1
Arcitecta Pty Ltd, Victoria, Australia
2
Florey Neuroscience Institutes, University of Melbourne, Victoria, Australia
3
Centre for Neuroscience, University of Melbourne, Victoria, Australia
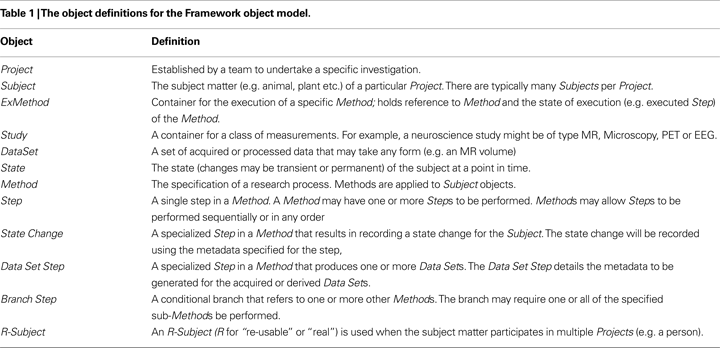
A framework that integrates an object model, research methods (workflows), the capture of experimental data sets and the provenance of those data sets for subject-centric research is presented. The design of the Framework object model draws on and extends pre-existing object models in the public domain. In particular the Framework tracks the state and life cycle of a subject during an experimental method, provides for reusable subjects, primary, derived and recursive data sets of arbitrary content types, and defines a user-friendly and practical scheme for citably identifying information in a distributed environment. The Framework is currently used to manage neuroscience Magnetic Resonance and microscopy imaging data sets in both clinical and basic neuroscience research environments. The Framework facilitates multi-disciplinary and collaborative subject-based research, and extends earlier object models used in the research imaging domain. Whilst the Framework has been explicitly validated for neuroimaging research applications, it has broader application to other fields of subject-centric research.
Research groups worldwide are facing data management challenges
1
. Not only is the volume of data rising dramatically, but also the processes that a researcher follows to analyze and manage research data are increasingly complex. Of crucial importance for a data management system is the way in which information is organized. A common method of data organization is use of an object model that is motivated by the processes and protocols of the specific research domain. These include how the data are acquired, what the relationships between data are, and how the data will be distributed, analyzed and interpreted.
Neuroimaging is a rapidly developing research domain in which enormous quantities of data are acquired. Identification of an appropriate object model for neuroimaging involves firstly identifying the particular class of research that it belongs to. Neuroimaging is an example of “subject-centric” research, which refers to well-defined, persistent subject matter for which data are being acquired over some (perhaps extended) period of time. For example, a subject might be an animal (human, mouse etc.), chemical or mineral sample with a number of data acquisitions undertaken for each subject over time.
A second important aspect of data management is to recognize that research data are often obtained through a well-defined, and sometimes complex workflow. Although organization of information with an object model is an established methodology, the method (or workflow) is less commonly captured along with the data.
An object model captures domain-specific data and metadata. It is a very significant challenge to develop metadata (and data) standards within domains, let alone across domains. Our approach is to develop a framework that represents the essential components of subject-centric research without prescribing particular metadata. The Framework then requires the domain specialists to define the appropriate metadata for their research.
Research is increasingly distributed through collaborations involving researchers at different institutions. The location of objects associated with data and metadata should be largely transparent to the researcher and accessible from anywhere through a number of mechanisms including distributed queries, remote access and replication.
In this paper, we describe a framework that defines an object model to explicitly represent research methods and the resulting acquired and derivative data for subject-centric research. The Framework captures the core relationships required for auditable and reproducible research. The Framework is extended with metadata that is specific to the type of subject and domain of research and it explicitly provides an identification scheme that supports distributed objects. The Framework has been implemented and is used to manage distributed neuroimaging data.
Background
Neuroscience research increasingly involves scientific collaborations across sub-domains that acquire, share and analyze multi-modal data (e.g. Gardner et al., 2003
; Martone et al., 2004
; Toga, 2002
). For example, neuroscience research may include types of data such as: magnetic resonance imaging (MR) and spectroscopy (MRS), optical and electron microscopy (OM and EM), positron-electron tomography (PET), computed tomography (CT), electrophysiological, genotype, electroencephalogram (EEG) and event related potential (ERP) data types. This list will undoubtedly continue to lengthen, particularly as new forms of collaborative research emerge over time. Various neuroimaging and related groups worldwide have developed applications to provide data management and application capabilities (examples include Keator et al., 2008
; Marcus et al., 2007
; Marenco et al., 2003
; SenseLab
2
, LONI Image Data Archive
3
and fMRIDC
4
).
