- Laboratory of Molecular Membrane Biology, Department of Cell Biology, Nencki Institute of Experimental Biology of the Polish Academy of Sciences, Warsaw, Poland
S-palmitoylation is a reversible, enzymatic posttranslational modification of proteins in which palmitoyl chain is attached to a cysteine residue via a thioester linkage. S-palmitoylation determines the functioning of proteins by affecting their association with membranes, compartmentalization in membrane domains, trafficking, and stability. In this review, we focus on S-palmitoylation of proteins, which are crucial for the interactions of pathogenic bacteria and viruses with the host. We discuss the role of palmitoylated proteins in the invasion of host cells by bacteria and viruses, and those involved in the host responses to the infection. We highlight recent data on protein S-palmitoylation in pathogens and their hosts obtained owing to the development of methods based on click chemistry and acyl-biotin exchange allowing proteomic analysis of protein lipidation. The role of the palmitoyl moiety present in bacterial lipopolysaccharide and lipoproteins, contributing to infectivity and affecting recognition of bacteria by innate immune receptors, is also discussed.
Introduction
Palmitic acid (C16:0) is a long-chain saturated fatty acid, and a component of various lipids playing important roles in cell membrane organization, signal transduction, and energy storage. Moreover, the palmitoyl chain can be attached to proteins in a process called palmitoylation (S-palmitoylation), which modification affects their localization and functioning.
In the human body palmitic acid is synthetized in a process called de novo lipogenesis. It takes place mainly in adipocytes, hepatocytes, and cells of lactating mammary glands (1). Palmitic acid is used for the synthesis of phospholipids and sphingolipids, may undergo elongation and/or desaturation into other fatty acids (e.g., stearic acid or oleic acid, respectively), and can be esterified to form storage lipids—triacylglycerols. Apart from de novo synthesis, palmitic acid is also provided to the human body with food. Since palmitic acid is universally found in natural fats, its consumption exceeds the consumption of other saturated fatty acids and, in the USA it accounts for about 60% of the total intake of saturated fatty acids (2). A growing body of experimental and clinical evidence points to a link between a westernized diet, including a high intake of saturated fatty acids, and chronic inflammatory diseases (3–5). As dietary saturated and unsaturated fatty acids apparently modulate activity of immune cells, their influence on the immune responses triggered upon infection is also beginning to be investigated (6). These facts drive the interest in palmitic acid with an aim of elucidating the molecular mechanisms of its immunomodulatory properties.
In this review, we focus on S-palmitoylation of proteins crucial for the interactions of pathogenic bacteria and viruses with the host. We emphasize novel data on the role of S-palmitoylated proteins in the invasion of host cells by pathogens and those involved in the host innate immune responses to the infection, which have been obtained thanks to the application of new technical approaches. Recently, substantial progress in the understanding of protein palmitoylation was made possible by the development of methods allowing high-throughput analysis of cellular/tissue palmitoyl proteomes. We begin, however, by showing how unique protein S-palmitoylation is among other protein lipidations.
The Many Faces of Fatty Acylation of Proteins
S-Palmitoylation of Proteins and Its Influence on Protein Localization, Trafficking, and Stability
S-palmitoylation is a posttranslational modification of proteins consisting in a potentially reversible covalent attachment of palmitoyl chain to a cysteine residue(s) of proteins through a thioester bond (Table 1). Thus, S-palmitoylation resembles other reversible regulatory posttranslational protein modifications, including phosphorylation or acetylation, well-established factors affecting protein structure and functions. In particular, S-palmitoylation modifies cellular localization of proteins and their stability. The most dramatic changes of localization concern cytosolic proteins which upon S-palmitoylation acquire a hydrophobic anchor facilitating their docking into membranes (Figure 1). However, several integral membrane proteins also undergo S-palmitoylation. It often occurs on cysteine residue(s) located in the proximity of the junction of the transmembrane and cytoplasmic domains of the protein. S-palmitoylated transmembrane proteins occupy various cellular compartments, such as endoplasmic reticulum, Golgi apparatus, and the plasma membrane. In accordance, for some proteins, such as transmembrane adaptor proteins in leukocytes, S-palmitoylation was found secondary to the length and hydrophobicity of the transmembrane domain as a determinant of plasma membrane destination (7).
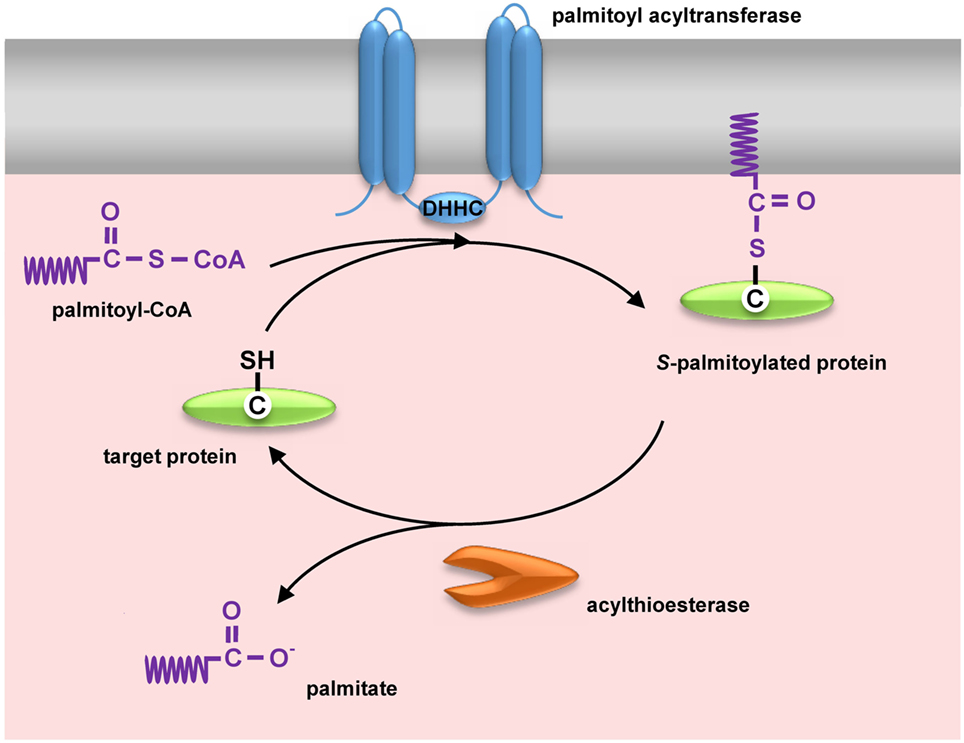
Figure 1. Dynamic protein S-palmitoylation. Palmitate is transferred to the thiol group of cysteine from palmitoyl-CoA by integral membrane zDHHC-family palmitoyl acyltransferases. Upon S-palmitoylation cytosolic proteins gain a hydrophobic moiety allowing their anchoring in the membrane. Proteins are depalmitoylated by acylthioesterases [acyl-protein thioesterase (APT) 1/APT2 and ABHD17] and translocate to the cytosol.
S-palmitoylation also contributes to the compartmentalization of proteins to distinct domains of membranes—rafts and tetraspanin-rich microdomains. In fact, the interest in S-palmitoylation was boosted when it was found to be required for the targeting of some signaling proteins to rafts. Rafts are nanodomains of the plasma membrane and some intracellular membranes, mainly of the trans-Golgi apparatus, rich in sphingolipids, glycerophospholipids with saturated fatty acid chains, and cholesterol (32). The plasma membrane nanodomains are sites of signal transduction by distinct receptors of immune cells involved in both acquired immune reactions, such as T cell receptor (TCR), Fcε receptor I, Fcγ receptor II, and in innate immune responses, such as toll-like receptor 4 (TLR4) (33, 34). Rafts are also sites of virion assembly and budding, as established, e.g., for influenza A virus and human immunodeficiency virus-1 (HIV-1) (35, 36). Peripheral membrane proteins acylated with saturated fatty acids are likely to anchor preferentially between the ordered saturated lipids of rafts rather than between the disordered lipids of the surrounding membrane. It has been shown that, owing to their raft localization, S-palmitoylated kinases of the Src family interact with raft-associating plasma membrane immunoreceptors and initiate signaling cascades fundamental to acquired immunity (15, 37, 38). It is worth noting that also the acyl chains attached to proteins can affect the membrane structure. Studies on model membranes have revealed that palmitic and myristic acids facilitate formation of ordered lamellar membrane regions (39, 40). In accordance, S-palmitoylation of erythrocyte peripheral membrane protein called membrane-palmitoylated protein 1 (MPP1) was found to be required for the proper lateral organization and fluidity of erythrocyte membrane. In the absence of MPP1 S-palmitoylation, raft assembly was disturbed and erythrocyte functioning compromised leading to hemolytic anemia in patients deficient in the enzyme catalyzing this reaction (41, 42). Preferential raft association is a feature of some S-palmitoylated transmembrane proteins, e.g., adaptor proteins PAG, LAT, and NTAL, which collaborate with the abovementioned immunoreceptors. In fact, palmitoylation is required for the raft association of most integral raft proteins (8, 43, 44).
On the other hand, S-palmitoylation does not obligatorily confer raft localization on transmembrane proteins. Certain S-palmitoylated proteins, such as transferrin receptor, glycoprotein G of vesicular stomatitis virus (VSV), and anthrax toxin receptor, tumor endothelial marker 8 (TEM8), are actually excluded from rafts. Apparently, a combination of S-palmitoylation and the properties of the transmembrane domain of the protein contribute to its destination to the raft or non-raft environment (43, 45). It has also been proposed that the attachment of a fatty acyl chain at the juxtamembrane cysteine(s) of a protein can induce tilting of its transmembrane fragment, determining in which part of the membrane it will accommodate to avoid a hydrophobic mismatch potentially caused by the thickness of the bilayer (46).
That not all S-palmitoylated proteins associate with rafts has been shown convincingly for macrophage-like RAW264 cells, where only about half of those proteins were found in the Triton X-100-resistant membrane fraction enriched in rafts (47, 48). In accordance, proteomic data on the distribution of S-palmitoylated proteins in prostate cancer cells have revealed that several such proteins are recovered in the non-raft (Triton X-100-soluble) fraction and are likely localized to microdomains enriched in scaffold proteins called tetraspanins (49). The tetraspanins are small integral membrane proteins found in the plasma membrane and other cellular membranes, having four transmembrane helices and undergoing S-palmitoylation at several conserved cysteine residues. The tetraspanins interact with each other and with various transmembrane and cytosolic partners, often also S-palmitoylated, forming microdomains (“tetraspanin web”) (50). It has been suggested that the amino acid composition of the S-palmitoylation site in some transmembrane proteins, such as the adaptor proteins involved in acquired immune responses, determines the association of those S-palmitoylated proteins with rafts or with the tetraspanin-enriched microdomains (44). An intriguing and still poorly addressed question concerns the relation between rafts and the tetraspanin-enriched microdomains, apparently of functional significance, e.g., during virus budding from host cells (35). This uncertainty stems partially from the fact that S-palmitoylation of tetraspanins governs their interactions with cholesterol and gangliosides leading at certain conditions to the recovery of tetraspanins in detergent-resistant membrane fractions enriched in rafts (51, 52). Besides its involvement in targeting proteins to rafts or tetraspanin-enriched microdomain, S-palmitoylation has been found to govern accumulation of the transmembrane chaperone protein calnexin in the perinuclear domain of endoplasmic reticulum (53).
S-palmitoylation also affects protein stability through its interplay with ubiquitination or phosphorylation, as found for the anthrax toxin receptor TEM8, antiviral interferon-induced transmembrane protein IFITM1, calnexin, and zDHHC6, one of palmitoyl acyltransferases described below (54–57).
Possibly the most intriguing is the reversible character of S-palmitoylation. Enzymes catalyzing palmitoylation and depalmitoylation of proteins have been characterized (58, 59). Palmitate is transferred onto the thiol group of cysteine from cytosolic palmitoyl-CoA by palmitoyl acyltransferases, enzymes containing the zinc finger DHHC domain named after the highly conserved Asp–His–His–Cys peptide (Figure 1). This is a two-step reaction comprising transient autoacylation of zDHHC enzymes and transfer of the fatty acyl chain from this intermediate to a protein substrate (60). In mammals, the zDHHC enzyme family consists of 24 proteins, and zDHHC proteins are also found in other eukaryotes but not in bacteria nor are they encoded by viral genomes. Mammalian zDHHC enzymes, each having at least four transmembrane helices, are located in the plasma membrane, endoplasmic reticulum, and Golgi apparatus (58). They display some specificity toward their protein substrates and also selectivity toward fatty acyl moieties other than palmitate, which contributes to the heterogeneity of lipids attached to proteins, such as viral glycoproteins described below (61). In the opposite process, the thioester bond is cleaved by acyl-protein thioesterases (APTs) (APT1 and APT2) and palmitoyl protein thioesterases (PPTs) (PPT1 and PPT2), which are localized in the cytosol and in lysosomes, respectively. APT1 and APT2 likely govern the dynamic functional changes of S-acylation of proteins (62) while PPT1 and PPT2 depalmitoylate proteins during their degradation (63, 64). Recently, serine hydrolases of the ABHD17 family have also been identified as depalmitoylating enzymes, and their specific substrate proteins determined (65, 66). Of note, the zDHHCs, APT1/APT2, and ABHD17 proteins are S-palmitoylated themselves, and palmitoylation of zDHHCs and depalmitoylation of APT1/2 can occur in a cascade manner (57, 62).
The dynamic cycles of palmitoylation/depalmitoylation detected for several peripheral membrane proteins are often synchronized with intracellular trafficking of those proteins. They circulate between the plasma membrane and the Golgi apparatus or endosomes, as exemplified by N- and H-Ras, R7-regulator of G protein and APTs. In fact, it is proposed that palmitoylation-dependent anchoring of APT1 in the plasma membrane allows it to depalmitoylate H-Ras at this location, while subsequent auto-depalmitoylation releases APT1 guiding it, alongside H-Ras, for another round of palmitoylation at the Golgi apparatus (62, 67–69). Cycles of palmitoylation/depalmitoylation are crucial for signaling by distinct plasma membrane receptors and for their distribution (69–71). Activation of TCR receptor or Fas receptor in T cells was found to trigger quick and transient palmitoylation of Lck kinase of the Src family (72, 73), but the exact meaning of the dynamic protein S-palmitoylation for processes triggered during the host–pathogen interaction awaits elucidation.
It is worth mentioning that although the zDDHC enzymes catalyze bulk protein palmitoylation in eukaryotic cells (74), some proteins have a unique autopalmitoylation activity. These include Bet3, a component of a multisubunit transport protein particle complex involved in vesicular trafficking, TEA domain transcription factors, and also bacterial Evf protein (75–78). The palmitic acid residue is attached constitutively to a specific cysteine residue of those proteins, remains buried inside a hydrophobic pocked in their core thereby affecting the tertiary structure and, thus, interactions with other proteins (75, 77). An exhaustive discussion on the physiology of S-palmitoylated proteins in eukaryotic cells can be found in several recent reviews (46, 79, 80).
