.1186/2046-0481-67-8
4. World Health Organization. Global Action Plan on Antimicrobial Resistance. (2019). Available online at: https://www.who.int/antimicrobial-resistance/global-action-plan/en/ (accessed January 13, 2020).
5. Hidasi HW, Neto JH, Moraes DMC, Linhares GFC, de Sá Jayme V, Andrade MA. Enterobacterial detection and Escherichia coli antimicrobial resistance in parrots seized from the illegal wildlife trade. J Zoo Wildl Med. (2013) 44:1–7. doi: 10.1638/1042-7260-44.1.1
6. Roca I, Akova M, Baquero F, Carlet J, Cavaleri M, Coenen S, et al. The global threat of antimicrobial resistance: science for intervention. New Microb New Infect. (2015) 6:22–9. doi: 10.1016/j.nmni.2015.02.007
7. Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol. (2009) 7:887–94. doi: 10.1038/nrmicro2245
8. D'Argenio V, Salvatore F. The role of the gut microbiome in the healthy adult status. Clin Chim Acta. (2015) 451:97–102. doi: 10.1016/j.cca.2015.01.003
9. Radhouani H, Igrejas G, Pinto L, Gonçalves A, Coelho C, Rodrigues J, et al. Molecular characterization of antibiotic resistance in enterococci recovered from seagulls (Larus cachinnans) representing an environmental health problem. J Environ Monit. (2011) 13:2227–33. doi: 10.1039/c0em00682c
10. Sacristán I, Esperón F, Acuña F, Aguilar E, García S, López MJ, et al. Antibiotic resistance genes as landscape anthropization indicators: using a wild felid as sentinel in Chile. Sci Total Environ. (2020) 703:134900. doi: 10.1016/j.scitotenv.2019.134900
11. Oro D, Martinez-Abrain A. Ecology and behaviour of seabirds. In: Duarte CM, editor. The Encyclopedia of Life Support Systems (EOLSS). Oxford: Eolss Publishers UNESCO (2004). p. 364–89.
12. Paleczny M, Hammill E, Karpouzi V, Pauly D. Population trend of the World's Monitored Seabirds, 1950–2010. PloS ONE. (2015) 10:e0129342. doi: 10.1371/journal.pone.0129342
13. Rabinowitz PM, Scotch ML, Conti LA. Animals as sentinels: using comparative medicine to move beyond the laboratory. ILAR J. (2010) 51:262–7. doi: 10.1093/ilar.51.3.262
14. Atterby C, Ramey AM, Hall GG, Järhult J, Börjesson S, Bonnedahl J. Increased prevalence of antibiotic-resistant E. coli in gulls sampled in Southcentral Alaska is associated with urban environments. Infec Ecol Epidemiol. (2016) 6:32334. doi: 10.3402/iee.v6.32334
15. Atterby C, Borjesson S, Ny S, Jarhult JD, Byfors S, Bonnedahl J. ESBL-producing Escherichia coli in Swedish gulls - A case of environmental pollution from humans? PloS ONE. (2017) 12:e0190380. doi: 10.1371/journal.pone.0190380
16. Ewbank AC, Esperón F, Sacristán C, Sacristán I, Krul R, Cavalcante de Macedo E., et al. Seabirds as anthropization indicators in two different tropical biotopes: a One Health approach to the issue of antimicrobial resistance genes pollution in oceanic islands. Sci. Total Environ. (2021) 754:142141. doi: 10.1016/j.scitotenv.2020.142141
17. Ahlstrom CA, Bonnedahl J, Woksepp H, Hernandez J, Olsen B, Ramey AM. Acquisition and dissemination of cephalosporin-resistant E. coli in migratory birds sampled at an Alaska landfill as inferred through genomic analysis. Sci Rep. (2018) 8:1–11. doi: 10.1038/s41598-018-25474-w