The need in our own research environment to manage many different types of data using a consistent model was the catalyst to seek a generic object model that supports: (i) project-based virtual organizations, (ii) representation of the subject of a study, (iii) recording the state changes in a subject, (iv) representation of the experimental method (process or workflow), (v) participation by subjects in multiple research projects, (vi) disassembling of subjects into constituent parts, (vii) controlled access to all information and especially the identity of a subject, (viii) capture and storage of all types of data, and (ix) the capability to manage raw and processed data.
The requirement to record state arises because the subject may undergo a number of procedures in an experimental process. These state changes might may be transient (e.g. anesthesia) or permanent (e.g. death) and affect the subsequent acquisition of data. A given subject may be disassembled (e.g. removal of the brain) into constituent parts for subsequent study. There may be parallel studies on different “parts”, each with a separate procedure and life cycle.
Rather than create yet another object model, we investigated whether an existing model would satisfy our main requirements. Consideration was given to: (i) the Digital Imaging and Communications in Medicine (DICOM
5
) model, (ii) the XML-based Clinical and Experimental Data Exchange (XCEDE
6
and see also Keator et al., 2006
) model, (iii) a Project-Subject-Study (PSS) model (our own earlier generation object model) and (iv) the Council for the Central Laboratory of the Research Councils (CCLRC
7
). As will be demonstrated, none of these object models fully met the requirements, but all provided valuable components that have been used and extended.
Dicom Object Model
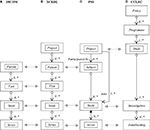
The DICOM standard includes formatting, communications and object modeling components. DICOM is ubiquitous in medical imaging and was originally created for clinically oriented studies conducted with patients although it can be utilized for other studies. The DICOM object model is complex – the key objects that are relevant to neuroimaging research are shown in a Unified Modeling Language (UML) object diagram
8
(Figure 1
A). Briefly, the Patient represents the subject of the investigation, and may undertake a number of Visits over time to imaging facilities. Each Visit results in a number of Studies that represent a particular imaging setup and procedure. Each Study generates a number of actual acquisitions of a particular type (e.g. MR image volumes) that are called Series.
Figure 1. UML object diagrams for the (A) DICOM, (B) XCEDE, (C) PSS and (D) CCLRC object models. In addition to the UML notation the horizontal arrows indicate equivalence of objects between the different models. The UML notation can be summarized as follows: Objects (or classes) are shown in rectangles and named relationships are shown between objects that are qualified by their cardinality (* means infinity, 0..* means 0 to infinity). The relationship direction is indicated via an arrow. Filled diamonds indicate that the relationship is containment (also called composition) and open diamonds indicate an aggregation (has) relationship.
The DICOM object model is limited in that it lacks the concept of a project consisting of many subjects, is unable to record the experimental method, nor represent the state of a subject. In addition, the DICOM standard requires the data sets to be encapsulated in the DICOM file format.
Biomedical Informatics Research Network (BIRN) XCEDE Schema
The XCEDE metadata schema (and implicit object model) is intended for the exchange of clinical and research imaging studies. The objects in the XCEDE model are Project, Subject, Visit, Study and Series (Figure 1
B), with the XCEDE objects equivalent to the DICOM objects from the Subject level. The XCEDE object model, and the associated metadata hierarchy described in the XCEDE XML schema are highly specific to image-based analysis and cannot be easily applied more generally. However, the model contains a number of interesting and useful concepts related to experimental method. For example, the provenance of any object may be used to describe the data processing protocol that was used to generate a sub-set of data. The inclusion of provenance information at any level in the object model hierarchy is an advantage of the XCEDE schema.
PSS Object Model
The project subject study (PSS) object model (Figure 1
C) was derived from the DICOM object model with two key extensions. Firstly, the Project object at the top of the hierarchy (like XCEDE) corresponds to the virtual project team collaborating on a specific scientific experiment. Secondly, the Subject object may be decomposed into two parts: the project-specific attributes of the subject, and the project-invariant aspects that are common to all projects. The ability to re-use Subjects in multiple Projects required a relationship to be specified between the Project and Study objects. The PSS model also removed the DICOM Visit object.
The PSS model was used in research using MR imaging data and although not mandated by the PSS model, only DICOM format data were included. While the PSS model had a number of improvements over the DICOM model, additional key requirements including the ability to capture the experimental method and track subject state were not met.