S-Palmitoylation Is a Special Case of S-Acylation of Proteins
It has been established that, in addition to palmitate, various other fatty acyl moieties, such as saturated stearate (C18:0) or monounsaturated palmitoleate (C16:1), and oleate (C18:1) can be attached via the thioester linkage to proteins. The early reports on the heterogeneity of the fatty acyl moieties attached to cysteines obtained by analysis of selected immunoprecipitated proteins (15, 16, 81–83) have recently been complemented by a comprehensive proteomic analysis of fatty-acylated proteins of macrophage-like RAW264 cells (14). The latter study showed that an enrichment of culture medium of cells with monounsaturated fatty acids leads to their incorporation into a similar set of proteins as those normally modified with palmitate. Among them, several proteins relevant to innate immune responses were found. All these data justify the use of a broader term S-acylation rather than S-palmitoylation (Table 1). The physiological consequences of S-acylation of proteins with individual fatty acids are slowly being revealed. Modification of Fyn kinase with polyunsaturated fatty acid residue, such as arachidonate (C20:4), disturbed its raft localization and, thereby, TCR signaling (15). A heterogeneity of S-acylation was also found in viral spike proteins, such as hemagglutinin (HA) of influenza A virus, and E1 and E2 of Semliki Forest virus, which are modified in host eukaryotic cells by attachment of both palmitate and stearate (9). In HA, stearate is attached at the transmembrane cysteine while palmitate is attached to two cysteine residues in a membrane-proximal region of the protein. The stearoyl chain seems to accommodate into a groove formed by amino acids of the transmembrane helix shaping the domain in a way that facilitates its fitting into rafts (84). S-stearoylation of human transferrin receptor 1 at the juxtamembrane cysteine residues(s) is a key factor of the signaling cascade controlling mitochondrial morphology and functioning (13). Of interest, the latter study also showed that dietary supplementation of stearic acid reversed the deleterious effects of a genetically determined mitochondria dysfunction in Drosophila. Taking into account that unsaturated fatty acids affect the profile of S-acylation of proteins in vitro (14, 15), it is of outmost interest whether a similar effect of unsaturated and saturated (palmitic) fatty acids could be achieved in vivo with respect to proteins of immune cells.
N- and O-Acylation of Proteins
Beside S-acylation, less frequently palmitate can also be attached to the amine group of various amino acids (glycine, cysteine, and lysine) giving N-palmitoylation or to the hydroxyl group of serine or threonine in a process called O-palmitoylation (Table 1). As during S-palmitoylation, also other fatty acids can be utilized in these processes named then N- and O-acylation. Thus, a type of protein N-acylation is N-myristoylation, a frequent modification contributing to membrane anchoring of peripheral proteins. The saturated myristate (C14:0) is transferred to the protein from myristoyl-CoA by N-myristoyl transferase (two isozymes in mammals). In a vast majority of cases, myristate is attached co-translationally to the N-terminal glycine residue (after removal of the initiator methionine) via an amide linkage (Table 1). Like most lipidations, this modification is irreversible. Several viral proteins are N-myristoylated, such as Gag of HIV-1 crucial for budding of newly formed virions from plasma membrane rafts of host cells, and proteins of parasitic protozoa Plasmodium falciparum, Trypanosoma brucei, and Leishmania donovani (causing malaria, African sleeping sickness, and leishmaniosis, respectively). For this reason, N-myristoyl transferase is considered a potential drug target in the therapy of these diseases (17–19, 85, 86).
Data on the N- and O-palmitoylation of proteins involved in the host–pathogen interactions are limited, but interesting conclusions can be drawn from the information concerning proteins taking part in other processes. N-palmitoylation of the N-terminal glycine of the α-subunit of a heterotrimeric G protein (Gαs) has been described (20) (Table 1) besides the well-known S-palmitoylation of this pivotal signaling protein. The N-palmitoylation of Gαs is irreversible, and the enzyme responsible for this modification is unknown. It has been speculated that S- to N-palmitoyl migration can occur both in vivo and also in vitro during mass spectrometry analysis (20, 87). This suggests that caution is needed in interpreting results of this methodological approach, which is used with increasing frequency to study fatty acylation of proteins in immune cells (see next sections). Probably the best-characterized is the N-palmitoylation of the N-terminal cysteine residue of hedgehog proteins (sonic, Indian, and desert in mammals). It determines secretion of these proteins, which regulate embryonic patterning (Table 1). Secreted Wnt and ghrelin proteins are examples of O-acylation of serine residues with unusual fatty acid residues such as palmitoleate (C16:1) and octaonoate (C8:0) (Table 1). The fatty acylation of hedgehog, Wnt, and ghrelin is catalyzed by enzymes from the multipass membrane-bound O-acyl transferases family (31). Besides these unusual fatty acid residues, attachment of palmitate to serine and threonine residues is found in secreted venom toxins of the spider Plectreurys tristis, which selectively target neuronal ion channels (88). Also histone H4 is O-palmitoylated at a serine residue in the nucleus by acyl-CoA:lysophosphatidylcholine acyltransferase (25) (Table 1). The latter is of special interest in the context of innate immune responses since histone H4 O-palmitoylation regulates transcriptional activity, which is the final outcome of the pro-inflammatory signaling pathways triggered by receptors of the innate immune system.
Special attention should be devoted to ε-N-acylation consisting in the attachment of a fatty acid residue to the side chain of lysine by amide linkage (Table 1). ε-N-myristoylated are interleukin 1α (IL-1α) and tumor necrosis factor α (TNFα), the pro-inflammatory cytokines crucial in combating bacterial infections (22). The enzyme(s) catalyzing this reaction is unknown, but it has been established that sirtuins reverse this modification (89). The ε-N-acylation affects the release of TNFα by immune cells (90, 91). Surprisingly, this rare modification is also found in toxins of so-called RTX (repeats-in-toxin) class released by some pathogenic Gram-negative bacteria (23, 24). We describe these cases in more detail in the following sections.
S-Prenylation, Another Common Lipidation of Proteins
Besides S-palmitoylation and N-myristoylation, S-prenylation is another common lipidation that endows proteins with a hydrophobic moiety and contributes to their association with membranes. This modification relies on the posttranslational and irreversible attachment of either farnesyl or geranylgeranyl chains to a cysteine residue in the C-terminal CAAX box (alternatively also CC and CXC motifs) via a thioether linkage. The process is catalyzed by protein prenyl transferases that use polyprenylpyrophosphate as the donor of the isoprenoid group (Table 1). In peripheral membrane proteins, the S-palmitoylation site is often located in proximity of N-myristoylation or S-prenylation sites or a polybasic motif, which all are likely to mediate initial weak binding of a protein to a membrane and thereby facilitate subsequent attachment of palmitate to the protein by the integral membrane zDHHC enzymes (31). In contrast to S-palmitoylation, data on the role of S-prenylation of proteins key to the host–pathogen interactions are scarce (92). However, since S-prenylation is typical for the ubiquitous small GTPases of Ras superfamily, it is vital for proper functioning of B and T cells (93, 94).
A glance at Table 1 indicates that palmitate can be covalently bound via oxyester, amide, and thioester linkages to respective amino acid residues creating an array of possible modifications. O- and N-palmitoylation of proteins seems to be stable, resembling in this regard the other common protein lipidations, N-myristoylation and S-prenylation. By contrast, there exist enzymes cleaving the thioester bond formed during S-palmitoylation. For a long time, our understanding of protein S-palmitoylation and its dynamics was poor in comparison with other reversible protein modifications due to technical difficulties. Only recently have these difficulties been overcome with the introduction of methods allowing high-throughput identification of palmitoylated proteins, also those involved in the immune response to microbial pathogens, as discussed in the next sections.
Methodological Progress Facilitates Detection of Protein Palmitoylation
One of the basic problems hindering studies on protein S-palmitoylation lies in the fact that there is no identifiable consensus sequence for the palmitoylation site that could facilitate its prediction. From the technical point of view, the progress in a comprehensive survey of protein S-palmitoylation was also hampered by a lack of antibodies detecting this modification, with the sole exception of an antibody specific to palmitoylated PSD-95 (95). A classical method used to demonstrate protein palmitoylation is based on metabolic labeling of living cells with [3H]-palmitic acid, subsequent immunoprecipitation of a selected protein and detection of the incorporated tritiated fatty acid by autoradiography (96). A major disadvantage of this method is its low sensitivity. Only a minute fraction of the radioactive palmitate is bound to proteins, the majority being incorporated into lipids, which requires lengthy film exposure (counting in days).
A methodological breakthrough in the identification of palmitoylated proteins came with the development of two non-radioactive methods based on so-called click chemistry (97–99) and acyl-biotin exchange (ABE) (74, 100). These techniques have paved the way for high-throughput mass spectrometry-based proteomic analysis of protein palmitoylation in various cells and tissues and facilitated identification of new palmitoylated proteins of both pathogens and host cells involved in the innate immune responses.
The Click Chemistry-Based Method of Analysis of Protein Fatty Acylation
In the click chemistry-based method, cells are metabolically labeled with a palmitic acid analog bearing an alkyne group at the ω carbon of the fatty acyl chain, such as 17-octadecynoic acid (17ODYA) or alk-16 (Figure 2A), and this step resembles the classic labeling of cells with [3H]-palmitic acid. However, in the click chemistry-based assay, the labeling and lysis of cells is followed by in vitro coupling of the function group of the palmitic acid analog to a reporter tag, which greatly enhances the sensitivity of detection of labeled proteins (98, 99). Thus, after cell lysis, the labeled proteins are subjected to Cu(I)-catalyzed cycloaddition known as “click” reaction with an azide-bearing detection tag. In this step, a triazol is formed between the alkyne group in the palmitic acid analog and the azide of the tag (Figure 2A). The azide-bearing tags can be either fluorescent, such as tetramethylrhodamine or dyes with infrared fluorescence, or carry a biotin moiety. Depending on the tag used, subsequent SDS-PAGE separation of proteins allows global visualization of palmitoylated proteins by simple in-gel fluorescence or by blotting with a streptavidin-conjugated reporter (98, 101, 102). Notably, proteins biotinylated via the click reaction can also be enriched on streptavidin-coated beads and then subjected to on-bead tryptic digestion (or in-gel digestion if eluted from the beads) followed by identification by mass spectrometry. Such comprehensive click chemistry-based proteomic analysis has brought about identification of an array of palmitoylated proteins in dendritic cells (10, 103), macrophage-like RAW264 cells (14), and T cells (99, 104, 105). Some of the S-palmitoylated proteins newly identified in those studies, such as IFITM3 and TLR2, are involved in the host–pathogen interactions regulating innate immune responses (10, 103), while many others are known to contribute to adaptive immunity (99, 105), as described below. Recently, global profiling of Toxoplasma gondii (the causative agent of toxoplasmosis) has been performed revealing that many components of the parasite’s motility complex are palmitoylated (106). Similar studies on Cryptococcus neoformans (the fungus causing cryptococcal meningitis) have revealed a contribution of specific zDHHC palmitoyl acyltransferase, called Pfa4, to its virulence (107). Moreover, application of analogs of various saturated and unsaturated fatty acids confirmed the heterogeneous nature of the fatty acylation of proteins in RAW264 cells and suggested that dietary unsaturated fatty acids, after incorporation to proteins, can change their properties and thereby affect the functioning of immune cells (14).
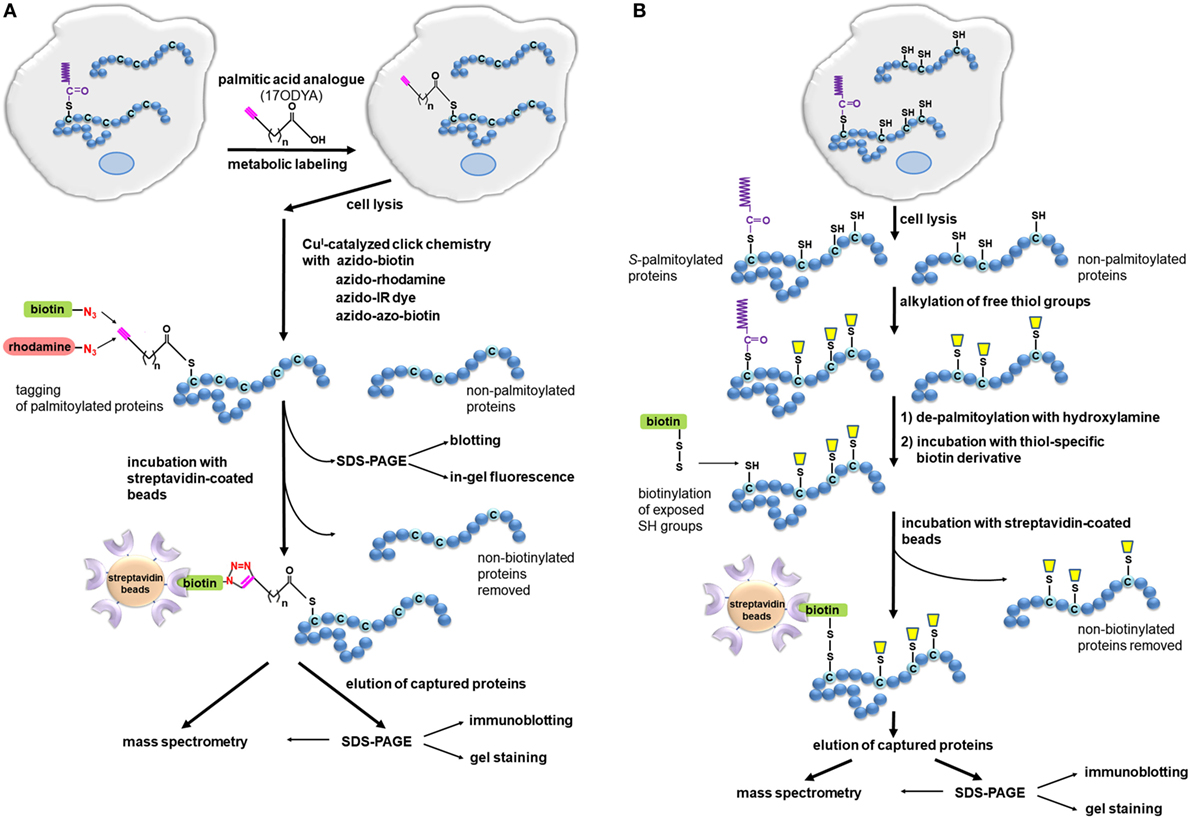
Figure 2. Detection of S-palmitoylated proteins using click chemistry and acyl-biotin exchange (ABE). (A) Click chemistry-based method. Cells are metabolically labeled with an alkyne-functionalized palmitic acid analog, such as 17-octadecynoic acid (17ODYA), and after cell lysis, the click reaction is conducted with azido-tagged biotin or fluorescent probes allowing enrichment and detection of labeled proteins in various ways. Biotinylated proteins can be bound on a streptavidin resin and then released using, e.g., high concentrations of urea and SDS (108). When a cleavable derivative of biotin, azido-azo-biotin, is used the labeled proteins are eluted from streptavidin beads with sodium dithionite, which cleaves the diazobenzene moiety in the linker arm of azido-azo-biotin, and analyzed by mass spectrometry or immunoblotting (109). (B) ABE method. Cells or tissues are lysed, free thiol groups of proteins are blocked by alkylation, and palmitoyl moieties are released with hydroxylamine. The newly exposed protein thiol groups are subjected to labeling with biotin-HPDP allowing selective binding, elution, and analysis of the originally S-palmitoylated proteins. The proteins can also be captured without biotinylation through a direct interaction of their thiol residues with a thiol-reactive resin (acyl-RAC technique).