18. Mukerji S, Stegger M, Trustwell AV, Laird T, Jordan D, Abraham RJ, et al. Resistance to critically important antimicrobials in Australian silver gulls (Chroicocephalus novaehollandiae) and evidence of anthropogenic origins. J Antimicrob Chemother. (2019) 74:2566–74. doi: 10.1093/jac/dkz242
19. Dolejska M, Literak I. Wildlife is overlooked in the epidemiology of medically important antibiotic-resistant bacteria. Antimicrob Agents Chemother. (2019) 63:e01167-19. doi: 10.1128/AAC.01167-19
20. Marcelino VR, Wille M, Hurt AC, González-Acuña D, Klaassen M, Schlub TE, et al. Meta-transcriptomics reveals a diverse antibiotic resistance gene pool in avian microbiomes. BMC Biol. (2019) 17:1–11. doi: 10.1186/s12915-019-0649-1
21. Hamady M, Knight R. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res. (2009) 19:1141–52. doi: 10.1101/gr.085464.108
22. Monier J-M, Demanèche S, Delmont TO, Mathieu A, Vogel TM, Simonet P. Metagenomic exploration of antibiotic resistance in soil. Curr Opin Microbiol. (2011) 14:229–35. doi: 10.1016/j.mib.2011.04.010
23. Cevidanes A, Esperón F, Di Cataldo S, Neves E, Sallaberry-Pincheira N, Millán J. Antimicrobial resistance genes in Andean foxes inhabiting anthropized landscapes in central Chile. Sci Total Environ. (2020) 724:138247. doi: 10.1016/j.scitotenv.2020.138247
24. BirdLife International. Species factsheet: Larus dominicanus. (2020). Available online at: http://www.birdlife.org (accessed December 12, 2020).
25. Ruoppolo V, Vanstreels RET, Woehler EJ, Heredia SAR, Adornes AC, da Silva-Filho RP, et al. Survival and movements of Magellanic penguins rehabilitated from oil fouling along the coast of South America, 2000–2010. Mar Pollut Bull. (2012) 64:1309–17. doi: 10.1016/j.marpolbul.2012.05.009
26. Boersma PD, Frere E, Kane O, Pozzi LM, Pütz K, Rey AR, et al. Magellanic penguin. In: Borboroglu PG, Boersma PD, editors. Penguins: Natural History and Conservation. Seattle, WA: University of Washington Press. (2013). p. 233–63.
27. Peak N, Knapp CW, Yang RK, Hanfelt MM, Smith MS, Aga DS, et al. Abundance of six tetracycline resistance genes in wastewater lagoons at cattle feedlots with different antibiotic use strategies. Microb Ecol. (2007) 9:143–51. doi: 10.1111/j.1462-2920.2006.01123.x
28. Jiang L, Hu X, Xu T, Zhang H, Sheng D, Yin D. Prevalence of antibiotic resistance genes and their relationship with antibiotics in the Huangpu River and the drinking water sources, Shanghai, China. Sci Total Environ. (2013) 458–60:267–72. doi: 10.1016/j.scitotenv.2013.04.038
29. Esperón F, Sacristán C, Carballo M, de la Torre A. Antimicrobial resistance genes in animal manure, manure-amended and nonanthropogenically impacted soils in Spain. Adv Biosci Biotechnol. (2018) 9:469–80. doi: 10.4236/abb.2018.99032
30. Nieto-Claudin A, Esperón F, Blake S, Deem SL. Antimicrobial resistance genes present in the faecal microbiota of free-living Galapagos tortoises (Chelonoidis porteri). Zoonoses Pub Health. (2019) 66:900–8. doi: 10.1111/zph.12639
31. Devarajan N, Laffite A, Mulaji CK, Otamonga JP, Mpiana PT, Mubedi JI, et al. Occurrence of antibiotic resistance genes and bacterial markers in a tropical river receiving hospital and urban wastewaters. PloS ONE. (2016) 11:e0149211. doi: 10.1371/journal.pone.0149211