CCLRC Object Model
The CCLRC model was defined as a generic model for handling e-Science data (Figure 1
D). This model was examined to establish if it satsified the requirements for representing subject-centric, neuroimaging research studies. The CCLRC Study is sometimes referred to as a Project and each Investigation is directly linked with one Data Holding that contains the data generated by the investigation. A Data Holding is a hierarchy of Data Collections and/or atomic Data Objects. The CCLRC model has no concept of the “subject” of an investigation (and associated state), nor the method of research and thus does not meet the requirements for neuroimaging research without extension. However, novel parts of the model, such as the hierarchy of Data Holding objects, provide useful elements for inclusion into subject-centric data models.
Framework Description
Overview
A subject-centric research object model that includes details of research experimental methods has been developed. The model can be applied to studies involving subjects such as people, animals, plants or minerals. The model does not prescribe any particular domain-specific metadata, but instead the domain of research defines specific metadata and semantic interpretation through associated ontologies. The model is independent of a particular implementation technology.
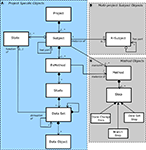
The Framework has a number of characteristics including: (i) objects may have location independent Citable Identifiers that allow objects to be referenced in a distributed environment; (ii) objects are primarily organized into a hierarchy of Project, Subject, ExMethod, Study and DataSet (see below); (iii) the R-Subject object allows subjects to be used in multiple projects; (iv) the research Method (i.e. the set of steps in a workflow where each step may have meta-data and/or produce data) can be encoded; (v) all state changes for a subject are recorded; any data set produced is a function of the state of the subject at that point in time; and (vi) DataSets may be further organized into a hierarchy of DataSet(s) and DataObject(s).
Citable Identification
The ability to cite research data and data sets is an important part of research publication, allowing peer access, review and reuse of raw and derived data. Citation requires the assignment of unique and long lived identifiers (see Brase, 2004
; Klump et al., 2006
, 2008
) to each citable entity.
In this model, objects are identified using a hierarchical identification scheme that supports unique identity in a distributed environment. The citable identifier scheme is a human-friendly, arbitrary depth hierarchy of positive integer numbers (
NA.ORG.r.n1.n2…nk). Citable identifiers are used for all objects (see below for an example) within the object model that may be externally cited to allow collections to be distributed across many repositories. Once assigned, an identifier is immutable although replicas of the same object may exist in multiple locations. These identifiers are compatible with other identification schemes, such as DOI
9
and HANDLE
10
(see also PILIN
11
).These identifiers should be interpreted as follows: (i) an identifier has depth N (the number of dot characters (“.”) plus one), (ii) the identifier part at depth 1 is the Naming Authority, (iii) the identifier part at depth 2 is the Organization that can resolve the location of a resource, (iv) the pair (NA.ORG) is unique and (v) the naming authority must be able to reference the organization. The third digit, which follows the NA.ORG part of the identifier provides root namespace separation (e.g. to separate collections of Projects, R-Subjects and Methods).
Objects with the same parent are considered to be in the same collection. These collection semantics allow the members of a collection to be easily located (including any replicas) in a distributed system without requiring more complex centralized registries or cross-repository references.
Object Hierarchy
The object hierarchy (Figure 2
) and the objects (Table 1
) can be used in two ways. Firstly, a subject may exist only in a single project (Figure 2
A). Secondly, a subject may exist in multiple projects (e.g. people, a calibration reference) in which case it may be represented by the (real) R-Subject (Figure 2
B).
Figure 2. UML object diagram of the Framework object model (see Table 1
for definitions). (A) When subjects are not re-used across multiple projects, only the project specific objects are used. (B) If subjects participate in multiple projects then additional objects are required. (C) The Method object contains Steps, each of which is comprised of a possible State Change, production of a DataSet and a Branch Point.
A Subject is Project based and so has attributes of particular interest to that Project. The subject matter of an investigation may be disassembled into sub-parts. That is, parts may be removed (e.g. the brain removed from the skull of a mouse) and become independent entities for investigation. When a subject participates in more than one project, both R-Subject and Subject objects will represent it. The R-Subject captures time invariant characteristics, and, like the Subject, which is the subject’s manifestation in a project, an R-Subject may be an assembly of discrete parts.
Where ethics requirements allow, an R-Subject can be used to identify all of the Projects in which a subject has participated. The discovery or measurement of new time-invariant characteristics, or recognition of existing and potentially significant characteristics, may be retrospectively important and inform any of the projects in which the subject has participated.