The major advantage of the click chemistry-based method is that it can reveal the time course of protein S-palmitoylation. By using click chemistry-based labeling in the pulse-chase mode, one can follow the dynamics of protein palmitoylation. With such an approach, it was found that the palmitate turnover on Lck, an Src-family tyrosine kinase, is accelerated by T cell activation (72). Additional introduction of stable isotope labeling by amino acids in cells (SILAC) has provided quantitative proteomic data on the dynamics of protein palmitoylation in the cell (104, 110). This approach revealed, rather unexpectedly, that in unstimulated T cell hybridoma, the palmitoylation of most protein species does not undergo turnover (104). Another advantage of the click chemistry-based assay is its high specificity, because the alkyne group introduced in the analog of palmitic acid is not normally found in cells (98, 102). The click chemistry-based methods can also be used to follow the cellular localization of palmitoylated proteins by immunofluorescence when combined with the proximity ligation technique (111, 112). Palmitoylation of individual proteins can also be studied after their immunoprecipitation (11, 72, 73, 98).
Despite its unquestionable success, the click chemistry-based methods have limitations. They will detect only those proteins that undergo palmitoylation during the period of the metabolic labeling of cells. One should also bear in mind that the palmitic acid analog can be incorporated at S-, N-, and O-palmitoylation sites alike (111, 112). In addition, although 17ODYA (alk-16) is preferentially used to mimic palmitoylation of proteins, it can also be incorporated with low efficiency at N-myristoylation sites of proteins (98, 99). Another group of proteins that will be labeled with the palmitic acid analog but are not S-palmitoylated are those bearing the glycosylphosphatidylinositol (GPI) anchor (85, 113). Most of these limitations can be overcome using various fatty acid reporters, inhibitors, and by exploiting the sensitivity of the thioester bond to hydroxylamine treatment. Given the large variety of chemical reporters preferentially mimicking distinct fatty acids, recent years have witnessed a plethora of chemistry-based proteomic studies not only on palmitoylated but also myristoylated proteins and proteins bearing the GPI anchor, including those of pathogens and immune cells (10, 14, 85, 86, 114).
The ABE Method Reveals Protein S-Acylation
The ABE method can be used as a complement to the click chemistry-based approach in cell studies but unlike the latter it is uniquely suitable for studying whole tissues. ABE does not require metabolic labeling of proteins in living cells, thus some of the abovementioned limitations and difficulties do not apply. The ABE method relies on in vitro exchange of thioester-linked palmitate to a derivative of biotin which allows subsequent affinity purification of the resulting biotin-labeled proteins on streptavidin-coated beads (Figure 2B). The first step of the ABE involves lysis of cells or tissues followed by irreversible blockage of free thiol groups in the solubilized proteins by alkylation, most often with N-ethylmaleimide. Subsequently, the thioester bonds existing in S-palmitoylated proteins are broken with hydroxylamine, releasing palmitoyl moieties. The newly exposed thiol groups can now be tagged with sulfhydryl-reactive derivatives, such as biotin-HPDP, forming disulfide bonds with thiols. The biotinylated proteins are subsequently captured on streptavidin-coated beads and eluted with agents that reduce the disulfide bond between the protein and biotin-HPDP, such as β-mercapthoethanol, DTT, or TCEP (49, 74, 115, 116). As an alternative to biotinylation, in the so-called acyl-RAC technique, the newly exposed protein thiol groups in hydroxylamine-treated cell lysates are captured on a resin containing sulfhydryl-reactive groups (117). In both ABE and acyl-RAC, the eluted proteins can be separated by SDS-PAGE and visualized by gel staining or immunoblotting, or identified by mass spectrometry. Furthermore, when the hydroxylamine-released palmitoyl moieties are exchanged for a polyethylene glycol-maleimide derivative of a distinct molecular weight, a shift in-gel migration of tagged proteins is observed reflecting the number of fatty acyl residues originally S-bound to the protein (118, 119).
The ABE method has so far been used successfully for proteomic profiling of S-acylated proteins in immune cells, such as RAW264 cells (48), several types of blood cells, such as platelets, primary T cells, and immortalized B cells (120–122), pathogenic microorganisms such as T. brucei and T. gondii (123, 124), and tissues (125, 126). To quantify the aberrations in protein palmitoylation in a mouse model of Huntington’s disease, whole animal stable isotope labeling of mammals (SILAM) was applied followed by tissue isolation and ABE procedure (127). In another approach, for quantitative analysis of the T-cell palmitoylome, ABE was combined with labeling of proteins with various oxygen isotopes during their digestion with trypsin before mass spectrometry analysis (122). In addition, preselection of tryptic peptides obtained by ABE on streptavidin-coated or sulfhydryl-reactive resins greatly facilitates the identification of S-acylation sites by mass spectrometry (49, 110, 117).
Some aspects of the ABE method deserve a comment. Since the assay relies on the sensitivity of thioester bonds to hydroxylamine, ABE detects all S-acylation without distinguishing between S-palmitoylation and the other cases. Furthermore, there is a possibility of false-positive detection of proteins bearing a thioester linkage with compounds other than fatty acyl residues, such as ubiquitin in the E2 ubiquitin conjugase Ubc1 (115). Another source of false-positives is proteins in which free thiol groups were not completely alkylated before biotinylation. On the other hand, insufficient deacylation of bonafide fatty-acylated proteins with hydroxylamine results in their absence in the final sample (116).
In summary, the click chemistry-based method relies on metabolic labeling of cells with a palmitic acid analog which incorporates into proteins and next tagging it with reporter molecules greatly enhancing the sensitivity of detection. It only reveals proteins undergoing S-palmitoylation during metabolic labeling of cells and allows revealing turnover of this modification. By contrast, the ABE method is based on direct binding of sulfhydryl-reactive derivatives to thiol groups of cysteines unraveled by hydroxylamine treatment after lysis of cells or tissues. It allows the investigation of the whole but static palmitoylome. A comparative proteomic study of protein palmitoylation in P. falciparum found that the sets of proteins identified using these two approaches overlapped in 57.2% (113), indicating that they provide complementary data on the cellular palmitoyl proteome. Thanks to the application of the click chemistry- and ABE-based methods numerous new palmitoylated proteins have been identified. In 2015, a SwissPalm database was launched, (128) which provides an excellent, manually curated resource of information on palmitoylated proteins, palmitoylation sites, etc., available at http://swisspalm.epfl.ch/. All these efforts have greatly furthered our knowledge on molecular mechanisms regulating diverse aspects of cell functioning, including host–pathogen interactions and progress of infectious diseases, as highlighted below.
Palmitate as a Component of Proteins and Lipids Related to Bacterial Pathogenicity
Bacteria lack protein palmitoyl acyltransferases of the zDHHC family and, therefore, are essentially devoid of S-palmitoylated proteins. Yet, they have developed unique mechanisms utilizing fatty acids, such as palmitic acid, to modify their glycolipids and proteins. These modifications augment infectivity and help bacteria evade recognition by the host innate immune system. For example, the vast majority of Gram-negative bacteria produce lipopolysaccharide (LPS) as a part of their outer membrane. LPS is composed of the variable polysaccharide O-antigen and more-conserved lipid A containing two glucosamine residues hexa-acylated with hydroxymyristic, myristic, and lauric acid. Lipid A is recognized by CD14 protein and TLR4 receptor complexed with MD2 protein on the plasma membrane of the host immune and some non-immune cells. Activation of TLR4 triggers strong pro-inflammatory reactions aiming at eradication of the bacteria, but when exaggerated, eventually leading to sepsis (129). Incorporation of an additional palmitoyl chain into lipid A markedly diminishes its ability to activate TLR4 and to induce the host pro-inflammatory responses, which is correlated with an increased survival of bacteria forming a biofilm (130, 131). This strategy is utilized among others by Salmonella typhimurium, a causative agent of gastroenteritis, by Bordetella bronchiseptica, a respiratory pathogen of human and other mammals, and by Yersinia pestis causing plague (132, 133). The formation of the extra-acylated LPS relies on the transfer of palmitate from phospholipids onto the hydroxymyristate chain at position 2 of glucosamine of lipid A. The reaction is catalyzed by lipid A palmitoyltransferases (PagP in Salmonella and its homologs in other bacteria) localized in the outer membrane of these pathogens (134, 135). In addition to causing steric hindrance preventing the binding to the TLR4/MD2 complex, the hepta-acylation of LPS also protects bacteria from the lytic activity of cationic antimicrobial peptides, most likely by reducing the fluidity of the bacterial outer membrane (136, 137).
Apart from being incorporated into LPS in diverse bacteria, palmitate has also been found to modify a virulence factor of Gram-negative Erwinia carotovora, the Evf protein. The palmitoyl chain is linked via a thioester bond to the Cys209 residue at the center of Evf, plausibly by a self-palmitoylating activity of the protein. E. carotovora is a phytopatogen using insects such as Drosophila as vectors for dissemination between plants. The palmitoylation of Evf is required for infectivity of E. carotovora and its persistence in the insect gut, however, its mode of action of unknown. It has been speculated to be linked with an ability of Evf to associate with lipid bilayers, but the lack of similarities between Evf and any other bacterial protein of known function makes prediction on this subject difficult (79).
A number of bacterial toxins of so-called RTX class released during infection of mammals by pathogenic Gram-negative bacteria undergo ε-N-acylation of the side chain of internal lysines. These toxins include adenylate cyclase of Bordetella pertussis, acylated with palmitic acid, and α-hemolysin of extraintestinal (uropathogenic) Escherichia coli, acylated with myristic acid and also 15- and 17-carbon fatty acids. The acylation is catalyzed by an endogenous bacterial acyltransferase which, unlike its eukaryotic counterparts, transfers the acyl chain not from acyl-CoA but from acyl-carrier protein. The acylated toxins secreted by the bacteria bind to the plasma membrane of the host cells, oligomerize and form pores causing cell lysis. In the case of the toxin of B. pertussis, essential is also the delivery of the adenylate cyclase moiety to the cell interior. Acylation is required for virulence possibly being involved in oligomerization of the toxins (23, 24, 138).
Although lacking S-palmitoylated proteins (with the single known exception of Evf), bacteria express a wide range of membrane-bound proteins modified by a complex lipidation at the N-terminus, with palmitate frequently being a component of the lipid moiety (139, 140). The bacterial lipoproteins are synthesized in a multistep process catalyzed by a unique set of lipoprotein processing enzymes, Lgt, LspA, and Lnt, absent in eukaryotic cells. The formation of these lipoproteins begins with the attachment of a diacylglycerol via a thioester bond to a cysteine residue located in the so-called lipobox motif of the signal sequence of the transmembrane lipoprotein precursor. The signal sequence is then cleaved next to the lipid-modified cysteine leaving it at the N-terminus of the mature protein (141). In Gram-negative and less frequently also Gram-positive bacteria, a third fatty acid residue is additionally attached via an amide linkage to the amino group of the cysteine in a reaction analogous to the N-acylation of hedgehog proteins (see Table 1). This di- and tri-lipidation ensures membrane anchoring of the lipoproteins. All such lipoproteins of Gram-positive bacteria are exposed to the milieu while in Gram-negative bacteria some face the periplasm. The lipoproteins of Gram-positive bacteria, e.g., Streptococcus pneumoniae (causing pneumonia), Mycobacterium tuberculosis (tuberculosis), and Gram-negative bacteria, such as Neisseria meningitidis (meningitis), Y. pestis (plague), the spirochaete Borrelia burgdorferi (Lyme disease) and Treponema pallidum (syphilis) are crucial for their virulence. They control several aspects of the host–pathogen interactions, like adhesion and entry to host cells, protection against proteolysis and oxidative stress in the host cell, and regulation of expression of genes encoding cytokines both during initiation and progress of the disease (140–142). The surface exposure of the lipoproteins allows their involvement in the host cell invasion while on the other hand forming the so-called pattern signal recognized by the TLR2 receptor, which triggers the pro-inflammatory responses helping to combat the bacteria (143). Of interest, TLR2 is S-palmitoylated, as discussed below. The involvement of lipoproteins in pathogenesis fuels studies on their properties. One such recent work employing click chemistry to profile the lipoproteins of E. coli identified 88 lipoproteins with high/medium confidence, 70% of them predicted before by bioinformatics analysis (144). Notably, in that study a 14-carbon alkynyl fatty acid analog alk-14 rather than alk-16 was preferentially incorporated into the lipoproteins, contradicting earlier studies using gas chromatography and TLC, which found that palmitate was predominantly used for bacterial protein modification (139). Further studies are required to establish whether the fatty acid found in lipoproteins varies depending on culture conditions or is species specific. For example, 17ODYA labeling for click reaction confirmed incorporation of palmitate into pallilysin (Tp0751), a lipoprotein of T. pallidum. Pallilysin is a metalloprotease that degrades human fibrinogen and laminin. It is suggested that its exposure on the bacteria surface enables degradation of host structural proteins to facilitate rapid dissemination of this highly invasive pathogen (140).
Bacteria occasionally high-jack the palmitoylation machinery of host cells to modify the environment so as to favor their internalization, survival, and replication inside the cells. Bacillus anthracis (the causative agent of anthrax) is an example of such bacteria that modify S-palmitoylation of host proteins to their ends. The anthrax toxin produced by this pathogen binds to the TEM8 and CMG2 (capillary morphogenesis protein-2) proteins which, under physiological conditions, are involved in cell–cell and cell–extracellular matrix interactions. They are S-palmitoylated at multiple (two to four) cysteines (54). The S-palmitoylation of TEM8 was found to inhibit its association with plasma membrane rafts preventing its ubiquitination by the raft-associated E3 ubiquitin ligase Cbl. The binding of anthrax toxin drives association of the receptor-toxin complexes with rafts possibly correlated with depalmitoylation of the receptor. This allows subsequent ubiquitination of the receptor, an uptake of the receptor/toxin complexes in a clathrin-dependent manner and eventual delivery of the toxin to endosomes. These events are facilitated by S-palmitoylation of partner(s) of the receptors, most likely including kinases of the Src family (54, 145, 146).
While B. anthracis utilizes palmitoylated host proteins to induce its internalization, a growing body of data suggests that also bacterial proteins can undergo S-palmitoylation inside the host cells. This type of modification concerns so-called effectors, bacterial proteins that are injected into the host cell cytoplasm either across the plasma membrane or the membrane of vesicles enclosing internalized pathogens, with the help of their secretion systems. These are secretion systems type III and type IV, homologs of which have been described for pathogens and symbionts of mammals, insects, and plants (147, 148). The bacterial effectors can be S-palmitoylated to reach host cell membranes and thereby accumulate at a location most suitable for their activity. Application of the click chemistry-based method utilizing an analog of palmitic acid (alk-16) for cell labeling has revealed S-palmitoylation of two effector proteins of Salmonella enterica, such as SspH2 and SseI (149). S. enterica invades gut endothelial cells and is a leading cause of gastroenteritis and typhoid fever. SspH2 carries an E3 ubiquitin ligase domain while SseI shows sequence homology to bacterial proteins that have a deamidase activity, and inhibits migration of Salmonella-infected cells. The latter activity requires S-palmitoylation of SseI. Both proteins are stably S-palmitoylated, most likely by zDHH3 and zDHH7 of the host and bind to the plasma membrane in a palmitoylation-dependent manner (149). Also two effector proteins of the IpaH family of Shigella spp. were found to be S-palmitoylated in that study, suggesting that this modification can control the activity of effector proteins of other pathogens as well (149). Indeed, GobX and LpdA, effector proteins of Legionella pneumophila, the causative agent of Legionnaires’ disease invading macrophages and lung endothelial cells, are S-palmitoylated as was found recently using click chemistry. LpdA is a phospholipase hydrolyzing various phosphatidylinositols while GobX is an E3 ubiquitin ligase. GobX is targeted in a palmitoylation-dependent manner to the Golgi apparatus, and LpdA to the plasma membrane and a subset of intracellular vesicles (150, 151). Thus, the diversified subcellular localization of bacterial effector proteins reflects that of eukaryotic proteins.