32. Wang FH, Qiao M, Su JQ, Chen Z, Zhou X, Zhu YG. High throughput profiling of antibiotic resistance genes in urban park soils with reclaimed water irrigation. Environ Sci Technol. (2014) 48:9079–85. doi: 10.1021/es502615e
33. Chen J, Yu Z, Michel FC Jr, Wittum T, Morrison M. Development and application of real-time PCR assays for quantification of erm genes conferring resistance to macrolides-lincosamides-streptogramin B in livestock manure and manure management systems. Appl Environ Microbiol. (2007) 73:4407–16. doi: 10.1128/AEM.02799-06
34. Cummings DE, Archer KF, Arriola DJ, Baker PA, Faucett KG, Laroya JB, et al. Broad dissemination of plasmid-mediated quinolone resistance genes in sediments of two urban coastal wetlands. Environ Sci Technol. (2011) 45:447–54. doi: 10.1021/es1029206
35. Marti E, Balcázar JL. Real-time PCR assays for quantification of qnr genes in environmental water samples and chicken feces. Appl Environ Microbiol. (2013) 79:1743–5. doi: 10.1128/AEM.03409-12
36. Tsuru A, Setoguchi T, Kawabata N, Hirotsu M, Yamamoto T, Nagano S, et al. Enrichment of bacteria samples by centrifugation improves the diagnosis of orthopaedics-related infections via real-time PCR amplification of the bacterial methicillin-resistance gene. BMC Res Notes. (2015) 8:288. doi: 10.1186/s13104-015-1180-2
37. Sacristán C, Esperón F, Herrera-León S, Iglesias I, Neves E, Nogal V, et al. Virulence genes, antibiotic resistance and integrons in Escherichia coli strains isolated from synanthropic birds from Spain. Avian Pathol. (2014) 43:172–5. doi: 10.1080/03079457.2014.897683
38. Burger J, Gochfeld M. Family Laridae (gulls). In: del Hoyo J, Elliott A, Sartagal J, editors. Handbook of the Birds of the World, Vol. 3, Hoatzin to Auks. Barcelona: Lynx Editions. (1996). p. 572–623.
39. Jiguet F, Capainolo P, Tennyson A. Taxonomy of the Kelp gull Larus dominicanus Lichtenstein revisited with sex-separated analyses of biometrics and wing tip patterns. Zool Stud. (2012) 51:881–92.
40. Yorio P, Branco JO, Lenzi J, Luna-Jorquera G, Zavalaga C. Distribution and trends in kelp gull (Larus dominicanus) coastal breeding populations in South America. Waterbirds. (2016) 39:114–35. doi: 10.1675/063.039.sp103
41. Ludynia K, Garthe S, Luna-Jorquera G. Seasonal and regional variation in the diet of the Kelp gull in Northern Chile. Waterbirds. (2005) 28:359–65. doi: 10.1675/1524-4695(2005)028[0359:SARVIT]2.0.CO;2
42. Silva-Costa A, Bugoni L. Feeding ecology of Kelp Gulls (Larus dominicanus) in marine and limnetic environments. Aquat Ecol. (2013) 47:211–24. doi: 10.1007/s10452-013-9436-1
43. Liakopoulos A, Olsen B, Geurts Y, Artursson K, Berg C, Mevius DJ, et al. Molecular characterization of extended-spectrum-cephalosporin-resistant Enterobacteriaceae from wild Kelp Gulls in South America. Antimicrob Agents Chemother. (2016) 60:6924–7. doi: 10.1128/AAC.01120-16
44. Bonnedahl J, Drobni M, Gauthier-Clerc M, Hernandez J, Granholm S, Kayser Y, et al. Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the south of France. PLoS ONE. (2009) 4:e5958. doi: 10.1371/journal.pone.0005958