A subject will have one or more identities. Access to the identity (and other attributes) may be restricted by the implementation.
Subjects need not have a direct physical manifestation. They may represent derived entities, such as a probabilistic calculation from multiple input subjects (e.g. an atlas) or a computed model based on data sets from other subjects.
The ExMethod object represents the execution of a specific Method (which codifies a workflow and is discussed further below). The ExMethod object contains a reference to the specific Method that is being executed and specifies the state (e.g. “incomplete”, “complete”) of each step of the Method being executed. Subjects may have multiple Methods executed on them, and therefore may have multiple ExMethod objects. DataSets may be original (measured) or computed (processed). A computed data set may be derived from one or more other data sets.
The object model indicates containment by the filled diamonds. Therefore deleting a parent object will also delete all children objects. For example, deleting a Project will delete all contained Subject, ExMethod, Study, DataSet and DataObject objects. However, deleting a Subject does not delete any disassembled Subjects that were previously part of that Subject, since they are autonomous objects, nor would it delete any associated R-Subjects.
The following objects typically have citable identification: Project, Subject, ExMethod, Study, DataSet, R-Subject and Method. Although a DataSet is a member of the Subject collection based on the semantics of the assigned citable identifiers, there is an explicit relationship to the Subject to identify the state of the subject at the time of acquisition. Note that the identifier scheme can be used to allow for different identifier roots. For example, using
r = 1 (see above) for collections of Projects and r = 2 for collections of R-Subjects results in NA.ORG.1.10.23.2.12 referring to Study 12 of ExMethod 2 of Subject 23 of Project 10, whereas NA.ORG.2.17 refers to R-Subject 17.Methods
A Method is comprised of a number of Steps (Figure 2
C), with each step uniquely identified within the scope of the Method. A Method can utilize a specialized step to prescribe the metadata required to create a Subject (and optionally R-Subject) as well as the metadata for each workflow step. A Method object should not be confused with an ExMethod. A Method is simply the specification of a process. When a Method is actually executed, then an ExMethod object is instantiated for the Subject executing the Method. This object holds the citable identifier of the Method, the number of the current step, as well as containing the Studies generated as a result of executing certain steps.
A step may affect a change of state in the subject, or result in the generation of a Study, or branch to another step or method. Branching may be qualified as “any” or “all” if there are multiple options. A step may pre-define metadata or define metadata that must be entered by the researcher. An example of a multi-step Method that acquires MR and MicroscopyStudies is show in the Section “Results”. Note that the Method and definition of metadata can be used to dynamically drive user interfaces.
A Project may have one or more prescribed Methods (selectable by the researcher) which are applied to a Subject and which may result in the generation of Studies. All subjects may require the same Method, or there may be different Methods for different subjects. For example, there could be N control subjects, and M non-control subjects each with different research Methods. In addition, Figure 2
shows that Subjects may contain one or more ExMethods providing research flexibility. For example, subsequent Methods may refine an experimental process, or allow simple ad-hoc capture of data without prescriptive specification of process or metadata.
Methods are identified using citable identifiers so they may be referenced and re-used within a distributed environment. For example, an organization may have “standard” Methods that can be used directly or incorporated into more complex methods.
Life-Cycle and State
A subject’s state may be altered (transiently or permanently; e.g. application of chemicals, death, etc.) prior to the acquisition of data. An acquisition of data at a point in time reflects the state of the subject at that point in time. The conditions that cause a state change are fully recorded in metadata associated with the Subject. A state change is uniquely identified within the context of a Subject and the pair (Subject, State) is unique. Permanent changes should be recorded with the R-Subject, if there is one, or the Subject otherwise.
DataSets and DataObjects
A DataSet contains the acquired or derived data and may hold data directly or be comprised of one or more DataSets and/or DataObjects (the smallest addressable item in our object model). We have made use of concepts in the CCLRC’s DataHolding object model in this design. The definition of “small” is a matter of agreement, since, for example, the smallest unit of data might be a pixel within an image rather than an image.
DataSets may hold content directly, or they may be comprised of a number of smaller DataSets as well as zero or more DataObjects (Figure 2
A). For example, many measurements involve the acquisition of calibration data followed by a series of measurements. The calibration data constitute a DataSet in their own right, but they are also directly associated with the subsequent measurement DataSets. As well as storing primary data, as in the above example, the object model provides for derived DataSets that are the transformation of one or more other DataSets. The method of transformation (e.g. a series or analysis applications) must be recorded in metadata attached to the DataSet. The DataSet object may store the transformed data, or may simply maintain the method for the generation of the data, which may be computed dynamically. The ability to precisely record the method for generating a DataSet then allows the method of construction to be peer reviewed, and the data can be discarded (e.g. to release storage resources) and re-created on demand.