It is worth noting that global profiling of acylated proteins with the application of click chemistry and an alkyne-functionalized analog of myristic acid, alk-14, for cell labeling was effective in reveling the mechanism of action of Shigella flexneri effector protein IpaJ of type III secretion system. This is a unique protease that cleaves off the N-terminal myristoylated glycine. This proteolytic demyristoylation activity of IpaJ is specific toward Golgi-associated ARF/ARL family of GTPases regulating cargo transport through the Golgi apparatus, inhibition of which is apparently pivotal for virulence of the bacteria causing diarrhea in humans (152).
In addition to the S-palmitoylation of the effectors of pathogenic bacteria of mammals mentioned earlier, double acylation, N-myristoylation and S-palmitoylation, has been reported of the so-called avirulence (Avr) proteins (effectors of type III secretion system) of Pseudomonas syringae, a causative agent of diverse plant diseases. Among them, AvrRpm1 and ArvB are N-myristoylated and S-palmitoylated by host acyltransferases at neighboring glycine and cysteine residues localized at the N-terminus of the proteins (similarly to eukaryotic kinases of the Src family), while in AvrPphB and two AvrPphB-like effectors—ORF4 and NopT, the double acylation motif is exposed after auto-cleavage of the proteins (similarly to some eukaryotic proteins cleaved by caspases). The acylation of the Avr proteins ensures their anchoring in the host plasma membrane, which is required for their functioning. In disease-susceptible plants Avr proteins contribute to successful infection; however, in plants expressing host resistance (R) genes they trigger plant defense signals, in both cases engaging plasma membrane-associated host proteins (153, 154).
The importance of palmitoylation of bacterial effector proteins for their infectivity is only beginning to be uncovered, in no small part owing to the development of the click chemistry-based method for detection of this protein modification. However, the strategy of high-jacking the host palmitoylation machinery to modify own proteins seems to be much more commonly employed by viruses.
Protein Palmitoylation in Viral Infections
Viruses do not encode palmitoyl acyltransferases but exploit extensively the host palmitoylation machinery to modify their proteins essential for infection of host cells and own replication. In fact, S-palmitoylation of proteins was discovered in 1979 as a modification of envelope glycoproteins of Sindbis virus and VSV. In those studies [3H]-palmitic acid was used for metabolic labeling of virus-infected cells and labeled proteins were identified by autoradiography (12, 155). Subsequently, a number of other viral proteins have been found to be palmitoylated using this approach.
The most-studied group of viral palmitoylated proteins is those found in enveloped viruses, i.e., viruses covered by a lipid bilayer obtained during their replication from a membrane of the host cell, such as the plasma membrane or endoplasmic reticulum. Influenza virus, HIV-1, hepatitis C virus (HCV), and herpes simplex virus (HSV) are the best known enveloped viruses. The envelope is rich in transmembrane, often S-palmitoylated, glycoproteins called spikes, which can bind to cognate receptors on the host cell plasma membrane triggering endocytosis of the virion, mediate subsequent fusion of the viral and cellular membranes allowing entry of the viral genome to the cytoplasm, and are also involved in the budding of newly formed virus particles from the cell. An example of such multifunctional palmitoylated transmembrane glycoproteins is HA present in the envelope of influenza virus together with another palmitoylated transmembrane protein, the matrix protein M2, which forms a proton channel earning the protein the name viroporin. As mentioned earlier, HA of influenza A virus is S-stearoylated and S-palmitoylated, respectively, at one cysteine residue located in the transmembrane domain of HA and two cysteines found in the cytoplasmic (intraviral) tail in close proximity to the membrane (156). On the other hand, M2 is S-palmitoylated on the amphiphilic helix located in the cytoplasmic part of the protein. Due to the S-palmitoylation and the presence of a cholesterol-binding motif the helix bends toward and associates with membranes (157, 158). During infection, HA binds to sialic acid residues of glycans localized on the surface of airway and alveolar epithelial cells. The bound virions are endocytosed and next the viral and endosome membranes fuse. The membrane fusion is driven by HA, which undergoes conformational changes induced by low pH of endosomes. Acidification of endosomes activates also the M2 proton channel activity, protons entering viral core facilitate dissociation of the viral genome which then moves to the nucleus where RNA replication occurs. The S-palmitoylation of HA is required for the fusion of the viral and endosome membranes at least in some subtypes of the virus while the ion channel activity of M2 is not dependent on its S-palmitoylation (159). Newly synthesized viral proteins and RNA are assembled into virions in the plasma membrane rafts which merge into lager platforms crucial for the virion assembly and budding off. The triple fatty acylation of HA is required for its targeting to plasma membrane rafts (160, 161). Besides S-palmitoylation, also the amino acid sequence of the transmembrane domain of HA determines its association with rafts (45). On the other hand, among the amino acids of the cytoplasmic tail of HA no other than the two S-palmitoylated cysteines are required for viral assembly and replication, although it is still not clear whether raft targeting (in cooperation with the transmembrane fragment) is the only mechanism of their participation. It is proposed that they affect conformation of the HA tail controlling its interaction with structural matrix protein M1 lying beneath the viral envelope (162, 163). The budding off of the virion is facilitated by M2 which localizes at the edges of rafts as a result of a combination of its S-palmitoylation, cholesterol binding, and properties of the transmembrane fragment. M2 protein can create a “wedge” altering membrane curvature thereby facilitating membrane scission and release of the virion (157, 164).
The influenza virus S-palmitoylated proteins are the archetype for many other viral proteins. Thus, S-palmitoylated spike glycoproteins include S-protein of coronaviruses (e.g., severe acute respiratory syndrome virus), the fusion (F) protein of paramyxoviruses (e.g., measles virus), Env of retroviruses [e.g., HIV-1, feline immunodeficiency virus (FIV)], and filoviruses (e.g., Ebola). Other viral proteins modified with palmitate are viroporins, such as E protein of coronaviruses, and also peripheral membrane proteins or nucleocapsid proteins absent in influenza virus. It has been found that S-palmitoylation of F13L, a peripheral protein of the envelope of vaccinia virus, controls the association of the protein with intracellular membranes, thereby the formation of the envelope (165). The core protein of the nucleocapsid of HCV resides on the surface of lipid droplets and binds in a palmitoylation-dependent manner to membranes of the droplet-associated endoplasmic reticulum. Subsequently, it recruits viral proteins and newly synthesized RNA for viral particle formation (166). Besides the interest in the role of viral protein S-palmitoylation for infectivity and possible use of host zDHHC enzymes as targets of anti-influenza drugs (167), viral proteins often serve as a model to study the consequences of fatty acylation for protein functioning and localization in distinct membrane domains (see S-Palmitoylation of Proteins and Its Influence on Protein Localization, Trafficking, and Stability of this review). Readers are referred to recent exhaustive reviews that consider these topics (36, 84, 168) while we will focus here on the recent advances in the field of viral protein palmitoylation brought about mainly by proteomic studies.
The click chemistry-based approach has led to the identification of S-palmitoylation in the cytoplasmic domain of the transmembrane spike protein Env of FIV, considered to be the cat equivalent of HIV-1. Env comprises three transmembrane gp41 glycoproteins and three associated gp120 which bind to CD4 receptor and coreceptors on the surface of T lymphocytes allowing fusion of the viral envelope and the plasma membrane and entry of viral capsid. Four cysteines in FIV Env are S-palmitoylated vis-a-vis two found in the Env of HIV-1. The two most membrane-proximal cysteines, 804 and 811, are required for the FIV membrane-fusion activity and incorporation of Env into virions (169), in agreement with the importance of Env S-palmitoylation for virion assembly of some HIV-1 strains (170–172). The assembly of HIV-1 virions takes place in plasma membrane rafts and is driven by N-myristoylated Gag protein which anchors and oligomerizes preferentially in these plasma membrane domains due to the presence of the fatty acyl chain (18).
The development of click chemistry-based methods allowed for the first time global profiling of acylated proteins in virus-infected cells. In addition to identifying acylated viral proteins this approach has also revealed how the viral infection modulates the acylation pattern of the host cell proteins. Thus far, click chemistry has been used to study protein myristoylation and palmitoylation in cells infected with HIV-1 and with HSV. In the latter case, the standard metabolic labeling with alkyne-functionalized myristic and palmitic acid analogs followed by click chemistry and mass spectrometry was combined with SILAC to discern between the changes in the extent of protein acylation and those in their abundance following viral infection. This approach allowed an elaborate quantitative analysis of host protein acylation and has revealed an overall downregulation of the level of both host protein modifications in infected cells. While the decreased content of myristoylated proteins resulted mainly from suppression of host protein synthesis, the drop in several S-palmitoylated proteins ensued from the inhibition of their palmitoylation in infected cells. The affected proteins were localized mainly to the plasma membrane and the Golgi apparatus and were involved in vesicle-mediated transport and ion transport. In addition, the study has expanded the list of HSV-encoded acylated (mostly palmitoylated) proteins that play different functions in the viral cycle, such as gE, gI, gK, US2, and US3 (110). Similar results pointing to global changes of host protein acylation were obtained upon analysis of protein myristoylation and palmitoylation in cells infected with HIV-1. In that study, the cells were labeled with analogs of palmitic or myristic acid tagged with an azide moiety for click chemistry reaction; however, the following mass spectrometry analysis did not address the relation between changes of protein acylation vs. alteration of protein level. The study identified 17 palmitoylated and 7 myristoylated proteins significantly differing in abundance between HIV-1 infected and uninfected cells. Several of the proteins affected by the infection were of host origin. The abundance of myristoylated proteins was in general increased while that of the palmitoylated ones—decreased in infected cells (173). In other words, the two studies have revealed that HSV and HIV-1 not only encode proteins that are acylated in the host cell but also alter the palmitoylation of host proteins, likely to adapt the cellular environment to favor their replication and budding. The majority of the acylated proteins affected by HIV-1 or HSV infection had not been described earlier in this context; therefore, further studies on these proteins could be crucial for better understanding of viral infection. Thus, the click chemistry-based approach has been highly effective in revealing changes of the host protein palmitoylation and opening new possibilities for the identification of novel antiviral drug targets.
Palmitoylation of Host Proteins Involved in Antibacterial and Antiviral Defense
The innate immune responses are the first line of active defense against microbial infections. The application of click chemistry-based and ABE methods and their use for large-scale analysis of protein palmitoylation in murine dendritic CD2.4 cells (10, 103), and murine macrophage-like RAW264 cells (14, 48) complemented by proteomic analysis of the raft fraction of those cells (47) have contributed significantly to the understanding of the role of palmitoylation of host receptors and signaling proteins involved in innate immune responses. Thus, the palmitoyl proteome analysis of murine dendritic cells unraveled S-palmitoylation of TLR2, a receptor expressed in cells of myeloidal lineage, which heterodimerizes with TLR1 or TLR6 to bind bacterial tri- or diacylated lipoproteins, respectively, and also other microbial components, such as glycolipids (e.g., lipoarabinomannan) of Mycobacterium and yeast zymosan (174). Besides TLR2, two other human TLRs out of 10 ectopically expressed in HEK293 cell, flagellin receptor TLR5, and TLR10, a unique TLR negatively regulating the pro-inflammatory activity of TLR2, were also found to be palmitoylated. The S-palmitoylation site of human TLR2 was mapped to Cys609 adjacent to its transmembrane domain. The modification was present in unstimulated cells and was linked with up-regulation of the cell surface localization of TLR2. Mutation of Cys609 abolished the ability of the receptor to induce pro-inflammatory signaling in response to microbial ligands of TLR2 (10). Further studies are needed to reveal whether S-palmitoylation of TLR2 controls its association with rafts as sites of TLR2 activation (175) and/or affects endocytosis of the receptor, as found for the anthrax toxin receptor (54).
One of the most extensively studied TLRs, TLR4 activated by bacterial LPS, is not palmitoylated. Yet, saturated fatty acids have been indicated to trigger pro-inflammatory signaling of TLR4. Thus, the TLR4/MD2 receptor complex is involved in the pro-inflammatory outcome of a diet rich in palmitic acid, as was found when analyzing markers of inflammation in the heart and adipose tissue of high fat diet-fed mice (176, 177). The molecular mechanisms underlying the pro-inflammatory properties of palmitic acid can involve its influence on the plasma membrane lipid order, hence raft organization, in a way that facilitates translocation of TLR4 (and TLR2) toward rafts (178, 179). Palmitic acid also directly binds to the TLR4-associated MD2 protein (177, 180). An influence of palmitic acid on sphingomyelin/ceramide metabolism, which enhances the LPS-induced responses, has also been considered (181). Recent proteomic studies based on 17ODYA labeling of RAW264 macrophage-like cells followed by click chemistry have revealed that stimulation of cells with LPS induces profound changes of the abundance of palmitoylated proteins (182). The data are in agreement with earlier findings showing that LPS induces accumulation of S-palmitoylated Lyn kinase in the raft-enriched fraction of cells, allowing it to downregulate TLR4 signaling (11). One of the upregulated S-palmitoylated proteins was type II phosphatidylinositol 4-kinase IIβ, which phosphorylates phosphatidylinositol to phosphatidylinositol 4-monophosphate. It was shown that palmitoylation determines the involvement of the kinase in LPS-induced signaling (182). These data suggest that S-palmitoylated proteins, including enzymes catalyzing phosphatidylinositol synthesis and turnover, are important factors affecting the pro-inflammatory responses triggered by LPS.
Notably LPS induces production of TNFα, a pro-inflammatory cytokine that is S-palmitoylated itself. TNFα is synthesized as a transmembrane 27-kDa precursor (tmTNFα) transported from the endoplasmic reticulum to the plasma membrane through the Golgi apparatus and recycling endosomes (183). Human tmTNFα is S-palmitoylated at Cys30 located at the boundary between its transmembrane and cytosolic fragments, as was found independently by radiolabeling and by labeling with 17ODYA followed by click chemistry (184, 185). Poggi et al. (185) arrived at a complex model explaining how the S-palmitoylation of TNFα affects its activity (Figure 3A). The modification was shown to favor the association of tmTNFα with rafts. Upon cell activation, the extracellular domain of tmTNF is cleaved by ADAM17 metalloproteinase whereupon the soluble TNFα (sTNFα) is released to the extracellular milieu and activates TNF receptor (TNFR) 1 and TNFR2. As ADAM17 localizes to both non-raft and raft regions of the plasma membrane, the S-palmitoylation of tmTNFα does not affect its cleavage and production of the soluble cytokine. However, S-palmitoylated tmTNFα interacts with TNFR1 in rafts thereby reducing the binding of sTNFα and consequently reducing the sensitivity of the cell to this cytokine. In addition, the fragment of tmTNFα which remains after the release of sTNFα in rafts if further processed by intramembrane SPPL2a and 2b proteases giving rise to ICD (intracellular domain) of an own biological activity. By contrast, the non-raft fragment of the ADAM17-cleaved tmTNFα is rapidly degraded (185).