45. Bonnedahl J, Stedt J, Waldenström J, Svensson L, Drobni M, Olsen B. Comparison of extended-spectrum β-lactamase (ESBL) CTX-M genotypes in Franklin Gulls from Canada and Chile. PLoS ONE. (2015) 10:e0141315. doi: 10.1371/journal.pone.0141315
46. Rahman MH, Sakamoto KQ, Kitamura SI, Nonaka L, Suzuki S. Diversity of tetracycline-resistant bacteria and resistance gene tet(M) in fecal microbial community of Adélie penguin in Antarctica. Polar Biol. (2015) 38:1775–81. doi: 10.1007/s00300-015-1732-x
47. Matias CAR, Pereira IA, Rodrigues DP, Siciliano S. Staphylococcus spp. isolated from wild birds apprehended in the local illegal trade in Rio de Janeiro, Brazil, and relevance in public health. Lett Appl Microbiol. (2018) 67:292–8. doi: 10.1111/lam.13035
48. Loncaric I, Stalder GL, Mehinagic K, Rosengarten R, Hoelzl F, Knauer F, et al. Comparison of ESBL- and AmpC producing Enterobacteriaceae and methicillin-resistant Staphylococcus aureus (MRSA) isolated from migratory and resident population of rooks (Corvus frugilegus) in Austria. PLoS ONE. (2013) 8:e84048. doi: 10.1371/journal.pone.0084048
49. Ruiz-Ripa L, Gómez P, Alonso CA, Camacho MC, de la Puente J, Fernández-Fernández R, et al. Detection of MRSA of lineages CC130-mecC and CC398-mecA and Staphylococcus delphini-lnu (A) in magpies and cinereous vultures in Spain. Microb Ecol. (2019) 78:409. doi: 10.1007/s00248-019-01328-4
50. Gómez P, Lozano C, Camacho MC, Lima-Barbero JF, Hernández JM, Zarazaga M, et al. Detection of MRSA ST3061-t843-mecC and ST398-t011-mecA in white stork nestlings exposed to human residues. J Antimicrob Chemother. (2016) 71:53–7. doi: 10.1093/jac/dkv314
51. Merkeviciene L, Ruzauskaite N, Klimiene I, Siugzdiniene R, Dailidaviciene J, Virgailis M, et al. Microbiome and antimicrobial resistance genes in microbiota of cloacal samples from European herring gulls (Larus argentatus). J Vet Res. (2017) 61:27–35. doi: 10.1515/jvetres-2017-0004
52. Ito T, Hiramatsu K, Tomasz A, De Lencastre H, Perreten V, Holden MT, et al. Guidelines for reporting novel mecA gene homologues. Antimicrob Agents Chemother. (2012) 56:4997–9. doi: 10.1128/AAC.01199-12
53. Laurent F, Chardon H, Haenni M, Bes M, Reverdy ME, Madec JY, et al. MRSA harboring mecA variant gene mecC, France. Emerg Infect Dis. (2012) 18:1465. doi: 10.3201/eid1809.111920
54. Figueiredo AMS, Ferreira FA. The multifaceted resources and microevolution of the successful human and animal pathogen methicillin-resistant Staphylococcus aureus. Mem Inst Oswaldo Cruz. (2014) 109:265–78. doi: 10.1590/0074-0276140016
55. Taubes G. The bacteria fight back. Science. (2008) 321:356–61. doi: 10.1126/science.321.5887.356
56. Guzmán-Blanco M, Mejía C, Isturiz R, Alvarez C, Bavestrello L, Gotuzzo E, et al. Epidemiology of meticillin-resistant Staphylococcus aureus (MRSA) in Latin America. Int J Antimicrob Agents. (2009) 34:304–8. doi: 10.1016/j.ijantimicag.2009.06.005
57. Paterson GK, Larsen AR, Robb A, Edwards GE, Pennycott TW, Foster G, et al. The newly described mecA homologue, mecA LGA251, is present in methicillin-resistant Staphylococcus aureus isolates from a diverse range of host species. J Antimicrob Chemother. (2012) 67:2809–13. doi: 10.1093/jac/dks329