DataSet identifiers are of two types either with all or none of the members having citable identifiers. A DataSet that contains members with citable identifiers (and can return the list of members upon request) is unordered and mutable. A DataSet that contains members that have no citable identification can identify the number of members and return the metadata and/or data for any member based on the ordinal position of that member.
A DataSet that is accessed by ordinal position must guarantee that the ordinal position of every member is immutable; members may only be appended. For example, a DataSet that contains other DataSets is unordered. Therefore, the members therein must also have citable identification. A DICOM Series is an example of an ordered DataSet with no requirement to cite individual members since it contains one or more images, each addressable by an ordinal (slice) position.
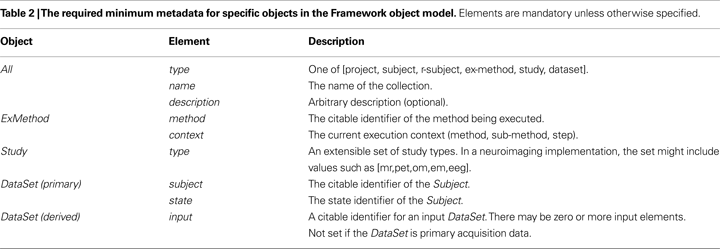
Metadata
The object model prescribes a minimum set of metadata elements for each object (Table 2
).
These are then extended with domain-specific metadata to fully describe the objects and the research being undertaken. For the purpose of hierarchical presentation, identifying metadata must be attached to each Project, Subject, R-Subject, ExMethod, Study and DataSet object. This will allow type independent presentation of each collection. The “type” is important for semantic interpretation and the “name” provides identifying information for users.
If the DataSet is derived from one or more other DataSets, then the provenance of the DataSet must be identified. In addition, the nature of the derivation should be defined, ideally using structured metadata (when that metadata can easily be captured). A precise description is required if the DataSet is to be computed/recomputed at any time. The definition of other provenance metadata is domain specific.
Augmenting the generic prescribed metadata, domain-specific metadata is placed on the objects according to the concept that they represent and the temporal scope of the object (Table 3 and see Results). For example, a Project object may hold metadata describing the project team accessing it as well as hold identifiers for Ethics documents. A Subject may hold demographical and identity information, medical and educational history (for humans), genetic breeding details (for animals) and so on. These choices are entirely driven by the needs of the research.
Where metadata standards are available for a domain, it is advantageous to follow those standards, or at least provide a means to transform metadata to those standards.
The Method may be used to define much of this metadata (it may utilize a specialized step to prescribe metadata needed to create a subject as well as that for workflow) but other agents (e.g. a DICOM server) may also add metadata to objects (e.g. Study and DataSet).
Controlled Access
Data in a repository must have controlled access. Explicit control over access to metadata and content is best provided by role-based authorization and we have defined four project-specific, hierarchical roles where each role inherits the rights of the subordinate roles. The roles are
ProjectAdministrator (“super-user” project permissions), SubjectAdministrator (administer subjects within the project), Member (read access to all research data and metadata generated by the project except protected identity information and Guest (can search the metadata only to find out what types of information are available). When an R-Subject is created, the Administrator roles have the ability to view the identity and update the details of the R-Subject. Alternatively, if an R-Subject is not utilized, the visibility of any sensitive identity information located on the Subject could be controlled via this role.These roles are further qualified by the citable identifier of the project to provide project-specific access control. For example, for the project with citable identifier
1.1.1.2, the ProjectAdministrator role would be named ProjectAdministrator_1.1.1.2.The Framework has been extensively tested through a functioning reference implementation applied to the neuroimaging research domain to manage research data.
Reference Implementation
A data repository has been built with a service-oriented Digital Asset Management system (Mediaflux™,
12
). A package of Mediaflux™ services implementing the Framework object model has been created. These services provide the basic interface to the data repository and allow a user to create, access and manage the objects of the model. As well as enabling the creation of the generic objects and metadata, the services also provide for the addition of domain-specific metadata and content, and the creation and use of Methods to manage experimental process and state.