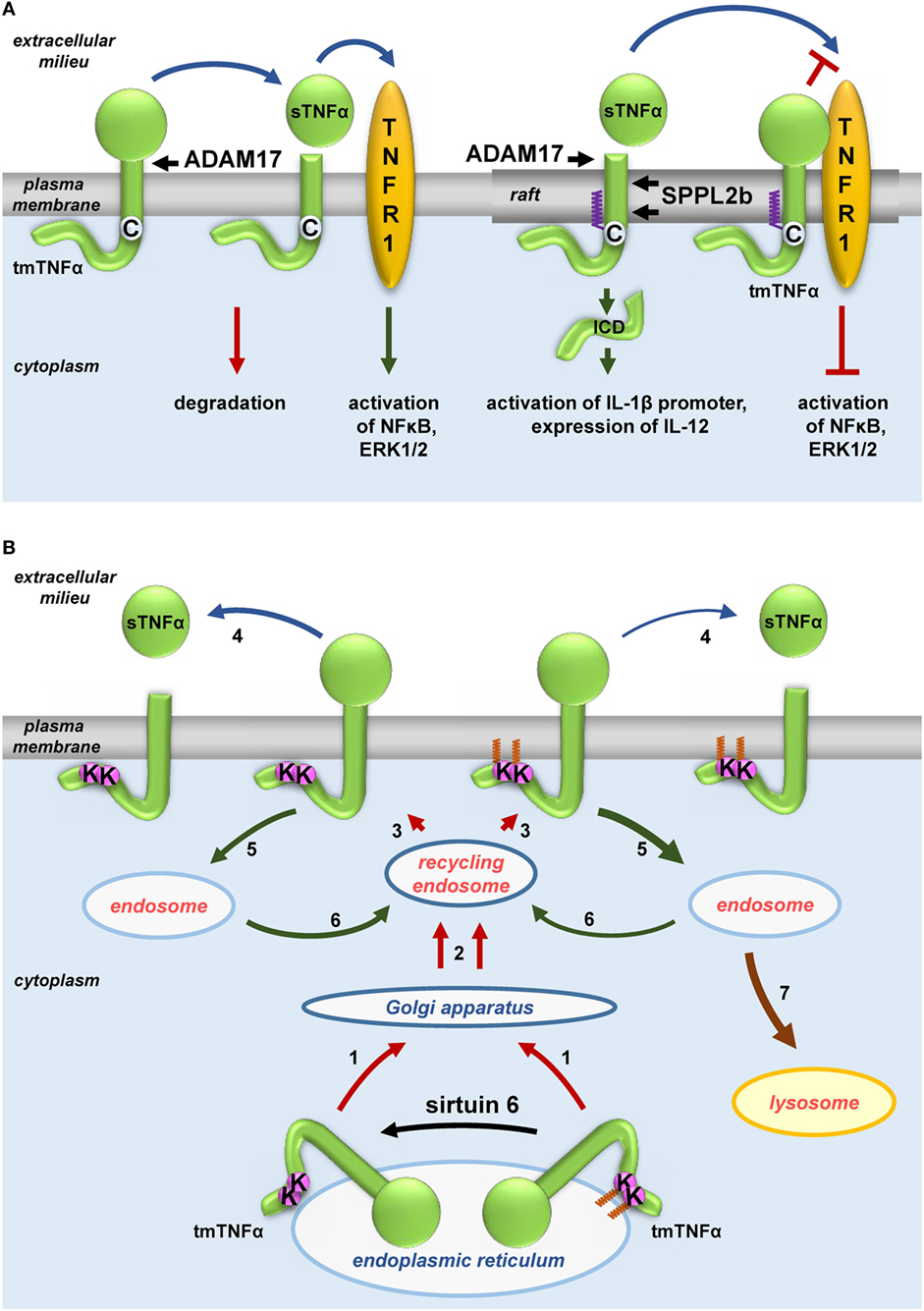
Figure 3. Influence of fatty acylation of transmembrane tumor necrosis factor α (TNFα) on production of soluble sTNFα. (A) S-palmitoylation and (B) ε-N-myristoylation of tmTNFα. (A) Non-palmitoylated tmTNFα is localized outside rafts while that S-palmitoylated on Cys30—in rafts of the plasma membrane. tmTNFα is cleaved by ADAM17 protease in both these plasma membrane environments giving rise to sTNFα, which subsequently activates TNF receptor (TNFR) 1 receptor leading to activation of NFκB and ERK1/2. However, only the raft-residing tmTNFα is further processed by SPPL2b protease to yield ICD, which activates the promoter of interleukin (IL)-1β and expression of IL-12. On the other hand, a pool of S-palmitoylated tmTNFα interacts in rafts with TNFR1 preventing its activation by sTNFα. (B) tmTNFα is transported from the endoplasmic reticulum via Golgi apparatus and recycling endosomes [1, 2] to the plasma membrane [3]. In the plasma membrane, TNFα is cleaved by ADAM17 giving rise to sTNFα [4] or is internalized [5] and either returns from the endosomes to the plasma membrane [6, 3] or is directed to lysosomes for degradation [7]. ε-N-myristoylation of tmTNFα at Lys19 and Lys20 facilitates its degradation [5, 7] at the expense of processing to sTNFα [4]. Oligomerization of tmTNFα and TNFR1 is not shown.
The transport and maturation of TNFα are also regulated by another posttranslational acylation, ε-N-myristoylation (22). As shown in Figure 3B, myristic acid residues are attached to two lysines (Lys19 and 20) of human tmTNFα. This modification is reversed by sirtuin 6 catalyzing the demyristoylation. Depletion of sirtuin 6 decreases the release of sTNFα since the ε-N-acylated TNFα precursor is redirected to and accumulates in lysosomes (90, 91). It is worth noting that exogenous palmitic acid stimulates the ε-N-myristoylation of tmTNFα, thereby reducing the release of sTNFα in favor of accumulation of tmTNFα in lysosomes (90, 91). This somehow surprising anti-inflammatory effect of palmitic acid can be explained by competitive binding between long-chain fatty acids (in this case, palmitic) and myristoylated substrates of sirtuin 6 found in vitro—(89) and adds a new dimension to the potential effects of palmitic acid.
S-palmitoylation of host proteins is also vital in antiviral defense. Viral nucleic acids, which are recognized by several TLRs and also cytoplasmic pattern-recognition receptors, induce robust production of type I interferons (IFNs), mainly INFα and IFNβ. The IFNα and IFNβ released from cells which first encounter viruses, e.g., dendritic cells, induce an antiviral reaction in an autocrine and paracrine manner upon binding to plasma membrane IFNα/β receptor (IFNAR) consisting of subunits 1 and 2. Both human IFNAR subunits are S-palmitoylated, as has been found by classical radiolabeling. The S-palmitoylation of IFNAR1 on Cys463, localized near the cytoplasmic end of the transmembrane domain, is required for downstream activation of STAT1 and STAT2 and the following transcription of IFNα-activated genes (186). Among the IFN-induced proteins, some have been shown to be palmitoylated, using click chemistry and ABE. They include the immunity-related GTPase Irgm1, BST2 also known as tetherin, and IFITM1 and 3 (10, 104). IFITMs are potent restriction factors against a wide range of enveloped viruses, e.g., influenza, West Nile, dengue, and Zika viruses (187, 188). IFITMs localize primarily to endolysosomal membranes where they inhibit viral replication by blocking their fusion with these membranes and also facilitate virus degradation (187). The exact mechanism of this antiviral activity is not clear, but it seems to rely on a perturbation of the organization of endolysosomal membranes. This can be linked with the intramembrane topology of IFITMs and their S-palmitoylation. IFITM1 and 3 likely possess two loops embedded in but not spanning the membrane with both the N- and C-termini facing the cytoplasm (55, 189). S-palmitoylation of conserved cysteine residues adjacent to these loops, Cys71, 72, and 105 in murine IFITM3, contributes to the membrane binding, similarly as found earlier for caveolins (119, 189). The S-palmitoylation also facilitates clustering of IFITM3 in the membranes, which is of potential significance for its antiviral activity (103). In support of the latter, the antiviral capacity was markedly reduced for non-palmitoylated mutant forms of IFITM3 (103, 119). However, S-palmitoylation did not affect the endolysosomal localization or stability of IFITM3. Subsequent studies have revealed that the localization and degradation of murine IFITM3, both shaping its antiviral capacity, are orchestrated by numerous posttranslational modifications comprising polyubiquitination, tyrosine phosphorylation by the Src-family kinase Fyn, and methylation (189, 190). By contrast, S-palmitoylation alone of the closely related murine IFITM1 endowed it with an antiviral activity and enhanced stability by preventing proteasomal degradation (55), which indicates diverse effects of this modification on individual IFITM isoforms.
The presented data are only beginning to fill the gap which existed in our understanding of the role of protein palmitoylation in innate immune responses. For a long time, it was lagging behind that on acquired immune responses, in which a plethora of S-palmitoylated proteins have long been known to be involved. They include receptors (CD4 and CD8), tyrosine kinases of the Src family, transmembrane adaptor proteins (e.g., LAT, NTAL, and PAG/Cbp), and α subunits of heterotrimeric G proteins. Their S-palmitoylation in most cases targets them to rafts and is a prerequisite for their involvement in the signaling pathways triggered by immunoreceptors [TCR, B cell receptor (BCR), and Fcγ and Fcε receptors] crucial for the acquired immune responses. An association of some components of these signaling pathways with tetraspanin-enriched domains has also been considered. These topics are discussed in several earlier reviews (44, 79, 191, 192). It is worth noting that large-scale proteomic analyses of fatty-acylated proteins of T cells (99, 104, 105, 122) and B cells (121), identifying numerous new palmitoylated proteins, have been published recently. Further studies will shed light on the possible engagement of those proteins in acquired immune responses and/or in the cross talk between the innate and the acquired immune system, in which phagocytic cells, such as macrophages and dendritic cells, are essential (193).
Concluding Remarks
Protein S-palmitoylation affects their localization, trafficking, and stability. It has long been known as an important factor controlling signal transduction by the BCR and TCR receptors involved in acquired immune responses. It is now becoming evident that palmitic acid is also a key lipid affecting the diverse processes at the host–pathogen encounter. Palmitate is a component of bacterial LPS and lipoproteins; S-palmitoylation of viral, some bacterial, and numerous host proteins is recognized as a crucial factor affecting both the virulence of pathogens and the innate immune reactions of the host. Our understanding of the latter has benefited greatly from the development of novel methods of detection of this protein modification. Their application has led to the identification of numerous proteins involved in the host–pathogen interaction. The methods have also allowed high-throughput proteomic analysis of palmitoylation of proteins in infected cells, showing widespread changes of the host cell palmitoylome. Future studies will tell whether complex feedback loops comprising palmitoyl acyltransferases and acylthioesterases, similar to those of kinases and phosphatases carrying out protein phosphorylation/dephosphorylation, are involved in controlling protein S-palmitoylation in infected cells. Revealing how the S-palmitoylation of particular proteins is regulated during the host–pathogen interactions should allow its modulation to favor the host defense.
Author Contributions
All authors contributed to writing and critically revised the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Prof. Andrzej Sobota from the Laboratory of Molecular Membrane Biology of the Nencki Institute (Warsaw, Poland) and Dr. Jan Fronk from the Faculty of Biology, University of Warsaw for helpful comments and critical discussion.
Funding
The work was supported by the National Science Centre, Poland, grant number DEC-2013/08/A/NZ3/00850 to KK.
Abbreviations
17ODYA, 17-octadecynoic acid; ABE, acyl-biotin exchange; APT, acyl-protein thioesterase; BCR, B cell receptor; FIV, feline immunodeficiency virus; GPI, glycosylphosphatidylinositol; HA, hemagglutinin; HCV, hepatitis C virus; HIV-1, human immunodeficiency virus-1; HSV, herpes simplex virus; IFITM, interferon-induced transmembrane protein; IFN, interferon; IFNAR, IFNα/β receptor; IL, interleukin; LPS, lipopolysaccharide; PPT, palmitoyl protein thioesterase; SILAC, stable isotope labeling by amino acids in cells; SILAM, stable isotope labeling of mammals; TCR, T cell receptor; TEM8, tumor endothelial marker 8; TNF, tumor necrosis factor; TNFR, TNF receptor; TLR, toll-like receptor; VSV, vesicular stomatitis virus; zDHHC, zinc finger DHHC domain containing.
References
1. Kersten S. Mechanisms of nutritional and hormonal regulation of lipogenesis. EMBO Rep (2001) 2:282–6. doi:10.1093/embo-reports/kve071
2. Fuster V, Topol EJ, Nabel EG. Atherothrombosis and Coronary Artery Disease. Philadelphia: Lippincott Williams & Wilkins (2005).