58. Wan MT, Chou CC. Spreading of β-lactam resistance gene (mecA) and methicillin-resistant Staphylococcus aureus through municipal and swine slaughterhouse wastewaters. Water Res. (2014) 64:288–95. doi: 10.1016/j.watres.2014.07.014
59. Papadopoulos P, Angelidis AS, Papadopoulos T, Kotzamanidis C, Zdragas A, Papa A, et al. Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) in bulk tank milk, livestock and dairy-farm personnel in north-central and north-eastern Greece: prevalence, characterization and genetic relatedness. Food Microbiol. (2019) 84:103249. doi: 10.1016/j.fm.2019.103249
60. Bonnedahl J, Hernandez J, Stedt J, Waldenström J, Olsen B, Drobni M. Extended-spectrum β-lactamases in Escherichia coli and Klebsiella pneumoniae in Gulls, Alaska, USA. Emerg Infec Dis. (2014) 20:897–9. doi: 10.3201/eid2005.130325
61. Dolejska M, Cizek A, Literak I. High prevalence of antimicrobial-resistant genes and integrons in Escherichia coli isolates from black-headed gulls in the Czech Republic. J Appl Microbiol. (2007) 103:11–9. doi: 10.1111/j.1365-2672.2006.03241.x
62. Poeta P, Radhouani H, Igrejas G, Gonçalves A, Carvalho C, Rodrigues J, et al. Seagulls of the Berlengas natural reserve of Portugal as carriers of fecal Escherichia coli harboring CTX-M and TEM extended-spectrum beta-lactamases. Appl Environ Microbiol. (2008) 74:7439–41. doi: 10.1128/AEM.00949-08
63. Radhouani H, Poeta P, Igrejas G, Goncalves A, Vinue L, Torres C. Antimicrobial resistance and phylogenetic groups in isolates of Escherichia coli from seagulls at the Berlengas nature reserve. Vet Rec. (2009) 165:138–42. doi: 10.1136/vr.165.5.138
64. Literak I, Dolejska M, Janoszowska D, Hrusakova J, Meissner W, Rzyska H, et al. Antibiotic-resistant Escherichia coli bacteria, including strains with genes encoding the extended-spectrum beta-lactamase and QnrS, in waterbirds on the Baltic Sea Coast of Poland. Appl Environ Microbiol. (2010) 76:8126–34. doi: 10.1128/AEM.01446-10
65. Mroczkowska JE, Barlow M. Fitness trade-offs in blaTEM evolution. Antimicrob Agents Chemother. (2008) 52:2340–5. doi: 10.1128/AAC.00018-08
66. Guenther S, Ewers C, Wieler LH. Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front Microbiol. (2011) 2:246. doi: 10.3389/fmicb.2011.00246
67. Sharland M, Pulcini C, Harbarth S, Zeng M, Gandra S, Mathur S, et al. Classifying antibiotics in the WHO Essential Medicines List for optimal use—be AWaRe. Lancet Infect Dis. (2018) 18:18–20. doi: 10.1016/S1473-3099(17)30724-7
68. Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Micobiol Mol Biol R. (2001) 65:232–60. doi: 10.1128/MMBR.65.2.232-260.2001
69. Daghrir R, Drogui P. Tetracycline antibiotics in the environment: a review. Environ Chem Lett. (2013) 11:209–27. doi: 10.1007/s10311-013-0404-8
70. Martiny AC, Martiny JBH, Weihe C, Field A, Ellis J. Functional metagenomics reveals previously unrecognized diversity of antibiotic resistance genes in gulls. Front Microbiol. (2011) 2:238. doi: 10.3389/fmicb.2011.00238