The implementation uses the citable identifiers described above as arguments to many services to identify specific objects. The implementation does not explicitly create a State object. Instead, the state is contained within the Subject object. The implementation uses a well-defined XML metadata structure for each object. For example, on Subject and R-Subject objects, the implementation allows public and private metadata. The visibility of the metadata contained within these elements then depends upon the user’s role (e.g.
ProjectAdministrator [can see private] or Member [cannot see private]) and their semantic interpretation.Sophisticated adaptive (to the metadata) graphical (“Web 2.0” and Java) interfaces that are driven by the object model (and especially the Method) have also been created (see below). These interfaces (which in turn use the above Mediaflux™ package) provide the primary interface to the system for research scientists. These interfaces are generic and domain independent.
Specific Neuroimaging Implementation
The Framework object model and implementation is currently being used to manage a data repository in the Neuroimaging domain. Services that are not explicitly part of the Framework implementation are used to upload the data (and some associated metadata) into the repository (e.g. a DICOM client). The repository manages over 60 projects that contain mainly MR data (human and small animal) in DICOM (and proprietary formats) and optical microscopy data in TIFF format. Thus we have defined modular (reusable) XML metadata documents and Methods specifically to handle these kinds of data in a neuroimaging research environment.
In this implementation, an authorized user first creates a Project object, defining the project goals, project context and the team members (and their roles). When the Project is created, pre-existing Method objects (one or more) are also registered for use with that Project. Subsequently, team members with the
SubjectAdministrator role for this project create Subjects (and possibly R-Subjects) as needed (ExMethod objects are auto-created in this process). Study objects are generally created as needed by the agents that upload data (although they can pre-created).A design principle of the implementation has been to enable the creation of adaptive user interfaces by providing services that: (i) retrieve the metadata required to create objects and (ii) retrieve metadata and data on existing objects for subsequent presentation. The implementation makes heavy use of Method objects. In particular, a Method object defines the metadata required to create Subject (and possibly R-Subject) objects; this can be thought of as a specialized Method step. The Method object also defines the metadata required per step of the Method during execution and this may include metadata for Study objects. The Method may pre-specify metadata values and whether it is immutable or not.
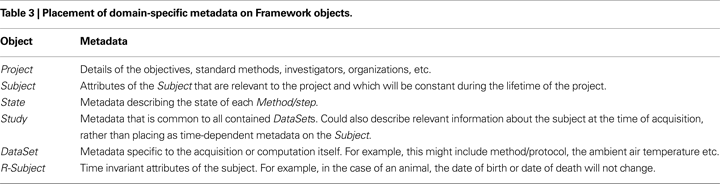
As an example, Figure 3
shows the metadata required to create a Subject for a specialized Method that combines MR, optical microscopy and electron microscopy image data acquired in translational research of mice (Wu et al., 2007
).
Figure 3. The metadata specified by a particular Method (developed for a particular Project) that is required to create a Subject. The adaptive graphical interface interrogates the Method to discover the required metadata. Metadata are presented in XML fragments. Some metadata are predefined and immutable (e.g. species) whereas other metadata requires entry.
This Method specifies that the subjects are a particular strain of mouse targeting a specific disease (and these metadata are immutable). Details such as birth date are entered by the user to complete the Subject creation. Other Methods may specify the use of an R-Subject, or different metadata for the creation of the Subject/R-Subject.
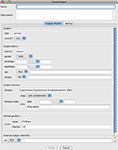
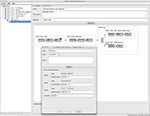
The ExMethod (the instantiation of the Method) object that was (auto) created for the above Subject is show in Figure 4
. This Method acquires MR (of the whole brain) and optical microscopy (of the removed optic nerve) images for mouse subjects. Each numbered step has a name and specifies metadata, the state, and whether a Study is created or not. The inset shows the metadata for the Perfusion step. These metadata are immutable and pre-specified by the Method so that entry by the user is not required.
Figure 4. The adaptive interface shows the object trees for the projects that the user is authorized to access. The Project with citable ID
1005.4.361 is opened and the ExMethod object 1005.4.361.1.1 is displayed. For presentation, this figure shows a simplified version of the ExMethod object (it has more steps in reality). The inset shows the (immutable) metadata for the Perfusion step. It can be seen that the overall Method (1005.5.388), from which this ExMethod is instantiated, was built from a number of Method fragments (1005.5.[384,385,386]).The subject undergoes distinct (permanent) state changes during the execution of the Method. When the imaging data are uploaded and the Study objects created, each Study is tagged with the relevant step of the Method. The Method branches can be executed in parallel or serially as the tissue specimens are imaged. Each removed tissue specimen could be represented as a new (dis-assembled) Subject.