3. Greer JB, O’Keefe SJ. Microbial induction of immunity, inflammation, and cancer. Front Physiol (2011) 1:168. doi:10.3389/fphys.2010.00168
4. Kirwan AM, Lenighan YM, O’Reilly ME, McGillicuddy FC, Roche HM. Nutritional modulation of metabolic inflammation. Biochem Soc Trans (2017) 45:979–85. doi:10.1042/BST20160465
5. Statovci D, Aguilera M, MacSharry J, Melgar S. The impact of Western diet and nutrients on the microbiota and immune response at mucosal interfaces. Front Immunol (2017) 8:838. doi:10.3389/fimmu.2017.00838
6. DeCoffe D, Quin C, Gill SK, Tasnim N, Brown K, Godovannyi A, et al. Dietary lipid type, rather than total number of calories, alters outcomes of enteric infection in mice. J Infect Dis (2016) 213:1846–56. doi:10.1093/infdis/jiw084
7. Chum T, Glatzová D, Kvíčalová Z, Malínský J, Brdička T, Cebecauer M. The role of palmitoylation and transmembrane domain in sorting of transmembrane adaptor proteins. J Cell Sci (2016) 129:95–107. doi:10.1242/jcs.175190
8. Levental I, Lingwood D, Grzybek M, Coskun U, Simons K. Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc Natl Acad Sci U S A (2010) 107:22050–4. doi:10.1073/pnas.1016184107
9. Kordyukova LV, Serebryakova MV, Baratova LA, Veit M. Site-specific attachment of palmitate or stearate to cytoplasmic versus transmembrane cysteines is a common feature of viral spike proteins. Virology (2010) 398:49–56. doi:10.1016/j.virol.2009.11.039
10. Chesarino NM, Hach JC, Chen JL, Zaro BW, Rajaram MV, Turner J, et al. Chemoproteomics reveals toll-like receptor fatty acylation. BMC Biol (2014) 12:91. doi:10.1186/s12915-014-0091-3
11. Borzęcka-Solarz K, Dembińska J, Hromada-Judycka A, Traczyk G, Ciesielska A, Ziemlińska E, et al. Association of Lyn kinase with membrane rafts determines its negative influence on LPS-induced signaling. Mol Biol Cell (2017) 28:1147–59. doi:10.1091/mbc.E16-09-0632
12. Schmidt MF, Schlesinger MJ. Fatty acid binding to vesicular stomatitis virus glycoprotein: a new type of post-translational modification of the viral glycoprotein. Cell (1979) 17:813–9. doi:10.1016/0092-8674(79)90321-0
13. Senyilmaz D, Virtue S, Xu X, Tan CY, Griffin JL, Miller AK, et al. Regulation of mitochondrial morphology and function by stearoylation of TFR1. Nature (2015) 525:124–8. doi:10.1038/nature14601
14. Thinon E, Percher A, Hang HC. Bioorthogonal chemical reporters for monitoring unsaturated fatty-acylated proteins. Chembiochem (2016) 17:1800–3. doi:10.1002/cbic.201600213
15. Liang X, Nazarian A, Erdjument-Bromage H, Bornmann W, Tempst P, Resh MD. Heterogeneous fatty acylation of Src family kinases with polyunsaturated fatty acids regulates raft localization and signal transduction. J Biol Chem (2001) 276:30987–94. doi:10.1074/jbc.M104018200
16. Webb Y, Hermida-Matsumoto L, Resh MD. Inhibition of protein palmitoylation, raft localization, and T cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J Biol Chem (2000) 275:261–70. doi:10.1074/jbc.275.1.261
17. Pal R, Reitz MS, Tschachler E, Gallo RC, Sarngadharan MG, Veronese FD. Myristoylation of gag proteins of HIV-1 plays an important role in virus assembly. AIDS Res Hum Retroviruses (1990) 6:721–30. doi:10.1089/aid.1990.6.721
18. Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci U S A (2006) 103:11364–9. doi:10.1073/pnas.0602818103
19. Wright MH, Paape D, Price HP, Smith DF, Tate EW. Global profiling and inhibition of protein lipidation in vector and host stages of the sleeping sickness parasite Trypanosoma brucei. ACS Infect Dis (2016) 2:427–41. doi:10.1021/acsinfecdis.6b00034
20. Kleuss C, Krause E. Gα(s) is palmitoylated at the N-terminal glycine. EMBO J (2003) 22:826–32. doi:10.1093/emboj/cdg095
21. Buglino JA, Resh MD. Hhat is a palmitoylacyltransferase with specificity for N-palmitoylation of Sonic Hedgehog. J Biol Chem (2008) 283:22076–88. doi:10.1074/jbc.M803901200
22. Stevenson FT, Bursten SL, Locksley RM, Lovett DH. Myristyl acylation of the tumor necrosis factor alpha precursor on specific lysine residues. J Exp Med (1992) 176:1053–62. doi:10.1084/jem.176.4.1053
23. Lim KB, Walker CR, Guo L, Pellett S, Shabanowitz J, Hunt DF, et al. Escherichia coli α-hemolysin (HlyA) is heterogeneously acylated in vivo with 14-, 15-, and 17-carbon fatty acids. J Biol Chem (2000) 275:36698–702. doi:10.1074/jbc.C000544200
24. Hackett M, Guo L, Shabanowitz J, Hunt DF, Hewlett EL. Internal lysine palmitoylation in adenylate cyclase toxin from Bordetella pertussis. Science (1994) 266:433–5. doi:10.1126/science.7939682
25. Zou C, Ellis BM, Smith RM, Chen BB, Zhao Y, Mallampalli RK. Acyl-CoA:lysophosphatidylcholine acyltransferase I (Lpcat1) catalyzes histone protein O-palmitoylation to regulate mRNA synthesis. J Biol Chem (2011) 286:28019–25. doi:10.1074/jbc.M111.253385
26. Gutierrez JA, Solenberg PJ, Perkins DR, Willency JA, Knierman MD, Jin Z, et al. Ghrelin octanoylation mediated by an orphan lipid transferase. Proc Natl Acad Sci U S A (2008) 105:6320–5. doi:10.1073/pnas.0800708105
27. Takada R, Satomi Y, Kurata T, Ueno N, Norioka S, Kondoh H, et al. Monounsaturated fatty acid modification of Wnt protein: its role in Wnt secretion. Dev Cell (2006) 11:791–801. doi:10.1016/j.devcel.2006.10.003
28. Couvertier SM, Zhou Y, Weerapana E. Chemical-proteomic strategies to investigate cysteine posttranslational modifications. Biochim Biophys Acta (2014) 1844:2315–30. doi:10.1016/j.bbapap.2014.09.024
29. Sou Y, Tanida I, Komatsu M, Ueno T, Kominami E. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. J Biol Chem (2006) 281:3017–24. doi:10.1074/jbc.M505888200
30. Zha J, Weiler S, Oh KJ, Wei MC, Korsmeyer SJ. Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science (2000) 290:1761–5. doi:10.1126/science.290.5497.1761
31. Resh MD. Fatty acylation of proteins: the long and the short of it. Prog Lipid Res (2016) 63:120–31. doi:10.1016/j.plipres.2016.05.002
32. Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science (2010) 327:46–50. doi:10.1126/science.1174621
33. Horejsi V, Hrdinka M. Membrane microdomains in immunoreceptor signaling. FEBS Lett (2014) 588:2392–7. doi:10.1016/j.febslet.2014.05.047
34. Płóciennikowska A, Hromada-Judycka A, Borzęcka K, Kwiatkowska K. Co-operation of TLR4 and raft proteins in LPS-induced pro-inflammatory signaling. Cell Mol Life Sci (2015) 72:557–81. doi:10.1007/s00018-014-1762-5
35. Ono A. Relationships between plasma membrane microdomains and HIV-1 assembly. Biol Cell (2010) 102:335–50. doi:10.1042/BC20090165
36. Veit M, Serebryakova MV, Kordyukova LV. Palmitoylation of influenza virus proteins. Biochem Soc Trans (2013) 41:50–5. doi:10.1042/BST20120210
37. Kabouridis PS, Magee AI, Ley SC. S-acylation of LCK protein tyrosine kinase is essential for its signalling function in T lymphocytes. EMBO J (1997) 16:4983–98. doi:10.1093/emboj/16.16.4983
38. Kwiatkowska K, Frey J, Sobota A. Phosphorylation of FcγRIIA is required for the receptor-induced actin rearrangement and capping: the role of membrane rafts. J Cell Sci (2003) 116:537–50. doi:10.1242/jcs.00254
39. Gonzalez-Horta A, Andreu D, Morrow MR, Perez-Gil J. Effects of palmitoylation on dynamics and phospholipid-bilayer-perturbing properties of the N-terminal segment of pulmonary surfactant protein SP-C as shown by 2H-NMR. Biophys J (2008) 95:2308–17. doi:10.1529/biophysj.108.132845
40. Casas J, Ibarguren M, Alvarez R, Teres S, Llado V, Piotto SP, et al. G protein-membrane interactions II: effect of G protein-linked lipids on membrane structure and G protein-membrane interactions. Biochim Biophys Acta (2017) 1859:1526–35. doi:10.1016/j.bbamem.2017.04.005
41. Łach A, Grzybek M, Heger E, Korycka J, Wolny M, Kubiak J, et al. Palmitoylation of MPP1 (membrane-palmitoylated protein 1)/p55 is crucial for lateral membrane organization in erythroid cells. J Biol Chem (2012) 287:18974–84. doi:10.1074/jbc.M111.332981
42. Biernatowska A, Podkalicka J, Majkowski M, Hryniewicz-Jankowska A, Augoff K, Kozak K, et al. The role of MPP1/p55 and its palmitoylation in resting state raft organization in HEL cells. Biochim Biophys Acta (2013) 1833:1876–84. doi:10.1016/j.bbamcr.2013.03.009
43. Diaz-Rohrer BB, Levental KR, Simons K, Levental I. Membrane raft association is a determinant of plasma membrane localization. Proc Natl Acad Sci U S A (2014) 111:8500–5. doi:10.1073/pnas.1404582111
44. Stepanek O, Draber P, Horejsi V. Palmitoylated transmembrane adaptor proteins in leukocyte signaling. Cell Signal (2014) 26:895–902. doi:10.1016/j.cellsig.2014.01.007
45. Scheiffele P, Roth MG, Simons K. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J (1997) 16:5501–8. doi:10.1093/emboj/16.18.5501
46. Blaskovic S, Blanc M, van der Goot FG. What does S-palmitoylation do to membrane proteins? FEBS J (2013) 280:2766–74. doi:10.1111/febs.12263
47. Dhungana S, Merrick BA, Tomer KB, Fessler MB. Quantitative proteomics analysis of macrophage rafts reveals compartmentalized activation of the proteasome and of proteasome-mediated ERK activation in response to lipopolysaccharide. Mol Cell Proteomics (2009) 8:201–13. doi:10.1074/mcp.M800286-MCP200
48. Merrick BA, Dhungana S, Williams JG, Aloor JJ, Peddada S, Tomer KB, et al. Proteomic profiling of S-acylated macrophage proteins identifies a role for palmitoylation in mitochondrial targeting of phospholipid scramblase 3. Mol Cell Proteomics (2011) 10:M110.006007. doi:10.1074/mcp.M110.006007
49. Yang W, Di Vizio D, Kirchner M, Steen H, Freeman MR. Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol Cell Proteomics (2010) 9:54–70. doi:10.1074/mcp.M800448-MCP200
50. van Deventer SJ, Dunlock V-ME, van Spriel AB. Molecular interactions shaping the tetraspanin web. Biochem Soc Trans (2017) 45:741–50. doi:10.1042/BST20160284
51. Claas C, Stipp CS, Hemler ME. Evaluation of prototype transmembrane 4 superfamily protein complexes and their relation to lipid rafts. J Biol Chem (2001) 276:7974–84. doi:10.1074/jbc.M008650200
52. Charrin S, Manié S, Thiele C, Billard M, Gerlier D, Boucheix C, et al. A physical and functional link between cholesterol and tetraspanins. Eur J Immunol (2003) 33:2479–89. doi:10.1002/eji.200323884
53. Lakkaraju AK, Abrami L, Lemmin T, Blaskovic S, Kunz B, Kihara A, et al. Palmitoylated calnexin is a key component of the ribosome-translocon complex. EMBO J (2012) 31:1823–35. doi:10.1038/emboj.2012.15
54. Abrami L, Leppla SH, van der Goot FG. Receptor palmitoylation and ubiquitination regulate anthrax toxin endocytosis. J Cell Biol (2006) 172:309–20. doi:10.1083/jcb.200507067
55. Hach JC, McMichael T, Chesarino NM, Yount JS. Palmitoylation on conserved and nonconserved cysteines of murine IFITM1 regulates its stability and anti-influenza A virus activity. J Virol (2013) 87:9923–7. doi:10.1128/JVI.00621-13
56. Dallavilla T, Abrami L, Sandoz PA, Savoglidis G, Hatzimanikatis V, van der Goot FG. Model-driven understanding of palmitoylation dynamics: regulated acylation of the endoplasmic reticulum chaperone calnexin. PLoS Comput Biol (2016) 12:e1004774. doi:10.1371/journal.pcbi.1004774
57. Abrami L, Dallavilla T, Sandoz PA, Demir M, Kunz B, Savoglidis G, et al. Identification and dynamics of the human ZDHHC16-ZDHHC6 palmitoylation cascade. Elife (2017) 6:e27826. doi:10.7554/eLife.27826
58. Ohno Y, Kihara A, Sano T, Igarashi Y. Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim Biophys Acta (2006) 1761:474–83. doi:10.1016/j.bbalip.2006.03.010
59. Blaskovic S, Adibekian A, Blanc M, van der Goot GF. Mechanistic effects of protein palmitoylation and the cellular consequences thereof. Chem Phys Lipids (2014) 180:44–52. doi:10.1016/j.chemphyslip.2014.02.001
60. Jennings BC, Linder ME. DHHC protein S-acyltransferases use similar ping-pong kinetic mechanisms but display different acyl-CoA specificities. J Biol Chem (2012) 287:7236–45. doi:10.1074/jbc.M111.337246
61. Greaves J, Munro KR, Davidson SC, Riviere M, Wojno J, Smith TK, et al. Molecular basis of fatty acid selectivity in the zDHHC family of S-acyltransferases revealed by click chemistry. Proc Natl Acad Sci U S A (2017) 114:E1365–74. doi:10.1073/pnas.1612254114
62. Kong E, Peng S, Chandra G, Sarkar C, Zhang Z, Bagh MB, et al. Dynamic palmitoylation links cytosol-membrane shuttling of acyl-protein thioesterase-1 and acyl-protein thioesterase-2 with that of proto-oncogene H-Ras product and growth-associated protein-43. J Biol Chem (2013) 288:9112–25. doi:10.1074/jbc.M112.421073
63. Camp LA, Verkruyse LA, Afendis SJ, Slaughter CA, Hofmann SL. Molecular cloning and expression of palmitoyl-protein thioesterase. J Biol Chem (1994) 269:23212–9.
64. Duncan JA, Gilman AG. A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein α subunits and p21RAS. J Biol Chem (1998) 273:15830–7. doi:10.1074/jbc.273.25.15830
65. Lin DT, Conibear E. ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization. Elife (2015) 4:e11306. doi:10.7554/eLife.11306
66. Yokoi N, Fukata Y, Sekiya A, Murakami T, Kobayashi K, Fukata M. Identification of PSD-95 depalmitoylating enzymes. J Neurosci (2016) 36:6431–44. doi:10.1523/JNEUROSCI.0419-16.2016
67. Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, et al. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science (2005) 307:1746–52. doi:10.1126/science.1105654
68. Greaves J, Chamberlain LH. Differential palmitoylation regulates intracellular patterning of SNAP25. J Cell Sci (2011) 124:1351–60. doi:10.1242/jcs.079095
69. Jia L, Chisari M, Maktabi MH, Sobieski C, Zhou H, Konopko AM, et al. A mechanism regulating G protein-coupled receptor signaling that requires cycles of protein palmitoylation and depalmitoylation. J Biol Chem (2014) 289:6249–57. doi:10.1074/jbc.M113.531475
70. El-Husseini AE-D, Schnell E, Dakoji S, Sweeney N, Zhou Q, Prange O, et al. Synaptic strength regulated by palmitate cycling on PSD-95. Cell (2002) 108:849–63. doi:10.1016/S0092-8674(02)00683-9
71. Dekker FJ, Rocks O, Vartak N, Menninger S, Hedberg C, Balamurugan R, et al. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat Chem Biol (2010) 6:449–56. doi:10.1038/nchembio.362
72. Zhang MM, Tsou LK, Charron G, Raghavan AS, Hang HC. Tandem fluorescence imaging of dynamic S-acylation and protein turnover. Proc Natl Acad Sci U S A (2010) 107:8627–32. doi:10.1073/pnas.0912306107
73. Akimzhanov AM, Boehning D. Rapid and transient palmitoylation of the tyrosine kinase Lck mediates Fas signaling. Proc Natl Acad Sci U S A (2015) 112:11876–80. doi:10.1073/pnas.1509929112
74. Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, et al. Global analysis of protein palmitoylation in yeast. Cell (2006) 125:1003–13. doi:10.1016/j.cell.2006.03.042
75. Kummel D, Heinemann U, Veit M. Unique self-palmitoylation activity of the transport protein particle component Bet3: a mechanism required for protein stability. Proc Natl Acad Sci U S A (2006) 103:12701–6. doi:10.1073/pnas.0603513103
76. Kummel D, Walter J, Heck M, Heinemann U, Veit M. Characterization of the self-palmitoylation activity of the transport protein particle component Bet3. Cell Mol Life Sci (2010) 67:2653–64. doi:10.1007/s00018-010-0358-y
77. Chan P, Han X, Zheng B, DeRan M, Yu J, Jarugumilli GK, et al. Autopalmitoylation of TEAD proteins regulates transcriptional output of the Hippo pathway. Nat Chem Biol (2016) 12:282–9. doi:10.1038/nchembio.2036
78. Quevillon-Cheruel S, Leulliot N, Muniz CA, Vincent M, Gallay J, Argentini M, et al. Evf, a virulence factor produced by the Drosophila pathogen Erwinia carotovora, is an S-palmitoylated protein with a new fold that binds to lipid vesicles. J Biol Chem (2009) 284:3552–62. doi:10.1074/jbc.M808334200
79. Resh MD. Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci STKE (2006) 2006:re14. doi:10.1126/stke.3592006re14
80. Chamberlain LH, Shipston MJ. The physiology of protein S-acylation. Physiol Rev (2015) 95:341–76. doi:10.1152/physrev.00032.2014
81. Bach R, Konigsberg WH, Nemerson Y. Human tissue factor contains thioester-linked palmitate and stearate on the cytoplasmic half-cystine. Biochemistry (1988) 27:4227–31. doi:10.1021/bi00412a004
82. Fujimoto T, Stroud E, Whatley RE, Prescott SM, Muszbek L, Laposata M, et al. P-selectin is acylated with palmitic acid and stearic acid at cysteine 766 through a thioester linkage. J Biol Chem (1993) 268:11394–400.