71. Toro M, Retamal P, Ayers S, Barreto M, Allard M, Brown EW, et al. Whole-genome sequencing analysis of Salmonella enterica serovar Enteritidis isolates in Chile provides insights into possible transmission between gulls, poultry, and humans. Appl Environ Microbiol. (2016) 82:6223–32. doi: 10.1128/AEM.01760-16
72. Antilles N, Garcia-Migura L, Joensen KG, Leekitcharoenphon P, Aarestrup FM, Cerda-Cuellar M, et al. Audouin's gull, a potential vehicle of an extended spectrum beta-lactamase producing Salmonella agona. FEMS Microbiol Lett. (2015) 362:1–4. doi: 10.1093/femsle/fnu039
73. Rahman MH, Sakamoto KQ, Nonaka L, Suzuki S. Occurrence and diversity of the tetracycline resistance gene tet(M) in enteric bacteria of Antarctic Adélie penguins. J Antimicrob Chemother. (2008) 62:627–8. doi: 10.1093/jac/dkn209
74. Prichula J, Pereira RI, Wachholz GR, Cardoso LA, Tolfo NCC, Santestevan NA, et al. Resistance to antimicrobial agents among enterococci isolated from fecal samples of wild marine species in the southern coast of Brazil. Mar Pollut Bull. (2016) 105:51–7. doi: 10.1016/j.marpolbul.2016.02.071
75. Parasca OM, Gheată F, Pânzariu A, Geangalău I, Profire L. Importance of sulfonamide moiety in current and future therapy. Rev Med Chir Soc Med Nat Iasi. (2013) 117:558–64.
76. Lamshöft M, Sukul P, Zuhlke S, Spiteller M. Metabolism of 14C-labelled and non-labelled sulfadiazine after administration to pigs. Anal Bioanal Chem. (2007) 388:1733–45. doi: 10.1007/s00216-007-1368-y
77. García-Galán MJ, Díaz-Cruz MS, Barceló D. Identification and determination of metabolites and degradation products of sulfonamide antibiotics. Trends Analyt Chem. (2008) 27:1008–22. doi: 10.1016/j.trac.2008.10.001
78. Kümmerer K. The presence of pharmaceuticals in the environment due to human use–present knowledge and future challenges. J Environ Manage. (2009) 90:2354–66. doi: 10.1016/j.jenvman.2009.01.023
79. Clark NC, Olsvik O, Swenson JM, Spiegel CA, Tenover FC. Detection of a streptomycin/spectinomycin adenylyltransferase gene (aadA) in Enterococcus faecalis. Antimicrob Agents Chemother. (1999) 43:157–60. doi: 10.1128/AAC.43.1.157
80. Magnet S, Blanchard JS. Molecular insights into aminoglycoside action and resistance. Chem Rev. (2005) 105:477–98. doi: 10.1021/cr0301088
81. Lundbäck IC, McDougall FK, Dann P, Slip DJ, Gray R, Power ML. Into the sea: antimicrobial resistance determinants in the microbiota of little penguins (Eudyptula minor). Infect Genet Evol. (2020) 88:104697. doi: 10.1016/j.meegid.2020.104697
82. Brahmi S, Dunyach-Rémy C, Touati A, Lavigne J-P. CTX-M-15-producing Escherichia coli and the pandemic clone O25b-ST131 isolated from wild fish in Mediterranean Sea. Clin Microbiol Infec. (2015) 21:18–20. doi: 10.1016/j.cmi.2014.09.019
83. Blyton MDJ, Pi H, Vangchhia B, Abraham S, Trott DJ, Johnson JR, et al. Genetic structure and antimicrobial resistance of Escherichia coli and cryptic clades in birds with diverse human associations. Appl Environ Microbiol. (2015) 81:5123–33. doi: 10.1128/AEM.00861-15
84. Haenni M, Métayer V, Jarry R, Drapeau A, Puech M-P, Madec J-Y, et al. Wide spread of blaCTX-M-9/mcr-9 IncHI2/ST1 plasmids and CTX-M-9-producing Escherichia coli and Enterobacter cloacae in rescued wild animals. Front Microbiol. (2020) 11:601317. doi: 10.3389/fmicb.2020.601317