Substantial effort from a number of groups has begun the development of biomedical ontological frameworks (e.g. the Unified Medical Language System
13
and the Open Biomedical Ontology
14
(Smith et al., 2007
). Specification of metadata in the system could adhere to existing domain standards either by direct use of metadata definitions, or by the ability to inter-operate through exchange processes (e.g. utilizing XSL and XSL Transformations
15
). The implementation of metadata also needs to remain flexible so that scientists can incorporate any metadata that they need, whilst still retaining standard components.
Because the PSSD framework enables project-specific Method specification, and because each Method specifies metadata independently, the system provides for flexibility and the adherence to standards.
Significance
Modern scientific research involves distributed collaborative teams, distributed data with distributed processing
16
,
17
; these are aspects of the e-Research paradigm. Whilst the need to organize information via an object model and the ability to federate information is of course not new, the Framework and methodology described in this paper have a number of significant advantages for e-Research applications. Firstly, the use of a distributed object model enables project teams to participate in a collaborative research project whilst using distributed data repositories and interfaces. Distributed object collections can be managed using the semantics of the citable identification scheme without requiring costly and potentially error prone distributed or centralized registries.
Secondly, codifying research processes into a Method means that: (i) Methods can be presented unambiguously and reviewed using simple diagrams, (ii) Methods can be re-used, (iii) application interfaces can be automatically constructed, (iv) researchers can define new research method(s) without requiring the development of new application interfaces to support the execution of those methods, and (v) the metadata for each class of experiments is derived from the relevant Method(s). Note that a Method can contain a super-set of any existing metadata standard. Importantly, by recording all state changes for a subject regardless of whether they are transient or permanent, the conditions the led to the acquisition of data can be identified, reviewed and reconstructed.
Thirdly, identification of “real” subjects (R-Subject) enables identification of all projects in which a particular subject has participated. For example, a genetic sequence may be identified in a subject that was not previously known. The state of the R-Subject could then be updated, with prior research conducted using that subject re-analyzed.
Finally, the Framework object model is extensible to accommodate new relevant information. For example, a human subject may enter into an agreement defining the terms and conditions under which their data may be used. That agreement may apply to all projects in which they have participated or alternatively may be project specific. The agreement may be scanned and associated with either the R-Subject or Subject objects, depending on the scope of the agreement. Similarly, a researcher may associate other information (via new objects) such as documents or data with any object.
Implementation Considerations
Our implementation of the Framework utilizes a service-oriented digital asset management platform which supports distributed citable identification and distributed repositories. All metadata are encoded using XML. Depending on the type of research, XML schemas for metadata are defined using existing standards where they exist, or defined specifically for the research method, or a combination of both. The Framework may be implemented with any service-oriented system utilizing most database technologies. A service-oriented approach, such as web-services, ensures user interfaces and other systems interact with the Framework’s interface, hiding the underlying method of implementation. The key capabilities supported are: (i) citable identifier allocation, (ii) object creation with the ability to associate metadata and arbitrary data with an object, (iii) metadata definitions (e.g. XML Schema) so that domain-specific metadata can be created for any type of object, and (iv) distributed data repositories where distributed projects are undertaken.
Limitations
The Framework has been developed for subject-centric research and thus is not necessarily optimal for other research domains. The number of objects in the object model has been minimized in order to improve accessibility of the model by researchers. However, a number of important aspects of information management are not included in the Framework. For example, many information models and metadata schema have been developed for the preservation of digital data (see OCLC working group report
18
). The development of a long-term information management capability requires the incorporation of aspects of these models and schemas. Since the Framework object model is extensible, future integration with other information object model components is possible.
The Framework includes the ability to notate and track subject state. In neuroimaging research the subject state changes slowly. However, this limitation could be overcome by acquiring vectors of metadata during the data acquisition process in order to measure rapid state changes. Whilst the Framework has broad applicability, limitations may arise from wider application of it to other domains of subject-centric research.
Future Work
Future developments of the Framework in the neuroimaging domain will include acquisition of data from different imaging modalities as well as increasingly complex workflows in distributed projects. Research outcomes should be enhanced by integration of the Framework with other resources such as application processing pipelines, brain atlases and publication portals. Finally, tools that support research uses of the model are being developed including a graphical user interface application to enable researchers to create Methods and define metadata themselves. The Framework will promote modularization of research processes and associated metadata, which in turn promote re-use and standardization. The unpredictable path of future research provides a significant challenge for identifying re-usable research specific metadata, but is important for interoperability and retrospective interpretation.