83. Liang X, Lu Y, Wilkes M, Neubert TA, Resh MD. The N-terminal SH4 region of the Src family kinase Fyn is modified by methylation and heterogeneous fatty acylation: role in membrane targeting, cell adhesion, and spreading. J Biol Chem (2004) 279:8133–9. doi:10.1074/jbc.M311180200
84. Veit M. Palmitoylation of virus proteins. Biol Cell (2012) 104:493–515. doi:10.1111/boc.201200006
85. Wright MH, Clough B, Rackham MD, Rangachari K, Brannigan JA, Grainger M, et al. Validation of N-myristoyltransferase as an antimalarial drug target using an integrated chemical biology approach. Nat Chem (2014) 6:112–21. doi:10.1038/nchem.1830
86. Wright MH, Paape D, Storck EM, Serwa RA, Smith DF, Tate EW. Global analysis of protein N-myristoylation and exploration of N-myristoyltransferase as a drug target in the neglected human pathogen Leishmania donovani. Chem Biol (2015) 22:342–54. doi:10.1016/j.chembiol.2015.01.003
87. Ji Y, Bachschmid MM, Costello CE, Lin C. S- to N-palmitoyl transfer during proteomic sample preparation. J Am Soc Mass Spectrom (2016) 27:677–85. doi:10.1007/s13361-015-1319-3
88. Zhou Y, Zhao M, Fields GB, Wu CF, Branton WD. δ/ω-Plectoxin-Pt1a: an excitatory spider toxin with actions on both Ca2+ and Na+ channels. PLoS One (2013) 8:e64324. doi:10.1371/journal.pone.0064324
89. Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem (2013) 288:31350–6. doi:10.1074/jbc.C113.511261
90. Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, et al. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature (2013) 496:110–3. doi:10.1038/nature12038
91. Jiang H, Zhang X, Lin H. Lysine fatty acylation promotes lysosomal targeting of TNF-α. Sci Rep (2016) 6:24371. doi:10.1038/srep24371
92. Reinicke AT, Hutchinson JL, Magee AI, Mastroeni P, Trowsdale J, Kelly AP. A Salmonella typhimurium effector protein SifA is modified by host cell prenylation and S-acylation machinery. J Biol Chem (2005) 280:14620–7. doi:10.1074/jbc.M500076200
93. Waiczies S, Bendix I, Zipp F. Geranylgeranylation but not GTP-loading of Rho GTPases determines T cell function. Sci Signal (2008) 1:t3. doi:10.1126/stke.112pt3
94. Shimabukuro-Vornhagen A, Zoghi S, Liebig TM, Wennhold K, Chemitz J, Draube A, et al. Inhibition of protein geranylgeranylation specifically interferes with CD40-dependent B cell activation, resulting in a reduced capacity to induce T cell immunity. J Immunol (2014) 193:5294–305. doi:10.4049/jimmunol.1203436
95. Fukata Y, Dimitrov A, Boncompain G, Vielemeyer O, Perez F, Fukata M. Local palmitoylation cycles define activity-regulated postsynaptic subdomains. J Cell Biol (2013) 202:145–61. doi:10.1083/jcb.201302071
96. Veit M, Ponimaskin E, Schmidt MF. Analysis of S-acylation of proteins. Methods Mol Biol (2008) 446:163–82. doi:10.1007/978-1-60327-084-7_12
97. Hang HC, Geutjes EJ, Grotenbreg G, Pollington AM, Bijlmakers MJ, Ploegh HL. Chemical probes for the rapid detection of fatty-acylated proteins in mammalian cells. J Am Chem Soc (2007) 129:2744–5. doi:10.1021/ja0685001
98. Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, Shamir E, et al. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J Am Chem Soc (2009) 131:4967–75. doi:10.1021/ja810122f
99. Martin BR, Cravatt BF. Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods (2009) 6:135–8. doi:10.1038/nmeth.1293
100. Drisdel RC, Alexander JK, Sayeed A, Green WN. Assays of protein palmitoylation. Methods (2006) 40:127–34. doi:10.1016/j.ymeth.2006.04.015
101. Yount JS, Zhang MM, Hang HC. Visualization and identification of fatty acylated proteins using chemical reporters. Curr Protoc Chem Biol (2011) 3:65–79. doi:10.1002/9780470559277.ch100225
102. Martin BR. Nonradioactive analysis of dynamic protein palmitoylation. Curr Protoc Protein Sci (2013) 73:Unit14.15. doi:10.1002/0471140864.ps1415s73
103. Yount JS, Moltedo B, Yang Y-Y, Charron G, Moran TM, López CB, et al. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat Chem Biol (2010) 6:610–4. doi:10.1038/nchembio.405
104. Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. Global profiling of dynamic protein palmitoylation. Nat Methods (2011) 9:84–9. doi:10.1038/nmeth.1769
105. Wilson JP, Raghavan AS, Yang YY, Charron G, Hang HC. Proteomic analysis of fatty-acylated proteins in mammalian cells with chemical reporters reveals S-acylation of histone H3 variants. Mol Cell Proteomics (2011) 10:M110.001198. doi:10.1074/mcp.M110.001198
106. Foe IT, Child MA, Majmudar JD, Krishnamurthy S, van der Linden WA, Ward GE, et al. Global analysis of palmitoylated proteins in Toxoplasma gondii. Cell Host Microbe (2015) 18:501–11. doi:10.1016/j.chom.2015.09.006
107. Santiago-Tirado FH, Peng T, Yang M, Hang HC, Doering TL. A single protein S-acyl transferase acts through diverse substrates to determine cryptococcal morphology, stress tolerance, and pathogenic outcome. PLoS Pathog (2015) 11:e1004908. doi:10.1371/journal.ppat.1004908
108. Rybak JN, Scheurer SB, Neri D, Elia G. Purification of biotinylated proteins on streptavidin resin: a protocol for quantitative elution. Proteomics (2004) 4:2296–9. doi:10.1002/pmic.200300780
109. Yang YY, Grammel M, Raghavan AS, Charron G, Hang HC. Comparative analysis of cleavable azobenzene-based affinity tags for bioorthogonal chemical proteomics. Chem Biol (2010) 17:1212–22. doi:10.1016/j.chembiol.2010.09.012
110. Serwa RA, Abaitua F, Krause E, Tate EW, O’Hare P. Systems analysis of protein fatty acylation in herpes simplex virus-infected cells using chemical proteomics. Chem Biol (2015) 22:1008–17. doi:10.1016/j.chembiol.2015.06.024
111. Gao X, Hannoush RN. Single-cell in situ imaging of palmitoylation in fatty-acylated proteins. Nat Protoc (2014) 9:2607–23. doi:10.1038/nprot.2014.179
112. Gao X, Hannoush RN. Method for cellular imaging of palmitoylated proteins with clickable probes and proximity ligation applied to Hedgehog, tubulin, and Ras. J Am Chem Soc (2014) 136:4544–50. doi:10.1021/ja410068g
113. Jones ML, Collins MO, Goulding D, Choudhary JS, Rayner JC. Analysis of protein palmitoylation reveals a pervasive role in Plasmodium development and pathogenesis. Cell Host Microbe (2012) 12:246–58. doi:10.1016/j.chom.2012.06.005
114. Thinon E, Serwa RA, Broncel M, Brannigan JA, Brassat U, Wright MH, et al. Global profiling of co- and post-translationally N-myristoylated proteomes in human cells. Nat Commun (2014) 5:ncomms5919. doi:10.1038/ncomms5919
115. Roth AF, Wan J, Green WN, Yates JR, Davis NG. Proteomic identification of palmitoylated proteins. Methods (2006) 40:135–42. doi:10.1016/j.ymeth.2006.05.026
116. Wan J, Roth AF, Bailey AO, Davis NG. Palmitoylated proteins: purification and identification. Nat Protoc (2007) 2:1573–84. doi:10.1038/nprot.2007.225
117. Forrester MT, Hess DT, Thompson JW, Hultman R, Moseley MA, Stamler JS, et al. Site-specific analysis of protein S-acylation by resin-assisted capture. J Lipid Res (2011) 52:393–8. doi:10.1194/jlr.D011106
118. Peng T, Thinon E, Hang HC. Proteomic analysis of fatty-acylated proteins. Curr Opin Chem Biol (2016) 30:77–86. doi:10.1016/j.cbpa.2015.11.008
119. Percher A, Ramakrishnan S, Thinon E, Yuan X, Yount JS, Hang HC. Mass-tag labeling reveals site-specific and endogenous levels of protein S-fatty acylation. Proc Natl Acad Sci U S A (2016) 113:4302–7. doi:10.1073/pnas.1602244113
120. Dowal L, Yang W, Freeman MR, Steen H, Flaumenhaft R. Proteomic analysis of palmitoylated platelet proteins. Blood (2011) 118:e62–73. doi:10.1182/blood-2011-05-353078
121. Ivaldi C, Martin BR, Kieffer-Jaquinod S, Chapel A, Levade T, Garin J, et al. Proteomic analysis of S-acylated proteins in human B cells reveals palmitoylation of the immune regulators CD20 and CD23. PLoS One (2012) 7:e37187. doi:10.1371/journal.pone.0037187
122. Morrison E, Kuropka B, Kliche S, Brügger B, Krause E, Freund C. Quantitative analysis of the human T cell palmitome. Sci Rep (2015) 5:11598. doi:10.1038/srep11598
123. Emmer BT, Nakayasu ES, Souther C, Choi H, Sobreira TJP, Epting CL, et al. Global analysis of protein palmitoylation in African trypanosomes. Eukaryot Cell (2011) 10:455–63. doi:10.1128/EC.00248-10
124. Caballero MC, Alonso AM, Deng B, Attias M, de Souza W, Corvi MM. Identification of new palmitoylated proteins in Toxoplasma gondii. Biochim Biophys Acta (2016) 1864:400–8. doi:10.1016/j.bbapap.2016.01.010
125. Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, Bailey A, et al. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature (2008) 456:904–9. doi:10.1038/nature07605
126. Shen LF, Chen YJ, Liu KM, Haddad ANS, Song IW, Roan HY, et al. Role of S-palmitoylation by ZDHHC13 in mitochondrial function and metabolism in liver. Sci Rep (2017) 7:2182. doi:10.1038/s41598-017-02159-4
127. Wan J, Savas JN, Roth AF, Sanders SS, Singaraja RR, Hayden MR, et al. Tracking brain palmitoylation change: predominance of glial change in a mouse model of Huntington’s Disease. Chem Biol (2013) 20:1421–34. doi:10.1016/j.chembiol.2013.09.018
128. Blanc M, David F, Abrami L, Migliozzi D, Armand F, Bürgi J, et al. SwissPalm: protein palmitoylation database. F1000Res (2015) 4:261. doi:10.12688/f1000research.6464.1
129. Poltorak A, Smirnova I, He X, Liu M-Y, Van Huffel C, McNally O, et al. Genetic and physical mapping of the Lps locus: identification of the toll-4 receptor as a candidate gene in the critical region. Blood Cells Mol Dis (1998) 24:340–55. doi:10.1006/bcmd.1998.0201
130. Kawasaki K, Ernst RK, Miller SI. 3-O-deacylation of lipid A by PagL, a PhoP/PhoQ-regulated deacylase of Salmonella typhimurium, modulates signaling through toll-like receptor 4. J Biol Chem (2004) 279:20044–8. doi:10.1074/jbc.M401275200
131. Chalabaev S, Chauhan A, Novikov A, Iyer P, Szczesny M, Beloin C, et al. Biofilms formed by gram-negative bacteria undergo increased lipid a palmitoylation, enhancing in vivo survival. MBio (2014) 5:e1116–1114. doi:10.1128/mBio.01116-14
132. Preston A, Maxim E, Toland E, Pishko EJ, Harvill ET, Caroff M, et al. Bordetella bronchiseptica PagP is a Bvg-regulated lipid A palmitoyl transferase that is required for persistent colonization of the mouse respiratory tract. Mol Microbiol (2003) 48:725–36. doi:10.1046/j.1365-2958.2003.03484.x
133. Kawasaki K, Teramoto M, Tatsui R, Amamoto S. Lipid A 3’-O-deacylation by Salmonella outer membrane enzyme LpxR modulates the ability of lipid A to stimulate toll-like receptor 4. Biochem Biophys Res Commun (2012) 428:343–7. doi:10.1016/j.bbrc.2012.10.054
134. Bishop RE, Gibbons HS, Guina T, Trent MS, Miller SI, Raetz CR. Transfer of palmitate from phospholipids to lipid A in outer membranes of Gram-negative bacteria. EMBO J (2000) 19:5071–80. doi:10.1093/emboj/19.19.5071
135. Bishop RE. The lipid A palmitoyltransferase PagP: molecular mechanisms and role in bacterial pathogenesis. Mol Microbiol (2005) 57:900–12. doi:10.1111/j.1365-2958.2005.04711.x
136. Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, et al. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell (1998) 95:189–98. doi:10.1016/S0092-8674(00)81750-X
137. Li Y, Powell DA, Shaffer SA, Rasko DA, Pelletier MR, Leszyk JD, et al. LPS remodeling is an evolved survival strategy for bacteria. Proc Natl Acad Sci U S A (2012) 109:8716–21. doi:10.1073/pnas.1202908109
138. Issartel JP, Koronakis V, Hughes C. Activation of Escherichia coli prohaemolysin to the mature toxin by acyl carrier protein-dependent fatty acylation. Nature (1991) 351:759–61. doi:10.1038/351759a0
139. Belisle JT, Brandt ME, Radolf JD, Norgard MV. Fatty acids of Treponema pallidum and Borrelia burgdorferi lipoproteins. J Bacteriol (1994) 176:2151–7. doi:10.1128/jb.176.8.2151-2157.1994
140. Houston S, Hof R, Francescutti T, Hawkes A, Boulanger MJ, Cameron CE. Bifunctional role of the Treponema pallidum extracellular matrix binding adhesin Tp0751. Infect Immun (2011) 79:1386–98. doi:10.1128/IAI.01083-10
141. Kovacs-Simon A, Titball RW, Michell SL. Lipoproteins of bacterial pathogens. Infect Immun (2011) 79:548–61. doi:10.1128/IAI.00682-10
142. Ponnusamy D, Fitts EC, Sha J, Erova TE, Kozlova EV, Kirtley ML, et al. High-throughput, signature-tagged mutagenic approach to identify novel virulence factors of Yersinia pestis CO92 in a mouse model of infection. Infect Immun (2015) 83:2065–81. doi:10.1128/IAI.02913-14