85. Bonnedahl J, Drobni P, Johansson A, Hernandez J, Melhus Å, Stedt J, et al. Characterization, and comparison, of human clinical and black-headed gull (Larus ridibundus) extended-spectrum β-lactamase-producing bacterial isolates from Kalmar, on the southeast coast of Sweden. J Antimicrob Chemother. (2010) 65:1939–44. doi: 10.1093/jac/dkq222
86. Hernandez J, Bonnedahl J, Eliasson I, Wallensten A, Comstedt P, Johansson A, et al. Globally disseminated human pathogenic Escherichia coli of O25b-ST131 clone, harbouring blaCTX-M-15, found in Glaucous-winged gull at remote Commander Islands, Russia. Environ Microbiol Rep. (2010) 2:329–32. doi: 10.1111/j.1758-2229.2010.00142.x
87. Hernandez J, Johansson A, Stedt J, Bengtsson S, Porczak A, Granholm S, et al. Characterization and comparison of extended-spectrum β-lactamase (ESBL) resistance genotypes and population structure of Escherichia coli isolated from Franklin's gulls (Leucophaeus pipixcan) and humans in Chile. PLoS ONE. (2013) 8:e76150. doi: 10.1371/journal.pone.0076150
88. Stedt J, Bonnedahl J, Hernandez J, Waldenström J, McMahon BJ, Tolf C, et al. Carriage of CTX-M type extended spectrum β-lactamases (ESBLs) in gulls across Europe. Acta Vet Scand. (2015) 57:74. doi: 10.1186/s13028-015-0166-3
89. Guenther S, Semmler T, Stubbe A, Stubbe M, Wieler LH, Schaufler K. Chromosomally encoded ESBL genes in Escherichia coli of ST38 from Mongolian wild birds. J Antimicrob Chemother. (2017) 72:1310–3. doi: 10.1093/jac/dkx006
90. Guenther S, Aschenbrenner K, Stamm I, Bethe A, Semmler T, Stubbe A, et al. Comparable high rates of extended-spectrum-beta-lactamase-producing Escherichia coli in birds of prey from Germany and Mongolia. PLoS ONE. (2012) 7:e53039. doi: 10.1371/journal.pone.0053039
Keywords: anthropization, marine pollution, antibiotic resistance, wildlife, gull, penguin, One Health
Citation: Ewbank AC, Esperón F, Sacristán C, Sacristán I, Neves E, Costa-Silva S, Antonelli M, Rocha Lorenço J, Kolesnikovas CKM and Catão-Dias JL (2021) Occurrence and Quantification of Antimicrobial Resistance Genes in the Gastrointestinal Microbiome of Two Wild Seabird Species With Contrasting Behaviors. Front. Vet. Sci. 8:651781. doi: 10.3389/fvets.2021.651781
Received: 11 January 2021; Accepted: 24 February 2021;
Published: 22 March 2021.
Edited by:
Alain Hartmann, Institut National de Recherche pour l'agriculture, l'alimentation et l'environnement (INRAE), FranceReviewed by:
Hazem Ramadan, Mansoura University, EgyptGetahun E. Agga, United States Department of Agriculture, United States
Copyright © 2021 Ewbank, Esperón, Sacristán, Sacristán, Neves, Costa-Silva, Antonelli, Rocha Lorenço, Kolesnikovas and Catão-Dias. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana Carolina Ewbank, carolewbank@yahoo.com.br
†These authors have contributed equally to this work and share first authorship
 Ana Carolina Ewbank
Ana Carolina Ewbank Fernando Esperón
Fernando Esperón Carlos Sacristán
Carlos Sacristán Irene Sacristán
Irene Sacristán Elena Neves2
Elena Neves2  Samira Costa-Silva
Samira Costa-Silva José Luiz Catão-Dias
José Luiz Catão-Dias