Conclusions
A Framework that incorporates an object model and research methods for distributed subject-centric research has been developed. The Framework facilitates multi-disciplinary and collaborative subject-based research, and extends earlier object models used in the research imaging domain. Whilst the Framework has been explicitly validated for neuroimaging research applications, it has broader applications to other fields of subject-centric research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Gavan McCarthy, Steve Melnikoff, Anna Shadbolt, Lyle Winton, Wilson Liu and Wee Siong-Soh for discussions and development that have helped us to validate and refine this work. We also acknowledge grants from the Australian Research Council (grants LE0561231, SR0564829) and the University of Melbourne (Cross-Faculty Fund 2006) that have in part supported this work.
- ^ http://www.nsf.gov.pubs/nsf0728/nsf0728.pdf
- ^ SenseLab: http://senselab.med.yale.edu/
- ^ http://ida.loni.ucla.edu/
- ^ The fMRI Data Center’s data management tools http://www.fmridc.org/f/fmridc/database/index.html
- ^ Digital Imaging and Communications in Medicine (DICOM). http://medical.nema.org
- ^ XML-based Clinical and Experimental Data Exchange (XCEDE). http://www.nbirn.net/tools/xcede/index.shtm
- ^ Sufi S, Mathews B. Council for the Central Laboratory of Research Councils (CCLRC) Scientific Metadata Model: Version 2. See http://epubs.cclrc.ac.uk
- ^ Unified Modeling Language. http://www.uml.org
- ^ Digital Object Identifier (DOI). http://www.doi.org
- ^ Unique persistent identifiers. http://www.handle.net
- ^ http://www.pilin.net.au
- ^ Mediaflux™ digital asset management platform. http://www.arcitecta.com
- ^ http://www.nlm.nih.gov/research/umls
- ^ http://www.obofoundry.org
- ^ http://www.w3.org/Style/XSL/
- ^ http://www.nsf.gov/pubs/2007/nsf0728/nsf0728.pdf
- ^ http://www.jisc.ac.uk
- ^ The Online Computer Library Centre & Research Libraries Group (OCLC/RLG) Working Group on Preservation Metadata. Preservation Metadata and the Open Archival Information System (OAIS) Information Model, 2002. http://www.oclc.org.research/pmwg .
Gardner, D., Toga, A. W., Ascoli, G. A., Beatty, J. T., Brinkley, J. F., Dale, A. M., Fox, P. T., Gardner, E. P., George, J. S., Goddard, N., Harris, K. M., Herskovits, E. H., Hines, M. L., Jacobs, G. A., Jacobs, R. E., Jones, E. G., Kennedy, D. N., Kimberg, D. Y., Mazziotta, J. C., Miller, P. L., Mori, S., Mountain, D. C., Reiss, A. L., Rosen, G. D., Rottenberg, D. A., Shepherd, G. M., Smalheiser, N. R., Smith, K. P., Strachan, T., Van Essen, D. C., Williams, R. W., and Wong, S. T. (2003). Towards effective and rewarding data sharing. Neuroinformatics 1, 289–296.
Keator, D. B., Grethe, J. S., Marcus, D., Ozyurt, B., Gadde, S., Murphy, S., Pieper, S., Greve, D., Notestine, R., Bockholt, H. J., Papadopoulos P, BIRN Function; BIRN Morphometry; and BIRN-Coordinating. (2008). A national human neuroimaging collaboratory enabled by the Biomedical Informatics Research Network (BIRN). IEEE Trans. Inf. Technol. Biomed. 12, 162–172.
Klump, J., Brase, J., Diepenbroek, M., Grobe, H., Hildenbrandt, B., Höck, H., Lautenschlager, M., and Sens, I. (2008). Use of Persistent Identifiers in the Publication and Citation of Scientific Data. AGU Fall Meeting, 5–19 December 2008, San Francisco, CA, USA. Available at: http://epic.awi.de/epic/Main?puid=31047&lang=en
.
Smith, B., Ashburner, M., Rosse, C., Bard, J., Bug, W., Ceusters, W., Goldberg, L. J., Eilbeck, K., Ireland, A., Mungall, C. J., The OBI Consortium, Leontis, N., Rocca-Serra, P., Ruttenberg, A., Sansone S-A., Scheuermann, R. H., Shah, N., Whetzel, P. L., and Lewis, S. (2007). The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 25, 1251–1255.