143. Henneke P, Dramsi S, Mancuso G, Chraibi K, Pellegrini E, Theilacker C, et al. Lipoproteins are critical TLR2 activating toxins in group B streptococcal sepsis. J Immunol (2008) 180:6149–58. doi:10.4049/jimmunol.180.9.6149
144. Rangan KJ, Yang YY, Charron G, Hang HC. Rapid visualization and large-scale profiling of bacterial lipoproteins with chemical reporters. J Am Chem Soc (2010) 132:10628–9. doi:10.1021/ja101387b
145. Abrami L, Bischofberger M, Kunz B, Groux R, van der Goot FG. Endocytosis of the anthrax toxin is mediated by clathrin, actin and unconventional adaptors. PLoS Pathog (2010) 6:e1000792. doi:10.1371/journal.ppat.1000792
146. Abrami L, Kunz B, van der Goot FG. Anthrax toxin triggers the activation of src-like kinases to mediate its own uptake. Proc Natl Acad Sci U S A (2010) 107:1420–4. doi:10.1073/pnas.0910782107
147. Coburn B, Sekirov I, Finlay BB. Type III secretion systems and disease. Clin Microbiol Rev (2007) 20:535–49. doi:10.1128/CMR.00013-07
148. Hicks SW, Galán JE. Exploitation of eukaryotic subcellular targeting mechanisms by bacterial effectors. Nat Rev Microbiol (2013) 11:316–26. doi:10.1038/nrmicro3009
149. Hicks SW, Charron G, Hang HC, Galán JE. Subcellular targeting of Salmonella virulence proteins by host-mediated S-palmitoylation. Cell Host Microbe (2011) 10:9–20. doi:10.1016/j.chom.2011.06.003
150. Lin YH, Doms AG, Cheng E, Kim B, Evans TR, Machner MP. Host cell-catalyzed S-palmitoylation mediates Golgi targeting of the Legionella ubiquitin ligase GobX. J Biol Chem (2015) 290:25766–81. doi:10.1074/jbc.M115.637397
151. Schroeder GN, Aurass P, Oates CV, Tate EW, Hartland EL, Flieger A, et al. Legionella pneumophila effector LpdA is a palmitoylated phospholipase D virulence factor. Infect Immun (2015) 83:3989–4002. doi:10.1128/IAI.00785-15
152. Burnaevskiy N, Peng T, Reddick LE, Hang HC, Alto NM. Myristoylome profiling reveals a concerted mechanism of ARF GTPase deacylation by the bacterial protease IpaJ. Mol Cell (2015) 58:110–22. doi:10.1016/j.molcel.2015.01.040
153. Nimchuk Z, Marois E, Kjemtrup S, Leister RT, Katagiri F, Dangl JL. Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell (2000) 101:353–63. doi:10.1016/S0092-8674(00)80846-6
154. Dowen RH, Engel JL, Shao F, Ecker JR, Dixon JE. A family of bacterial cysteine protease type III effectors utilizes acylation-dependent and -independent strategies to localize to plasma membranes. J Biol Chem (2009) 284:15867–79. doi:10.1074/jbc.M900519200
155. Schmidt MF, Bracha M, Schlesinger MJ. Evidence for covalent attachment of fatty acids to Sindbis virus glycoproteins. Proc Natl Acad Sci U S A (1979) 76:1687–91. doi:10.1073/pnas.76.4.1687
156. Kordyukova LV, Serebryakova MV, Baratova LA, Veit M. S acylation of the hemagglutinin of influenza viruses: mass spectrometry reveals site-specific attachment of stearic acid to a transmembrane cysteine. J Virol (2008) 82:9288–92. doi:10.1128/JVI.00704-08
157. Schroeder C, Heider H, Möncke-Buchner E, Lin TI. The influenza virus ion channel and maturation cofactor M2 is a cholesterol-binding protein. Eur Biophys J (2005) 34:52–66. doi:10.1007/s00249-004-0424-1
158. Thaa B, Levental I, Herrmann A, Veit M. Intrinsic membrane association of the cytoplasmic tail of influenza virus M2 protein and lateral membrane sorting regulated by cholesterol binding and palmitoylation. Biochem J (2011) 437:389–97. doi:10.1042/BJ20110706
159. Wagner R, Herwig A, Azzouz N, Klenk HD. Acylation-mediated membrane anchoring of avian influenza virus hemagglutinin is essential for fusion pore formation and virus infectivity. J Virol (2005) 79:6449–58. doi:10.1128/JVI.79.10.6449-6458.2005
160. Melkonian KA, Ostermeyer AG, Chen JZ, Roth MG, Brown DA. Role of lipid modifications in targeting proteins to detergent-resistant membrane rafts. Many raft proteins are acylated, while few are prenylated. J Biol Chem (1999) 274:3910–7. doi:10.1074/jbc.274.6.3910
161. Gerl MJ, Sampaio JL, Urban S, Kalvodova L, Verbavatz J-M, Binnington B, et al. Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J Cell Biol (2012) 196:213–21. doi:10.1083/jcb.201108175
162. Chen BJ, Takeda M, Lamb RA. Influenza virus hemagglutinin (H3 subtype) requires palmitoylation of its cytoplasmic tail for assembly: M1 proteins of two subtypes differ in their ability to support assembly. J Virol (2005) 79:13673–84. doi:10.1128/JVI.79.21.13673-13684.2005
163. Siche S, Brett K, Möller L, Kordyukova LV, Mintaev RR, Alexeevski AV, et al. Two cytoplasmic acylation sites and an adjacent hydrophobic residue, but no other conserved amino acids in the cytoplasmic tail of HA from influenza A virus are crucial for virus replication. Viruses (2015) 7:6458–75. doi:10.3390/v7122950
164. Rossman JS, Jing X, Leser GP, Lamb RA. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell (2010) 142:902–13. doi:10.1016/j.cell.2010.08.029
165. Grosenbach DW, Ulaeto DO, Hruby DE. Palmitylation of the vaccinia virus 37-kDa major envelope antigen. Identification of a conserved acceptor motif and biological relevance. Biol Chem (1997) 272:1956–64. doi:10.1074/jbc.272.3.1956
166. Majeau N, Fromentin R, Savard C, Duval M, Tremblay MJ, Leclerc D. Palmitoylation of hepatitis C virus core protein is important for virion production. J Biol Chem (2009) 284:33915–25. doi:10.1074/jbc.M109.018549
167. Veit M, Siche S. S-acylation of influenza virus proteins: are enzymes for fatty acid attachment promising drug targets? Vaccine (2015) 33:7002–7. doi:10.1016/j.vaccine.2015.08.095
168. Blanc M, Blaskovic S, van der Goot FG. Palmitoylation, pathogens and their host. Biochem Soc Trans (2013) 41:84–8. doi:10.1042/BST20120337
169. González SA, Paladino MG, Affranchino JL. Palmitoylation of the feline immunodeficiency virus envelope glycoprotein and its effect on fusion activity and envelope incorporation into virions. Virology (2012) 428:1–10. doi:10.1016/j.virol.2012.03.005
170. Rousso I, Mixon MB, Chen BK, Kim PS. Palmitoylation of the HIV-1 envelope glycoprotein is critical for viral infectivity. Proc Natl Acad Sci U S A (2000) 97:13523–5. doi:10.1073/pnas.240459697
171. Bhattacharya J, Peters PJ, Clapham PR. Human immunodeficiency virus type 1 envelope glycoproteins that lack cytoplasmic domain cysteines: impact on association with membrane lipid rafts and incorporation onto budding virus particles. J Virol (2004) 78:5500–6. doi:10.1128/JVI.78.10.5500-5506.2004
172. Yang P, Ai LS, Huang SC, Li HF, Chan WE, Chang CW, et al. The cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein gp41 harbors lipid raft association determinants. J Virol (2010) 84:59–75. doi:10.1128/JVI.00899-09
173. Colquhoun DR, Lyashkov AE, Ubaida Mohien C, Aquino VN, Bullock BT, Dinglasan RR, et al. Bioorthogonal mimetics of palmitoyl-CoA and myristoyl-CoA and their subsequent isolation by click chemistry and characterization by mass spectrometry reveal novel acylated host-proteins modified by HIV-1 infection. Proteomics (2015) 15:2066–77. doi:10.1002/pmic.201500063
174. Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity (2011) 34:637–50. doi:10.1016/j.immuni.2011.05.006
175. Triantafilou M, Manukyan M, Mackie A, Morath S, Hartung T, Heine H, et al. Lipoteichoic acid and toll-like receptor 2 internalization and targeting to the Golgi are lipid raft-dependent. J Biol Chem (2004) 279:40882–9. doi:10.1074/jbc.M400466200
176. Reynolds CM, McGillicuddy FC, Harford KA, Finucane OM, Mills KH, Roche HM. Dietary saturated fatty acids prime the NLRP3 inflammasome via TLR4 in dendritic cells-implications for diet-induced insulin resistance. Mol Nutr Food Res (2012) 56:1212–22. doi:10.1002/mnfr.201200058
177. Wang Y, Qian Y, Fang Q, Zhong P, Li W, Wang L, et al. Saturated palmitic acid induces myocardial inflammatory injuries through direct binding to TLR4 accessory protein MD2. Nat Commun (2017) 8:13997. doi:10.1038/ncomms13997
178. Ciesielska A, Kwiatkowska K. Modification of pro-inflammatory signaling by dietary components: the plasma membrane as a target. Bioessays (2015) 37:789–801. doi:10.1002/bies.201500017
179. Hwang DH, Kim JA, Lee JY. Mechanisms for the activation of toll-like receptor 2/4 by saturated fatty acids and inhibition by docosahexaenoic acid. Eur J Pharmacol (2016) 785:24–35. doi:10.1016/j.ejphar.2016.04.024
180. Nicholas DA, Zhang K, Hung C, Glasgow S, Aruni AW, Unternaehrer J, et al. Palmitic acid is a toll-like receptor 4 ligand that induces human dendritic cell secretion of IL-1β. PLoS One (2017) 12:e0176793. doi:10.1371/journal.pone.0176793
181. Jin J, Zhang X, Lu Z, Perry DM, Li Y, Russo SB, et al. Acid sphingomyelinase plays a key role in palmitic acid-amplified inflammatory signaling triggered by lipopolysaccharide at low concentrations in macrophages. Am J Physiol Endocrinol Metab (2013) 305:E853–67. doi:10.1152/ajpendo.00251.2013
182. Sobocińska J, Roszczenko-Jasińska P, Zaręba-Kozioł M, Hromada-Judycka A, Matveichuk OV, Traczyk G, et al. LPS upregulates palmitoylated enzymes of the phosphatidylinositol cycle. An insight from proteomic studies. Mol Cell Proteomics (2017). doi:10.1074/mcp.RA117.000050
183. Stow JL, Low PC, Offenhäuser C, Sangermani D. Cytokine secretion in macrophages and other cells: pathways and mediators. Immunobiology (2009) 214:601–12. doi:10.1016/j.imbio.2008.11.005
184. Utsumi T, Takeshige T, Tanaka K, Takami K, Kira Y, Klostergaard J, et al. Transmembrane TNF (pro-TNF) is palmitoylated. FEBS Lett (2001) 500:1–6. doi:10.1016/S0014-5793(01)02576-5
185. Poggi M, Kara I, Brunel JM, Landrier JF, Govers R, Bonardo B, et al. Palmitoylation of TNF alpha is involved in the regulation of TNF receptor 1 signalling. Biochim Biophys Acta (2013) 1833:602–12. doi:10.1016/j.bbamcr.2012.11.009
186. Claudinon J, Gonnord P, Beslard E, Marchetti M, Mitchell K, Boularan C, et al. Palmitoylation of interferon-α (IFN-α) receptor subunit IFNAR1 is required for the activation of Stat1 and Stat2 by IFN-α. J Biol Chem (2009) 284:24328–40. doi:10.1074/jbc.M109.021915
187. Bailey CC, Zhong G, Huang IC, Farzan M. IFITM-family proteins: the cell’s first line of antiviral defense. Annu Rev Virol (2014) 1:261–83. doi:10.1146/annurev-virology-031413-085537
188. Savidis G, Perreira JM, Portmann JM, Meraner P, Guo Z, Green S, et al. The IFITMs inhibit Zika virus replication. Cell Rep (2016) 15:2323–30. doi:10.1016/j.celrep.2016.05.074
189. Yount JS, Karssemeijer RA, Hang HC. S-palmitoylation and ubiquitination differentially regulate interferon-induced transmembrane protein 3 (IFITM3)-mediated resistance to influenza virus. J Biol Chem (2012) 287:19631–41. doi:10.1074/jbc.M112.362095
190. Chesarino NM, McMichael TM, Hach JC, Yount JS. Phosphorylation of the antiviral protein interferon-inducible transmembrane protein 3 (IFITM3) dually regulates its endocytosis and ubiquitination. J Biol Chem (2014) 289:11986–92. doi:10.1074/jbc.M114.557694
191. Levental I, Grzybek M, Simons K. Greasing their way: lipid modifications determine protein association with membrane rafts. Biochemistry (2010) 49:6305–16. doi:10.1021/bi100882y
192. Bijlmakers MJ. Protein acylation and localization in T cell signaling (review). Mol Membr Biol (2009) 26:93–103. doi:10.1080/09687680802650481
Keywords: acyl-biotin exchange, bacterial effector proteins, click chemistry, fatty acylation of proteins, hemagglutinin, IFITM, S-palmitoylation, tumor necrosis factor α
Citation: Sobocińska J, Roszczenko-Jasińska P, Ciesielska A and Kwiatkowska K (2018) Protein Palmitoylation and Its Role in Bacterial and Viral Infections. Front. Immunol. 8:2003. doi: 10.3389/fimmu.2017.02003
Received: 24 October 2017; Accepted: 26 December 2017;
Published: 19 January 2018
Edited by:
Olivier Neyrolles, Centre national de la recherche scientifique (CNRS), FranceReviewed by:
Michael Veit, Freie Universität Berlin, GermanyStefano Piotto, Università degli Studi di Salerno, Italy
Copyright: © 2018 Sobocińska, Roszczenko-Jasińska, Ciesielska and Kwiatkowska. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katarzyna Kwiatkowska, k.kwiatkowska@nencki.gov.pl
 Justyna Sobocińska
Justyna Sobocińska Paula Roszczenko-Jasińska
Paula Roszczenko-Jasińska Katarzyna Kwiatkowska
Katarzyna Kwiatkowska